Spatially Resolved Molecular Approaches for the Characterisation of Non-Invasive Follicular Tumours with Papillary-like Features (NIFTPs)
Abstract
1. Introduction
2. Results
2.1. Clinical and Pathological Characteristics of the Cohort
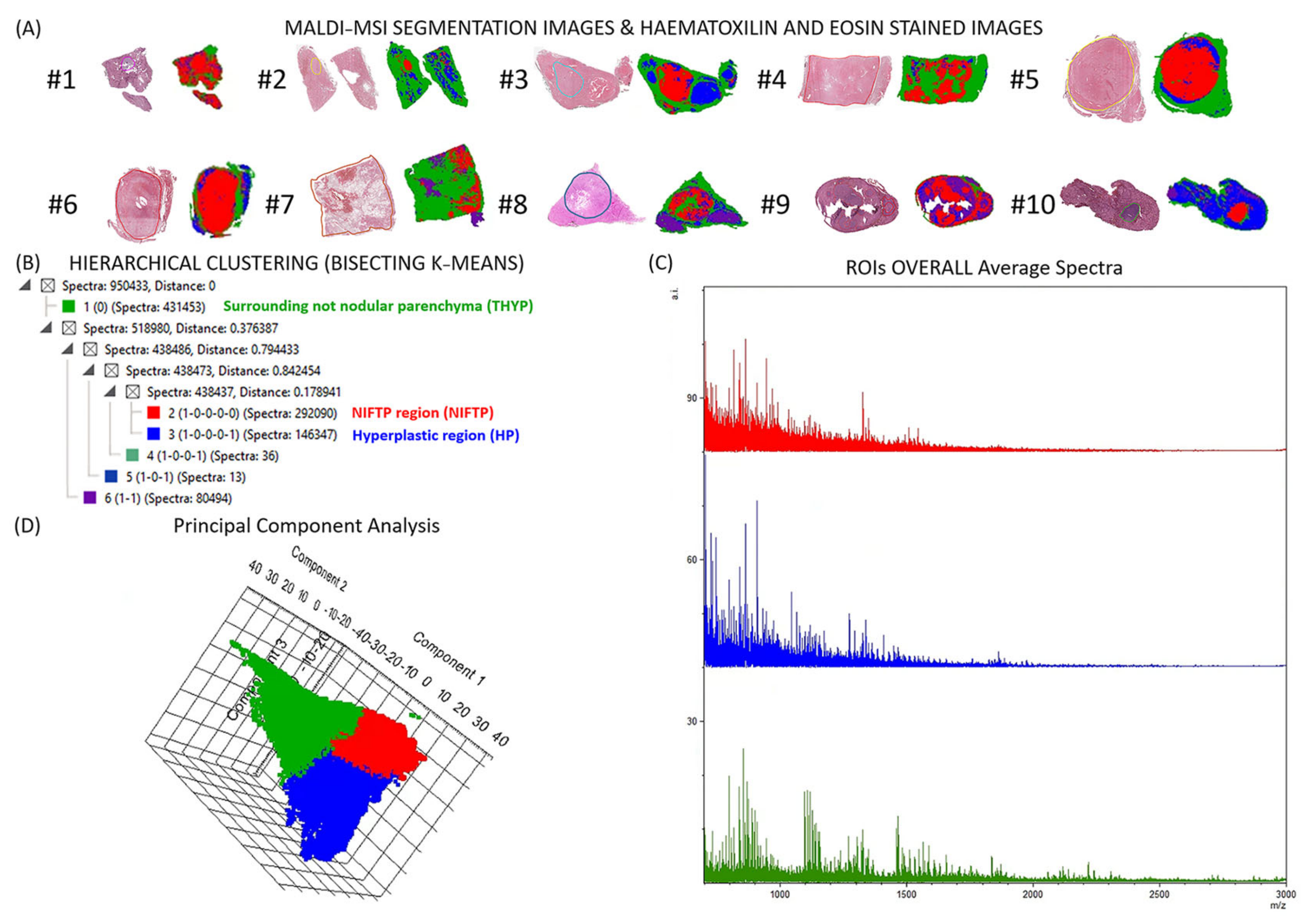
2.2. Detection and Characterization of NIFTPs with MALDI–MSI
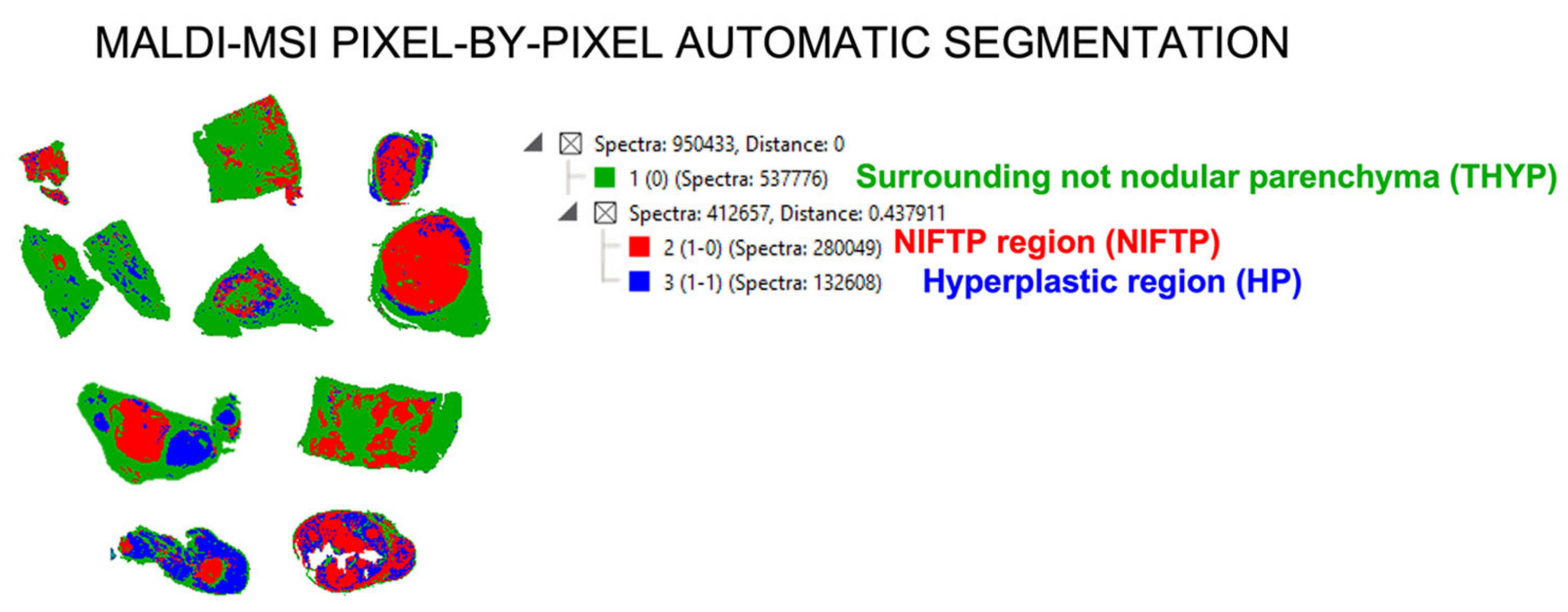
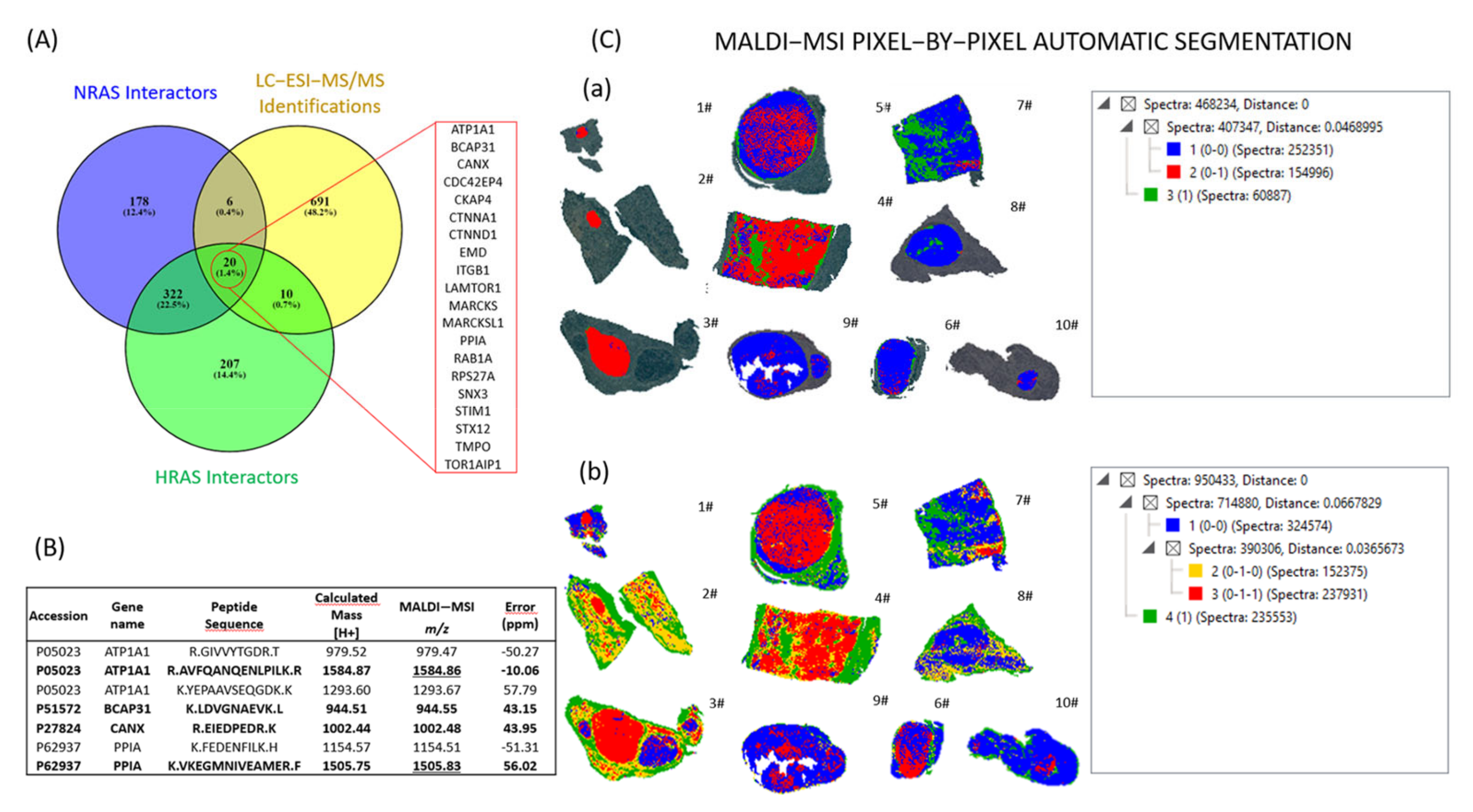
2.3. Automatic Segmentation and Proteogenomic Classification of NIFTPs
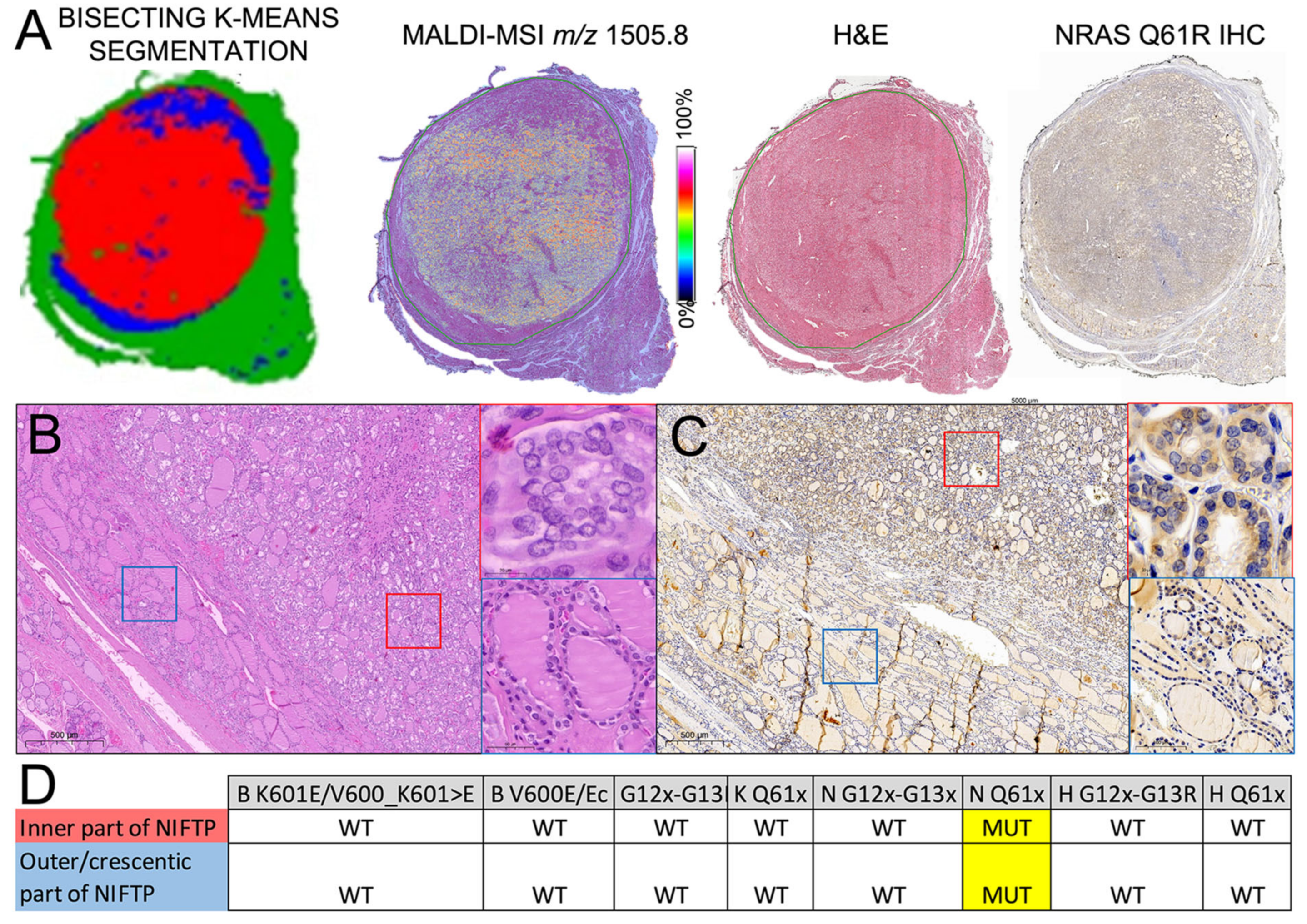
2.4. Heterogeneity of RAS-Mutant NIFTPs
3. Discussion
4. Materials and Methods
4.1. Pathology
4.2. Sample Preparation for MALDI–MSI
4.3. MALDI–MSI Analysis
4.4. MALDI–MSI Data Analysis
4.5. nLC–ESI-MS/MS Sample Preparation and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nikiforov, Y.E.; Seethala, R.R.; Tallini, G.; Baloch, Z.W.; Basolo, F.; Thompson, L.D.R.; Barletta, J.A.; Wenig, B.M.; Al Ghuzlan, A.; Kakudo, K.; et al. Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol. 2016, 2, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Ucal, Y.; Ozpinar, A. Proteomics in Thyroid Cancer and Other Thyroid-Related Diseases: A Review of the Literature. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2020, 1868, 140510. [Google Scholar] [CrossRef]
- Livhits, M.J.; Zhu, C.Y.; Kuo, E.J.; Nguyen, D.T.; Kim, J.; Tseng, C.-H.; Leung, A.M.; Rao, J.; Levin, M.; Douek, M.L.; et al. Effectiveness of Molecular Testing Techniques for Diagnosis of Indeterminate Thyroid Nodules: A Randomized Clinical Trial. JAMA Oncol. 2021, 7, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Kurczyk, A.; Gawin, M.; Chekan, M.; Wilk, A.; Łakomiec, K.; Mrukwa, G.; Frątczak, K.; Polanska, J.; Fujarewicz, K.; Pietrowska, M.; et al. Classification of Thyroid Tumors Based on Mass Spectrometry Imaging of Tissue Microarrays; a Single-Pixel Approach. Int. J. Mol. Sci. 2020, 21, 6289. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Ghossein, R.A. Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features (NIFTP): An Update. Head Neck Pathol. 2020, 14, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Capitoli, G.; Piga, I.; L’Imperio, V.; Clerici, F.; Leni, D.; Garancini, M.; Casati, G.; Galimberti, S.; Magni, F.; Pagni, F. Cytomolecular Classification of Thyroid Nodules Using Fine-Needle Washes Aspiration Biopsies. Int. J. Mol. Sci. 2022, 23, 4156. [Google Scholar] [CrossRef]
- Ucal, Y.; Tokat, F.; Duren, M.; Ince, U.; Ozpinar, A. Peptide Profile Differences of Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features, Encapsulated Follicular Variant, and Classical Papillary Thyroid Carcinoma: An Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Thyroid 2019, 29, 1125–1137. [Google Scholar]
- Piga, I.; Capitoli, G.; Clerici, F.; Brambilla, V.; Leni, D.; Scardilli, M.; Canini, V.; Cipriani, N.; Bono, F.; Valsecchi, M.G.; et al. Molecular Trait of Follicular-Patterned Thyroid Neoplasms Defined by MALDI-Imaging. Biochim. Biophys. Acta: Proteins Proteom. 2020, 1868, 140511. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. WHO Classification of Tumours Editorial Board: Endocrine and Neuroendocrine Tumours, 5th ed.; WHO: Geneva, Switzerland, 2022; Volume 8. [Google Scholar]
- Hung, Y.P.; Barletta, J.A. A User’s Guide to Non-Invasive Follicular Thyroid Neoplasm with Papillary-like Nuclear Features (NIFTP). Histopathology 2018, 72, 53–69. [Google Scholar] [CrossRef]
- Haugen, B.R.; Sawka, A.M.; Alexander, E.K.; Bible, K.C.; Caturegli, P.; Doherty, G.M.; Mandel, S.J.; Morris, J.C.; Nassar, A.; Pacini, F.; et al. American Thyroid Association Guidelines on the Management of Thyroid Nodules and Differentiated Thyroid Cancer Task Force Review and Recommendation on the Proposed Renaming of Encapsulated Follicular Variant Papillary Thyroid Carcinoma Without Invasion to Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features. Thyroid 2017, 27, 481–483. [Google Scholar]
- Chu, Y.-H.; Sadow, P.M. Noninvasive Follicular Thyroid Neoplasm with Papillary-like Nuclear Features (NIFTP): Diagnostic Updates and Molecular Advances. Semin. Diagn. Pathol. 2020, 37, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Canini, V.; Leni, D.; Pincelli, A.I.; Scardilli, M.; Garancini, M.; Villa, C.; Di Bella, C.; Capitoli, G.; Cimini, R.; Leone, B.E.; et al. Clinical-Pathological Issues in Thyroid Pathology: Study on the Routine Application of NIFTP Diagnostic Criteria. Sci. Rep. 2019, 9, 13179. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Lima, C.J.; Auh, S.; Thakur, S.; Zemskova, M.; Cochran, C.; Merkel, R.; Filie, A.C.; Raffeld, M.; Patel, S.B.; Xi, L.; et al. A Novel Risk Stratification System for Thyroid Nodules With Indeterminate Cytology-A Pilot Cohort Study. Front. Endocrinol. 2020, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Strickland, K.C.; Eszlinger, M.; Paschke, R.; Angell, T.E.; Alexander, E.K.; Marqusee, E.; Nehs, M.A.; Jo, V.Y.; Lowe, A.; Vivero, M.; et al. Molecular Testing of Nodules with a Suspicious or Malignant Cytologic Diagnosis in the Setting of Non-Invasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features (NIFTP). Endocr. Pathol. 2018, 29, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Denaro, M.; Ugolini, C.; Poma, A.M.; Borrelli, N.; Materazzi, G.; Piaggi, P.; Chiarugi, M.; Miccoli, P.; Vitti, P.; Basolo, F. Differences in miRNA Expression Profiles between Wild-Type and Mutated NIFTPs. Endocr. Relat. Cancer 2017, 24, 543–553. [Google Scholar] [CrossRef]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin A: A Key Player for Human Disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef]
- Yang, H.; Chen, J.; Yang, J.; Qiao, S.; Zhao, S.; Yu, L. Cyclophilin A Is Upregulated in Small Cell Lung Cancer and Activates ERK1/2 Signal. Biochem. Biophys. Res. Commun. 2007, 361, 763–767. [Google Scholar] [CrossRef]
- Qi, Y.-J.; He, Q.-Y.; Ma, Y.-F.; Du, Y.-W.; Liu, G.-C.; Li, Y.-J.; Tsao, G.S.W.; Ngai, S.M.; Chiu, J.-F. Proteomic Identification of Malignant Transformation-Related Proteins in Esophageal Squamous Cell Carcinoma. J. Cell. Biochem. 2008, 104, 1625–1635. [Google Scholar] [CrossRef]
- Wang, S.; Li, M.; Xing, L.; Yu, J. High Expression Level of Peptidylprolyl Isomerase A Is Correlated with Poor Prognosis of Liver Hepatocellular Carcinoma. Oncol. Lett. 2019, 18, 4691–4702. [Google Scholar] [CrossRef]
- Lewiński, A.; Brzeziańska, E.; Czarnecka, K.; Latek, J.; Koptas, W.; Cyniak-Magierska, A. Increased Expression of PIN1 Gene in Papillary Thyroid Carcinoma. Thyroid Res. 2011, 4, 4. [Google Scholar] [CrossRef]
- Jiang, L.; Chu, H.; Zheng, H. Pin1 Is Related with Clinical Stage of Papillary Thyroid Carcinoma. World J. Surg. Oncol. 2016, 14, 95. [Google Scholar] [CrossRef] [PubMed]
- Schulze, C.J.; Bermingham, A.; Choy, T.J.; Cregg, J.J.; Kiss, G.; Marquez, A.; Reyes, D.; Saldajeno-Concar, M.; Weller, C.E.; Whalen, D.M.; et al. Abstract PR10: Tri-Complex Inhibitors of the Oncogenic, GTP-Bound Form of KRASG12Covercome RTK-Mediated Escape Mechanisms and Drive Tumor Regressions in Vivo. Mol. Cancer Ther. 2019, 18, PR10. [Google Scholar] [CrossRef]
- Nichols, R.; Schulze, C.; Bermingham, A.; Choy, T.; Cregg, J.; Kiss, G.; Marquez, A.; Reyes, D.; Saldajeno-Concar, M.; Weller, C.; et al. A06 Tri-Complex Inhibitors of the Oncogenic, GTP-Bound Form of KRASG12C Overcome RTK-Mediated Escape Mechanisms and Drive Tumor Regressions in Preclinical Models of NSCLC. J. Thorac. Oncol. 2020, 15, S13–S14. [Google Scholar] [CrossRef]
- Guenter, R.; Patel, Z.; Chen, H. Notch Signaling in Thyroid Cancer. Adv. Exp. Med. Biol. 2021, 1287, 155–168. [Google Scholar]
- Lloyd, R.V.; Osamura, R.Y.; Klöppel, G.; Rosai, J. WHO Classification of Tumours of Endocrine Organs; International Agency for Research on Cancer: Lyon, France, 2017; ISBN 9789283244936. [Google Scholar]
- De Sio, G.; Smith, A.J.; Galli, M.; Garancini, M.; Chinello, C.; Bono, F.; Pagni, F.; Magni, F. A MALDI-Mass Spectrometry Imaging Method Applicable to Different Formalin-Fixed Paraffin-Embedded Human Tissues. Mol. Biosyst. 2015, 11, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Strohalm, M.; Kavan, D.; Novák, P.; Volný, M.; Havlícek, V. mMass 3: A Cross-Platform Software Environment for Precise Analysis of Mass Spectrometric Data. Anal. Chem. 2010, 82, 4648–4651. [Google Scholar] [CrossRef] [PubMed]
- Strohalm, M.; Hassman, M.; Kosata, B.; Kodícek, M. mMass Data Miner: An Open Source Alternative for Mass Spectrometric Data Analysis. Rapid Commun. Mass Spectrom. 2008, 22, 905–908. [Google Scholar] [CrossRef]
- Del Toro, N.; Shrivastava, A.; Ragueneau, E.; Meldal, B.; Combe, C.; Barrera, E.; Perfetto, L.; How, K.; Ratan, P.; Shirodkar, G.; et al. The IntAct Database: Efficient Access to Fine-Grained Molecular Interaction Data. Nucleic Acids Res. 2022, 50, D648–D653. [Google Scholar] [CrossRef]
- Hermjakob, H.; Montecchi-Palazzi, L.; Lewington, C.; Mudali, S.; Kerrien, S.; Orchard, S.; Vingron, M.; Roechert, B.; Roepstorff, P.; Valencia, A.; et al. IntAct: An Open Source Molecular Interaction Database. Nucleic Acids Res. 2004, 32, D452–D455. [Google Scholar] [CrossRef]

| Case No. | Age | Sex | No. Nodules | Diameter (cm) | Thyroid Lobe | Mutational Status | Other Thyroid Diseases | Serology | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RAS | Others | TSH (µIU/mL) | anti-TPO Ab (IU/mL) | anti-Tg Ab (IU/mL) | |||||||
| 1 | 65 | F | 3 | 0.3 | Left | NRAS Q61R | PTCH1, RAD51B, SMARCA4 | Hürthle cell carcinoma—Right lobe | 1.9 | 8 | 11 |
| 2 | 52 | F | 2 | 0.5 | Left | HRAS A59T | RICTOR, CHEK2, SLX4 | Multinodular goitre and C cell hyperplasia | 4.12 | 10 | 10 |
| 3 | 50 | F | multiple | 1.3 | Right | NRAS Q61R | - | PTC— Right lobe | 2.26 | 9 | 12 |
| 4 | 41 | F | 1 | 5.5 | Right | NRAS Q61R | - | - | 3.12 | 7 | 9 |
| 5 | 48 | M | 3 | 3 | Right | NRAS Q61R | NOTCH2 | - | 0.6 | 8 | 11 |
| 6 | 61 | F | 1 | 1.6 | Left | WT | NTRK1, FBXW7 | - | 1.5 | 10 | 12 |
| 7 | 58 | F | 1 | 9 | Left | WT | MDM4, ESR1, CDKN2A, AXL | microPTC—Right lobe | 1.16 | 9 | 11 |
| 8 | 53 | F | multiple | 1 | Right | WT | SLX4, ATM, ARID1A, AXL | Multinodular goitre | 4.4 | 172.9 | 2.83 |
| 9* | 56 | M | 2 | 2 | Left | HRAS Q61K | SLX4 | - | 0,85 | 5 | 7 |
| 10* | 56 | M | 2 | 2 | Right | WT | SLX4 | - | 0,85 | 5 | 7 |
| m/z | ROC Analysis | Gene Name |
|---|---|---|
| 815.39 | NIFTP/HP | RBMX |
| NIFTP/THYP | ||
| 836.48 | HP/THYP | COL1A1 |
| NIFTP/THYP | ||
| 868.39 | HP/THYP | COL1A2 |
| NIFTP/THYP | ||
| 894.42 | HP/THYP | PRDX1 |
| NIFTP/HP | ||
| NIFTP/THYP | ||
| 952.35 | HP/THYP | SFPQ |
| NIFTP/THYP | ||
| NIFTP/HP | ||
| 1063.57 | HP/THYP | HNRNPM |
| NIFTP/HP | ||
| NIFTP/THYP | ||
| 1095.62 | HP/THYP | COL1A2 |
| NIFTP/THYP | ||
| 1320.65 | HP/THYP | COL6A3 |
| NIFTP/THYP | ||
| 1505.83 | NIFTP/HP | PPIA |
| NIFTP RAS+/NIFTP RASWT | ||
| 1580.75 | HP/THYP | COL6A1 |
| NIFTP/THYP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piga, I.; L’Imperio, V.; Principi, L.; Bellevicine, C.; Fusco, N.; Maffini, F.; Venetis, K.; Ivanova, M.; Seminati, D.; Casati, G.; et al. Spatially Resolved Molecular Approaches for the Characterisation of Non-Invasive Follicular Tumours with Papillary-like Features (NIFTPs). Int. J. Mol. Sci. 2023, 24, 2567. https://doi.org/10.3390/ijms24032567
Piga I, L’Imperio V, Principi L, Bellevicine C, Fusco N, Maffini F, Venetis K, Ivanova M, Seminati D, Casati G, et al. Spatially Resolved Molecular Approaches for the Characterisation of Non-Invasive Follicular Tumours with Papillary-like Features (NIFTPs). International Journal of Molecular Sciences. 2023; 24(3):2567. https://doi.org/10.3390/ijms24032567
Chicago/Turabian StylePiga, Isabella, Vincenzo L’Imperio, Lucrezia Principi, Claudio Bellevicine, Nicola Fusco, Fausto Maffini, Konstantinos Venetis, Mariia Ivanova, Davide Seminati, Gabriele Casati, and et al. 2023. "Spatially Resolved Molecular Approaches for the Characterisation of Non-Invasive Follicular Tumours with Papillary-like Features (NIFTPs)" International Journal of Molecular Sciences 24, no. 3: 2567. https://doi.org/10.3390/ijms24032567
APA StylePiga, I., L’Imperio, V., Principi, L., Bellevicine, C., Fusco, N., Maffini, F., Venetis, K., Ivanova, M., Seminati, D., Casati, G., Pagani, L., Galimberti, S., Capitoli, G., Garancini, M., Gatti, A.-V., Magni, F., & Pagni, F. (2023). Spatially Resolved Molecular Approaches for the Characterisation of Non-Invasive Follicular Tumours with Papillary-like Features (NIFTPs). International Journal of Molecular Sciences, 24(3), 2567. https://doi.org/10.3390/ijms24032567













