Integrated Microarray-Based Data Analysis of miRNA Expression Profiles: Identification of Novel Biomarkers of Cisplatin-Resistance in Testicular Germ Cell Tumours
Abstract
1. Introduction
2. Results
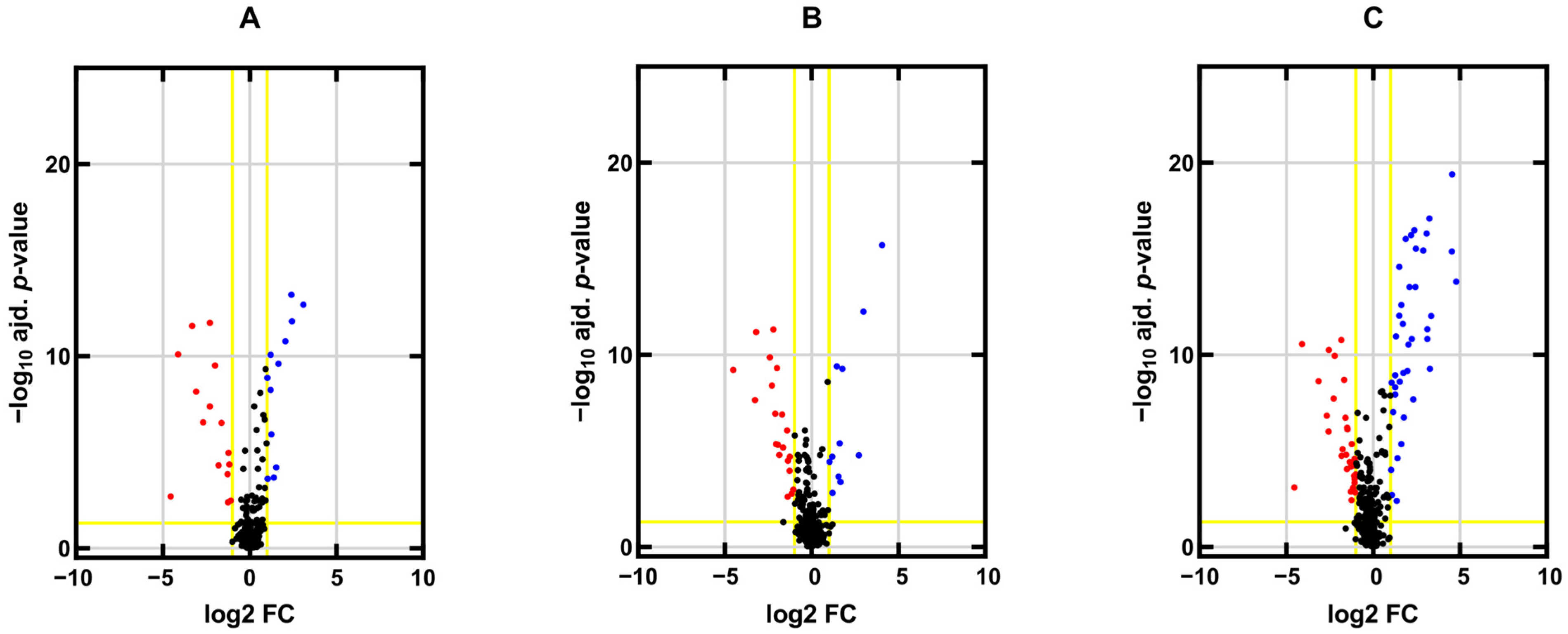
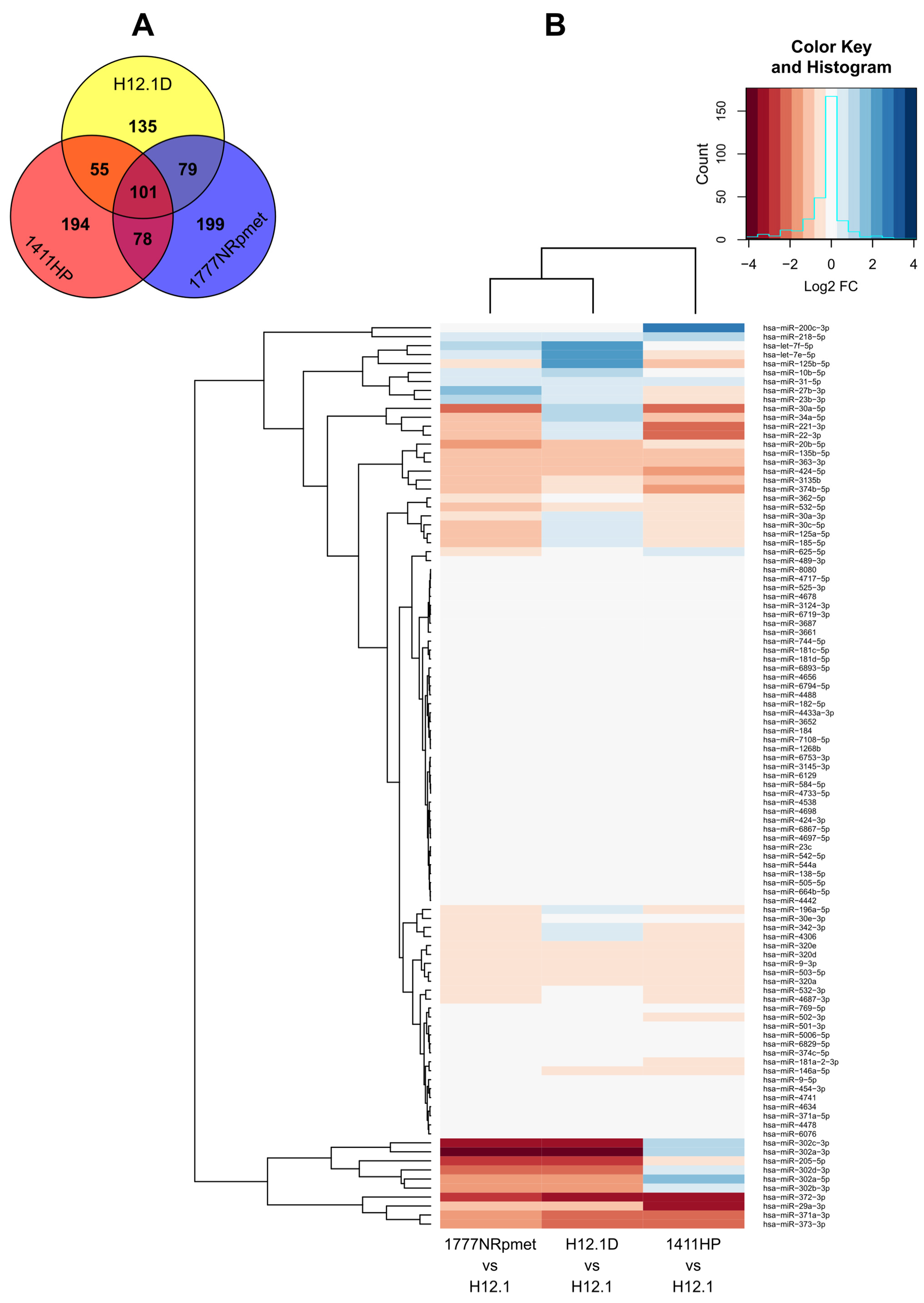
2.1. miRNA Expression Profiles in TGCT Cell Lines
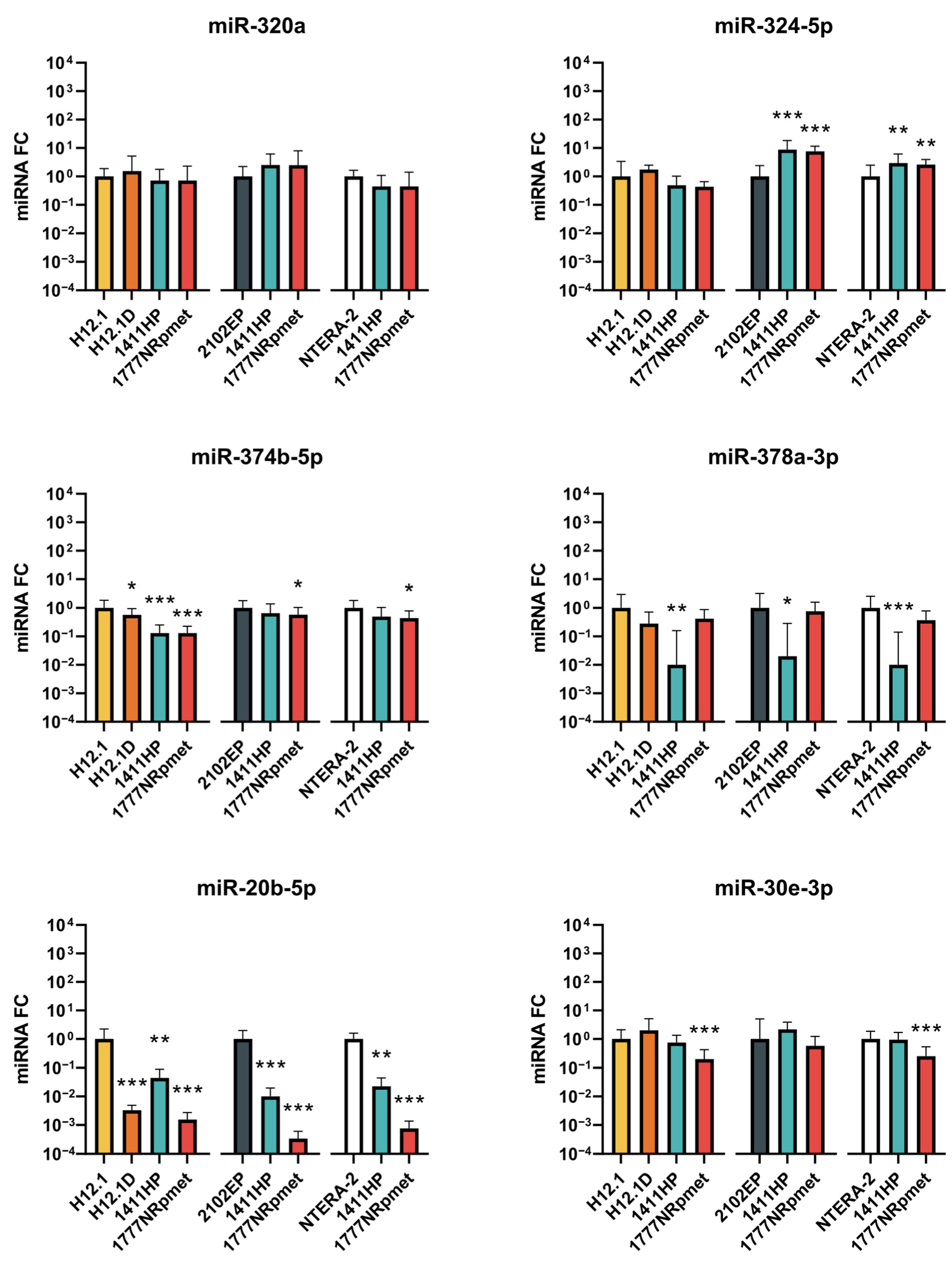
2.2. Candidate miRNAs Expression Validation
2.2.1. Up-Regulated miRNAs in TGCT Cell Lines
2.2.2. Down-Regulated miRNAs in TGCT Cell Lines
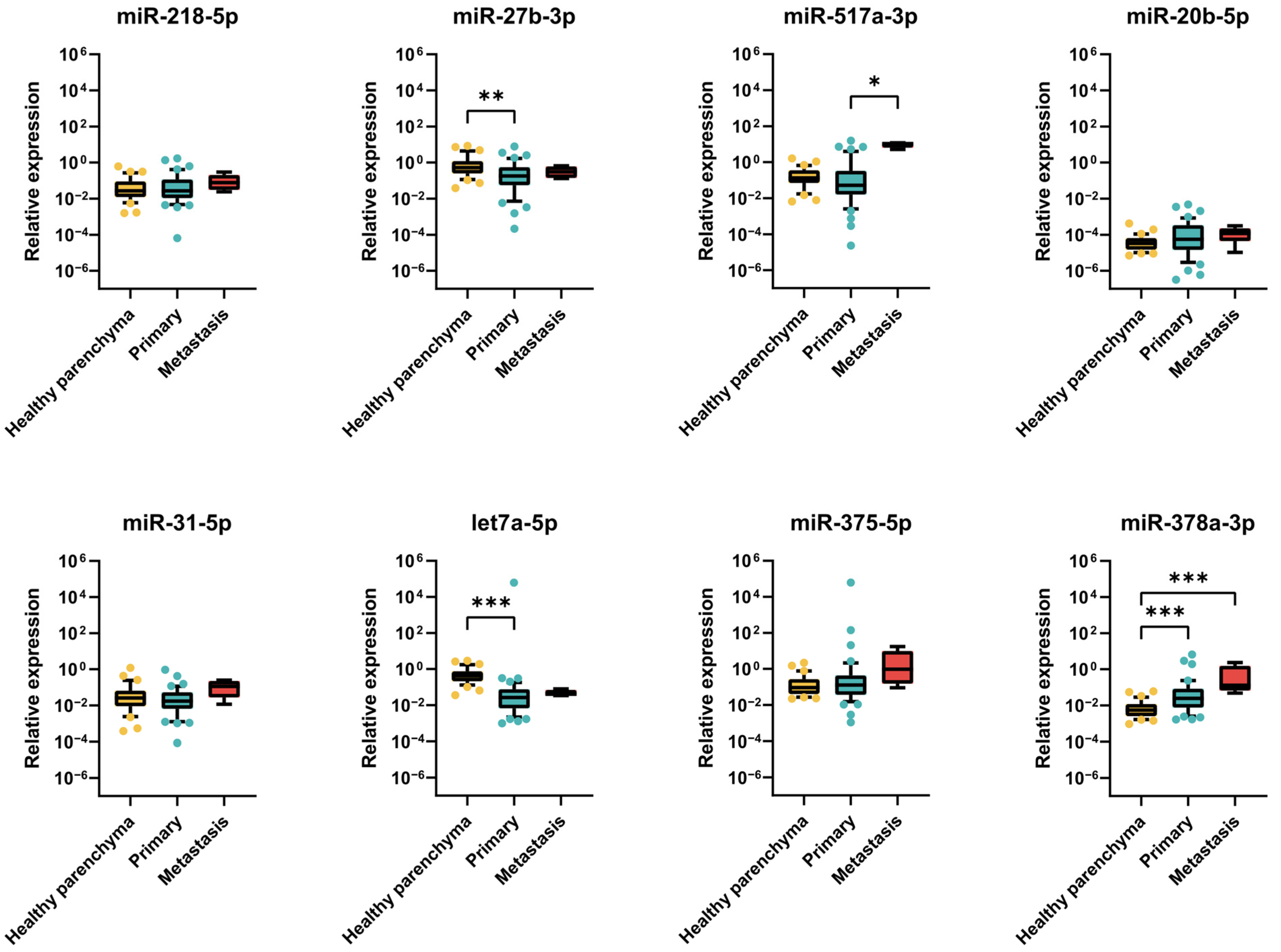
2.2.3. Expression of miRNAs in TGCT Patient Samples
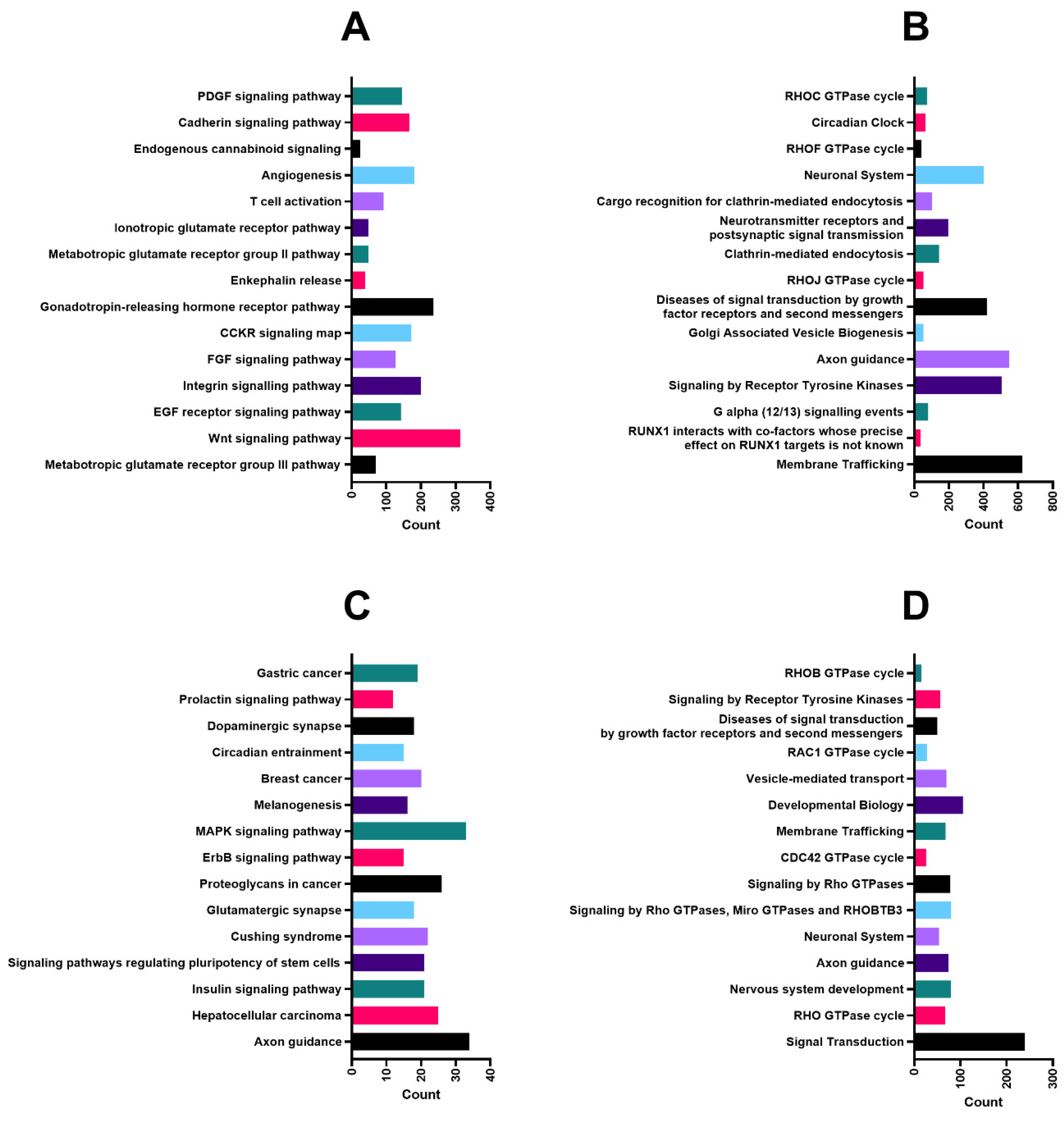
2.3. Predicted Targets and Functional and Regulatory Networks of Candidate miRNAs
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Patients
4.3. RNA Extraction
4.4. miRNA Microarray Profiling
4.5. RT-qPCR Validation
4.6. miRNA Target Prediction and Functional Annotation Analysis of Predicted Targets
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engholm, G.; Ferlay, J.; Christensen, N.; Bray, F.; Gjerstorff, M.L.; Klint, Å.; Køtlum, J.E.; Ólafsdóttir, E.; Pukkala, E.; Storm, H.H. NORDCAN—a Nordic Tool for Cancer Information, Planning, Quality Control and Research. Acta Oncol. 2010, 49, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Le Cornet, C.; Lortet-Tieulent, J.; Forman, D.; Béranger, R.; Flechon, A.; Fervers, B.; Schüz, J.; Bray, F. Testicular Cancer Incidence to Rise by 25% by 2025 in Europe? Model-Based Predictions in 40 Countries Using Population-Based Registry Data. Eur. J. Cancer 2014, 50, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Bleyer, A.; Ferrari, A.; Whelan, J.; Barr, R.D. Global Assessment of Cancer Incidence and Survival in Adolescents and Young Adults. Pediatr. Blood Cancer 2017, 64, e26497. [Google Scholar] [CrossRef] [PubMed]
- Hayes-Lattin, B.; Nichols, C.R. Testicular Cancer: A Prototypic Tumor of Young Adults. Semin. Oncol. 2009, 36, 432–438. [Google Scholar] [CrossRef]
- Horwich, A.; Shipley, J.; Huddart, R. Testicular Germ-Cell Cancer. Lancet 2006, 367, 754–765. [Google Scholar] [CrossRef]
- Znaor, A.; Skakkebæk, N.E.; Rajpert-De Meyts, E.; Laversanne, M.; Kuliš, T.; Gurney, J.; Sarfati, D.; McGlynn, K.A.; Bray, F. Testicular Cancer Incidence Predictions in Europe 2010–2035: A Rising Burden despite Population Ageing. Int. J. Cancer 2020, 147, 820–828. [Google Scholar] [CrossRef]
- Idrees, M.T.; Ulbright, T.M.; Oliva, E.; Young, R.H.; Montironi, R.; Egevad, L.; Berney, D.; Srigley, J.R.; Epstein, J.I.; Tickoo, S.K. The World Health Organization 2016 Classification of Testicular Non-Germ Cell Tumours: A Review and Update from the International Society of Urological Pathology Testis Consultation Panel. Histopathology 2017, 70, 513–521. [Google Scholar] [CrossRef]
- Williamson, S.R.; Delahunt, B.; Magi-Galluzzi, C.; Algaba, F.; Egevad, L.; Ulbright, T.M.; Tickoo, S.K.; Srigley, J.R.; Epstein, J.I.; Berney, D.M. The World Health Organization 2016 Classification of Testicular Germ Cell Tumours: A Review and Update from the International Society of Urological Pathology Testis Consultation Panel. Histopathology 2017, 70, 335–346. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Singh, R.; Fazal, Z.; Freemantle, S.J.; Spinella, M.J. Between a Rock and a Hard Place: An Epigenetic-Centric View of Testicular Germ Cell Tumors. Cancers 2021, 13, 1506. [Google Scholar] [CrossRef]
- Chovanec, M.; Kalavska, K.; Mego, M.; Cheng, L. Liquid Biopsy in Germ Cell Tumors: Biology and Clinical Management. Expert Rev. Mol. Diagn. 2020, 20, 187–194. [Google Scholar] [CrossRef]
- American Cancer Society. Risk Factors for Testicular Cancer. Available online: https://www.cancer.org/cancer/testicular-cancer/causes-risks-prevention/risk-factors.html (accessed on 5 January 2023).
- Looijenga, L.H.J.; Gillis, A.J.M.; Stoop, H.J.; Hersmus, R.; Oosterhuis, J.W. Chromosomes and Expression in Human Testicular Germ-Cell Tumors: Insight into Their Cell of Origin and Pathogenesis. Ann. N. Y. Acad. Sci. 2007, 1120, 187–214. [Google Scholar] [CrossRef]
- Vasdev, N.; Moon, A.; Thorpe, A.C. Classification, Epidemiology and Therapies for Testicular Germ Cell Tumours. Int. J. Dev. Biol. 2013, 57, 133–139. [Google Scholar] [CrossRef]
- Comiter, C.V.; Kibel, A.S.; Richie, J.P.; Nucci, M.R.; Renshaw, A.A. Prognostic features of teratomas with malignant transformation: A clinicopathological study of 21 cases. J. Urol. 1998, 159, 859–863. [Google Scholar] [CrossRef]
- Viatori, M. Testicular Cancer. Semin. Oncol. Nurs. 2012, 28, 180–189. [Google Scholar] [CrossRef]
- Calabrò, F.; Albers, P.; Bokemeyer, C.; Martin, C.; Einhorn, L.H.; Horwich, A.; Krege, S.; Schmoll, H.J.; Sternberg, C.N.; Daugaard, G. The Contemporary Role of Chemotherapy for Advanced Testis Cancer: A Systematic Review of the Literature. Eur. Urol. 2012, 61, 1212–1221. [Google Scholar] [CrossRef]
- Chovanec, M.; Vasilkova, L.; Petrikova, L.; Obertova, J.; Palacka, P.; Rejlekova, K.; Sycova-Mila, Z.; Kalavska, K.; Svetlovska, D.; Mladosievicova, B.; et al. Long-Term Sexual Functioning in Germ-Cell Tumor Survivors. BMC Cancer 2020, 20, 779. [Google Scholar] [CrossRef]
- Amidi, A.; Wu, L.M.; Pedersen, A.D.; Mehlsen, M.; Pedersen, C.G.; Rossen, P.; Agerbæk, M.; Zachariae, R. Cognitive Impairment in Testicular Cancer Survivors 2 to 7 Years after Treatment. Support. Care Cancer 2015, 23, 2973–2979. [Google Scholar] [CrossRef]
- Chovanec, M.; Vasilkova, L.; Setteyova, L.; Obertova, J.; Palacka, P.; Rejlekova, K.; Sycova-Mila, Z.; Kalavska, K.; Svetlovska, D.; Cingelova, S.; et al. Long-Term Cognitive Functioning in Testicular Germ-Cell Tumor Survivors. Oncologist 2018, 23, 617–623. [Google Scholar] [CrossRef]
- Cameron, A.C.; McMahon, K.; Hall, M.; Neves, K.B.; Rios, F.J.; Montezano, A.C.; Welsh, P.; Waterston, A.; White, J.; Mark, P.B.; et al. Comprehensive Characterization of the Vascular Effects of Cisplatin-Based Chemotherapy in Patients with Testicular Cancer. JACC CardioOncology 2020, 2, 443–455. [Google Scholar] [CrossRef]
- Gugic, J.; Zaletel, L.Z.; Oblak, I. Treatment-Related Cardiovascular Toxicity in Long-Term Survivors of Testicular Cancer. Radiol. Oncol. 2017, 51, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Van den Belt-Dusebout, A.W.; Nuver, J.; de Wit, R.; Gietema, J.A.; ten Bokkel Huinink, W.W.; Rodrigus, P.T.R.; Schimmel, E.C.; Aleman, B.M.P.; van Leeuwen, F.E. Long-Term Risk of Cardiovascular Disease in 5-Year Survivors of Testicular Cancer. J. Clin. Oncol. 2006, 24, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Panichpisal, K.; Kurtzman, N.; Nugent, K. Cisplatin Nephrotoxicity: A Review. Am. J. Med. Sci. 2007, 334, 115–124. [Google Scholar] [CrossRef]
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin Nephrotoxicity. Toxins 2010, 2, 2490–2518. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, F.; Curtis, L.M.; Truong, L.; Poss, K.; Visner, G.A.; Madsen, K.; Nick, H.S.; Agarwal, A. Heme Oxygenase-1 Gene Ablation or Expression Modulates Cisplatin-Induced Renal Tubular Apoptosis. Am. J. Physiol. Physiol. 2000, 278, F726–F736. [Google Scholar] [CrossRef]
- McWhinney, S.R.; Goldberg, R.M.; McLeod, H.L. Platinum Neurotoxicity Pharmacogenetics. Mol. Cancer Ther. 2009, 8, 10–16. [Google Scholar] [CrossRef]
- Dzagnidze, A.; Katsarava, Z.; Makhalova, J.; Liedert, B.; Yoon, M.-S.; Kaube, H.; Limmroth, V.; Thomale, J. Repair Capacity for Platinum-DNA Adducts Determines the Severity of Cisplatin-Induced Peripheral Neuropathy. J. Neurosci. 2007, 27, 9451–9457. [Google Scholar] [CrossRef]
- O’Sullivan, B.; Brierley, J.; Byrd, D.; Bosman, F.; Kehoe, S.; Kossary, C.; Piñeros, M.; Van Eycken, E.; Weir, H.K.; Gospodarowicz, M. The TNM Classification of Malignant Tumours—Towards Common Understanding and Reasonable Expectations. Lancet Oncol. 2017, 18, 849–851. [Google Scholar] [CrossRef]
- Gilligan, T.D.; Seidenfeld, J.; Basch, E.M.; Einhorn, L.H.; Fancher, T.; Smith, D.C.; Stephenson, A.J.; Vaughn, D.J.; Cosby, R.; Hayes, D.F. American Society of Clinical Oncology Clinical Practice Guideline on Uses of Serum Tumor Markers in Adult Males With Germ Cell Tumors. J. Clin. Oncol. 2010, 28, 3388–3404. [Google Scholar] [CrossRef]
- International Germ Cell Consensus Classification: A Prognostic Factor-Based Staging System for Metastatic Germ Cell Cancers. International Germ Cell Cancer Collaborative Group. J. Clin. Oncol. 1997, 15, 594–603. [CrossRef]
- Barlow, L.J.; Badalato, G.M.; McKiernan, J.M. Serum Tumor Markers in the Evaluation of Male Germ Cell Tumors. Nat. Rev. Urol. 2010, 7, 610–617. [Google Scholar] [CrossRef]
- Leão, R.; Ahmad, A.E.; Hamilton, R.J. Testicular Cancer Biomarkers: A Role for Precision Medicine in Testicular Cancer. Clin. Genitourin. Cancer 2019, 17, e176–e183. [Google Scholar] [CrossRef]
- Gutschner, T.; Richtig, G.; Haemmerle, M.; Pichler, M. From Biomarkers to Therapeutic Targets—The Promises and Perils of Long Non-Coding RNAs in Cancer. Cancer Metastasis Rev. 2018, 37, 83–105. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. MicroRNA and Cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef]
- Pichler, M.; Calin, G.A. MicroRNAs in Cancer: From Developmental Genes in Worms to Their Clinical Application in Patients. Br. J. Cancer 2015, 113, 569–573. [Google Scholar] [CrossRef]
- Barth, D.A.; Slaby, O.; Klec, C.; Juracek, J.; Drula, R.; Calin, G.A.; Pichler, M. Current Concepts of Non-Coding RNAs in the Pathogenesis of Non-Clear Cell Renal Cell Carcinoma. Cancers 2019, 11, 1580. [Google Scholar] [CrossRef]
- Klec, C.; Prinz, F.; Pichler, M. Involvement of the Long Noncoding RNA NEAT1 in Carcinogenesis. Mol. Oncol. 2019, 13, 46–60. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Pehserl, A.-M.; Ress, A.; Stanzer, S.; Resel, M.; Karbiener, M.; Stadelmeyer, E.; Stiegelbauer, V.; Gerger, A.; Mayr, C.; Scheideler, M.; et al. Comprehensive Analysis of MiRNome Alterations in Response to Sorafenib Treatment in Colorectal Cancer Cells. Int. J. Mol. Sci. 2016, 17, 2011. [Google Scholar] [CrossRef]
- Smolle, M.A.; Prinz, F.; Calin, G.A.; Pichler, M. Current Concepts of Non-Coding RNA Regulation of Immune Checkpoints in Cancer. Mol. Asp. Med. 2019, 70, 117–126. [Google Scholar] [CrossRef]
- Xu, L.; Yang, B.; Ai, J. MicroRNA Transport: A New Way in Cell Communication. J. Cell. Physiol. 2013, 228, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Junker, K.; Heinzelmann, J. Prognostic and Predictive MiRNA Biomarkers in Bladder, Kidney and Prostate Cancer: Where Do We Stand in Biomarker Development? J. Cancer Res. Clin. Oncol. 2016, 142, 1673–1695. [Google Scholar] [CrossRef] [PubMed]
- Constâncio, V.; Tavares, N.T.; Henrique, R.; Jerónimo, C.; Lobo, J. MiRNA Biomarkers in Cancers of the Male Reproductive System: Are We Approaching Clinical Application? Andrology 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.D.; Murray, M.J.; Saini, H.K.; van Dongen, S.; Abreu-Goodger, C.; Muralidhar, B.; Pett, M.R.; Thornton, C.M.; Nicholson, J.C.; Enright, A.J.; et al. Malignant Germ Cell Tumors Display Common MicroRNA Profiles Resulting in Global Changes in Expression of Messenger RNA Targets. Cancer Res. 2010, 70, 2911–2923. [Google Scholar] [CrossRef] [PubMed]
- Ernst, S.; Heinzelmann, J.; Bohle, R.M.; Weber, G.; Stöckle, M.; Junker, K.; Heinzelbecker, J. The Metastatic Potential of Seminomatous Germ Cell Tumours Is Associated with a Specific MicroRNA Pattern. Andrology 2020, 8, 1687–1698. [Google Scholar] [CrossRef]
- Burton, J.; Umu, S.U.; Langseth, H.; Grotmol, T.; Grimsrud, T.K.; Haugen, T.B.; Rounge, T.B. Serum RNA Profiling in the 10-Years Period Prior to Diagnosis of Testicular Germ Cell Tumor. Front. Oncol. 2020, 10, 574977. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. SnapShot: Unconventional MiRNA Functions. Cell 2018, 174, 1038. [Google Scholar] [CrossRef]
- Gillis, A.; Stoop, H.; Hersmus, R.; Oosterhuis, J.; Sun, Y.; Chen, C.; Guenther, S.; Sherlock, J.; Veltman, I.; Baeten, J.; et al. High-Throughput MicroRNAome Analysis in Human Germ Cell Tumours. J. Pathol. 2007, 213, 319–328. [Google Scholar] [CrossRef]
- Murray, M.J.; Saini, H.K.; van Dongen, S.; Palmer, R.D.; Muralidhar, B.; Pett, M.R.; Piipari, M.; Thornton, C.M.; Nicholson, J.C.; Enright, A.J.; et al. The Two Most Common Histological Subtypes of Malignant Germ Cell Tumour Are Distinguished by Global MicroRNA Profiles, Associated with Differential Transcription Factor Expression. Mol. Cancer 2010, 9, 290. [Google Scholar] [CrossRef]
- Rounge, T.; Furu, K.; Skotheim, R.; Haugen, T.; Grotmol, T.; Enerly, E. Profiling of the Small RNA Populations in Human Testicular Germ Cell Tumors Shows Global Loss of PiRNAs. Mol. Cancer 2015, 14, 153. [Google Scholar] [CrossRef]
- Murray, M.J.; Halsall, D.J.; Hook, C.E.; Williams, D.M.; Nicholson, J.C.; Coleman, N. Identification of MicroRNAs From the MiR-371∼373 and MiR-302 Clusters as Potential Serum Biomarkers of Malignant Germ Cell Tumors. Am. J. Clin. Pathol. 2011, 135, 119–125. [Google Scholar] [CrossRef]
- Leão, R.; Albersen, M.; Looijenga, L.H.J.; Tandstad, T.; Kollmannsberger, C.; Murray, M.J.; Culine, S.; Coleman, N.; Belge, G.; Hamilton, R.J.; et al. Circulating MicroRNAs, the Next-Generation Serum Biomarkers in Testicular Germ Cell Tumours: A Systematic Review. Eur. Urol. 2021, 80, 456–466. [Google Scholar] [CrossRef]
- Nappi, L.; Thi, M.; Lum, A.; Huntsman, D.; Eigl, B.J.; Martin, C.; O’Neil, B.; Maughan, B.L.; Chi, K.; So, A.; et al. Developing a Highly Specific Biomarker for Germ Cell Malignancies: Plasma MiR371 Expression Across the Germ Cell Malignancy Spectrum. J. Clin. Oncol. 2019, 37, 3090–3098. [Google Scholar] [CrossRef]
- Vilela-Salgueiro, B.; Barros-Silva, D.; Lobo, J.; Costa, A.L.; Guimarães, R.; Cantante, M.; Lopes, P.; Braga, I.; Oliveira, J.; Henrique, R.; et al. Germ Cell Tumour Subtypes Display Differential Expression of MicroRNA371a-3p. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170338. [Google Scholar] [CrossRef]
- Dieckmann, K.-P.; Radtke, A.; Spiekermann, M.; Balks, T.; Matthies, C.; Becker, P.; Ruf, C.; Oing, C.; Oechsle, K.; Bokemeyer, C.; et al. Serum Levels of MicroRNA MiR-371a-3p: A Sensitive and Specific New Biomarker for Germ Cell Tumours. Eur. Urol. 2017, 71, 213–220. [Google Scholar] [CrossRef]
- Henrique, R.; Jerónimo, C. Testicular Germ Cell Tumors Go Epigenetics: Will MiR-371a-3p Replace Classical Serum Biomarkers? Eur. Urol. 2017, 71, 221–222. [Google Scholar] [CrossRef]
- Regouc, M.; Belge, G.; Lorch, A.; Dieckmann, K.-P.; Pichler, M. Non-Coding MicroRNAs as Novel Potential Tumor Markers in Testicular Cancer. Cancers 2020, 12, 749. [Google Scholar] [CrossRef]
- Syring, I.; Bartels, J.; Holdenrieder, S.; Kristiansen, G.; Müller, S.C.; Ellinger, J. Circulating Serum MiRNA (MiR-367-3p, MiR-371a-3p, MiR-372-3p and MiR-373-3p) as Biomarkers in Patients with Testicular Germ Cell Cancer. J. Urol. 2015, 193, 331–337. [Google Scholar] [CrossRef]
- Mego, M.; Agthoven, T.; Gronesova, P.; Chovanec, M.; Miskovska, V.; Mardiak, J.; Looijenga, L.H.J. Clinical Utility of Plasma MiR-371a-3p in Germ Cell Tumors. J. Cell. Mol. Med. 2019, 23, 1128–1136. [Google Scholar] [CrossRef]
- Mueller, T.; Mueller, L.P.; Luetzkendorf, J.; Voigt, W.; Simon, H.; Schmoll, H.-J. Loss of Oct-3/4 Expression in Embryonal Carcinoma Cells Is Associated with Induction of Cisplatin Resistance. Tumor Biol. 2006, 27, 71–83. [Google Scholar] [CrossRef]
- Schaffrath, J.; Schmoll, H.J.; Voigt, W.; Muller, L.P.; Muller-Tidow, C.; Mueller, T. Efficacy of Targeted Drugs in Germ Cell Cancer Cell Lines with Differential Cisplatin Sensitivity. PLoS ONE 2017, 12, e0178930. [Google Scholar] [CrossRef]
- Roška, J.; Wachsmannová, L.; Hurbanová, L.; Šestáková, Z.; Mueller, T.; Jurkovičová, D.; Chovanec, M. Differential Gene Expression in Cisplatin-Resistant and -Sensitive Testicular Germ Cell Tumor Cell Lines. Oncotarget 2020, 11, 4735–4753. [Google Scholar] [CrossRef]
- Mathew, L.K.; Skuli, N.; Mucaj, V.; Lee, S.S.; Zinn, P.O.; Sathyan, P.; Imtiyaz, H.Z.; Zhang, Z.; Davuluri, R.V.; Rao, S.; et al. MiR-218 Opposes a Critical RTK-HIF Pathway in Mesenchymal Glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 291–296. [Google Scholar] [CrossRef]
- Wu, D.-W.; Cheng, Y.-W.; Wang, J.; Chen, C.-Y.; Lee, H. Paxillin Predicts Survival and Relapse in Non–Small Cell Lung Cancer by MicroRNA-218 Targeting. Cancer Res. 2010, 70, 10392–10401. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; How, C.; Chaudary, N.; Bruce, J.; Shi, W.; Hill, R.P.; Zahedi, P.; Yip, K.W.; Liu, F.-F. The MicroRNA-218~Survivin Axis Regulates Migration, Invasion, and Lymph Node Metastasis in Cervical Cancer. Oncotarget 2015, 6, 1090–1100. [Google Scholar] [CrossRef]
- Wu, D.-W.; Chuang, C.-Y.; Lin, W.-L.; Sung, W.-W.; Cheng, Y.-W.; Lee, H. Paxillin Promotes Tumor Progression and Predicts Survival and Relapse in Oral Cavity Squamous Cell Carcinoma by MicroRNA-218 Targeting. Carcinogenesis 2014, 35, 1823–1829. [Google Scholar] [CrossRef]
- Zanette, D.L.; Rivadavia, F.; Molfetta, G.A.; Barbuzano, F.G.; Proto-Siqueira, R.; Falcão, R.P.; Zago, M.A.; Silva, W.A., Jr. MiRNA Expression Profiles in Chronic Lymphocytic and Acute Lymphocytic Leukemia. Braz. J. Med. Biol. Res. 2007, 40, 1435–1440. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, L.; Zhou, J.; Zhou, S.; Li, L.; Guo, X.; Feng, G.; Ma, Z.; Huang, W.; Huang, F. Mir-218-2 Promotes Glioblastomas Growth, Invasion and Drug Resistance by Targeting CDC27. Oncotarget 2017, 8, 6304–6318. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Maeda, Y.; Taipaleenmaki, H.; Zhang, W.; Jafferji, M.; Gordon, J.A.R.; Li, Z.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; et al. MiR-218 Directs a Wnt Signaling Circuit to Promote Differentiation of Osteoblasts and Osteomimicry of Metastatic Cancer Cells. J. Biol. Chem. 2012, 287, 42084–42092. [Google Scholar] [CrossRef]
- Zhuang, Z.; Hu, F.; Hu, J.; Wang, C.; Hou, J.; Yu, Z.; Wang, T.T.; Liu, X.; Huang, H. MicroRNA-218 Promotes Cisplatin Resistance in Oral Cancer via the PPP2R5A/Wnt Signaling Pathway. Oncol. Rep. 2017, 38, 2051–2061. [Google Scholar] [CrossRef]
- Kalev, P.; Simicek, M.; Vazquez, I.; Munck, S.; Chen, L.; Soin, T.; Danda, N.; Chen, W.; Sablina, A. Loss of PPP2R2A Inhibits Homologous Recombination DNA Repair and Predicts Tumor Sensitivity to PARP Inhibition. Cancer Res. 2012, 72, 6414–6424. [Google Scholar] [CrossRef]
- Hsu, H.-H.; Kuo, W.-W.; Shih, H.-N.; Cheng, S.-F.; Yang, C.-K.; Chen, M.-C.; Tu, C.-C.; Viswanadha, V.P.; Liao, P.-H.; Huang, C.-Y. FOXC1 Regulation of MiR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers 2019, 11, 1576. [Google Scholar] [CrossRef]
- Oshima, S.; Asai, S.; Seki, N.; Minemura, C.; Kinoshita, T.; Goto, Y.; Kikkawa, N.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Identification of Tumor Suppressive Genes Regulated by MiR-31-5p and MiR-31-3p in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 6199. [Google Scholar] [CrossRef]
- He, J.; He, J.; Min, L.; He, Y.; Guan, H.; Wang, J.; Peng, X. Extracellular Vesicles Transmitted MiR-31-5p Promotes Sorafenib Resistance by Targeting MLH1 in Renal Cell Carcinoma. Int. J. Cancer 2020, 146, 1052–1063. [Google Scholar] [CrossRef]
- Shen, X.; Lei, J.; Du, L. MiR-31-5p May Enhance the Efficacy of Chemotherapy with Taxol and Cisplatin in TNBC. Exp. Ther. Med. 2019, 19, 375–383. [Google Scholar] [CrossRef]
- Hummel, R. MicroRNA Signatures in Chemotherapy Resistant Esophageal Cancer Cell Lines. World J. Gastroenterol. 2014, 20, 14904. [Google Scholar] [CrossRef]
- Lafin, J.T.; Kenigsberg, A.P.; Meng, X.; Abe, D.; Savelyeva, A.; Singla, N.; Woldu, S.L.; Lotan, Y.; Mauck, R.J.; Lewis, C.M.; et al. Serum Small RNA Sequencing and MiR-375 Assay Do Not Identify the Presence of Pure Teratoma at Postchemotherapy Retroperitoneal Lymph Node Dissection. Eur. Urol. Open Sci. 2021, 26, 83–87. [Google Scholar] [CrossRef]
- Lobo, J.; Gillis, A.J.M.; van den Berg, A.; Dorssers, L.C.J.; Belge, G.; Dieckmann, K.-P.; Roest, H.P.; van der Laan, L.J.W.; Gietema, J.; Hamilton, R.J.; et al. Identification and Validation Model for Informative Liquid Biopsy-Based MicroRNA Biomarkers: Insights from Germ Cell Tumor In Vitro, In Vivo and Patient-Derived Data. Cells 2019, 8, 1637. [Google Scholar] [CrossRef]
- Toffanin, S.; Hoshida, Y.; Lachenmayer, A.; Villanueva, A.; Cabellos, L.; Minguez, B.; Savic, R.; Ward, S.C.; Thung, S.; Chiang, D.Y.; et al. MicroRNA-Based Classification of Hepatocellular Carcinoma and Oncogenic Role of MiR-517a. Gastroenterology 2011, 140, 1618–1628. [Google Scholar] [CrossRef]
- Ward, A.; Shukla, K.; Balwierz, A.; Soons, Z.; König, R.; Sahin, Ö.; Wiemann, S. MicroRNA-519a Is a Novel Oncomir Conferring Tamoxifen Resistance by Targeting a Network of Tumour-suppressor Genes in ER + Breast Cancer. J. Pathol. 2014, 233, 368–379. [Google Scholar] [CrossRef]
- Flor, I.; Spiekermann, M.; Löning, T.; Dieckmann, K.-P.; Belge, G.; Bullerdiek, J. Expression of MicroRNAs of C19MC in Different Histological Types of Testicular Germ Cell Tumour. Cancer Genom. Proteom. 2016, 13, 281–289. [Google Scholar]
- Huang, X.; Yang, Y.; Yang, C.; Li, H.; Cheng, H.; Zheng, Y. Overexpression of LBX2 Associated with Tumor Progression and Poor Prognosis in Colorectal Cancer. Oncol. Lett. 2020, 19, 3751–3760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dai, S.; Zhen, T.; Shi, H.; Zhang, F.; Yang, Y.; Kang, L.; Liang, Y.; Han, A. Clinical and Biological Significance of MiR-378a-3p and MiR-378a-5p in Colorectal Cancer. Eur. J. Cancer 2014, 50, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yao, T.; Liu, W. MiR-378a-3p Sensitizes Ovarian Cancer Cells to Cisplatin through Targeting MAPK1/GRB2. Biomed. Pharmacother. 2018, 107, 1410–1417. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, J. MiR-378a-3p Regulates Glioma Cell Chemosensitivity to Cisplatin through IGF1R. Open Life Sci. 2021, 16, 1175–1181. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, J.-J.; Feng, Y.; Yang, J.-L.; Huang, H.; Chung, W.Y.; Hu, Y.-L.; Xue, W.-J. DNMT1-Induced MiR-378a-3p Silencing Promotes Angiogenesis via the NF-ΚB Signaling Pathway by Targeting TRAF1 in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 352. [Google Scholar] [CrossRef]
- Ding, C.; Ding, X.; Zheng, J.; Wang, B.; Li, Y.; Xiang, H.; Dou, M.; Qiao, Y.; Tian, P.; Xue, W. MiR-182-5p and MiR-378a-3p Regulate Ferroptosis in I/R-Induced Renal Injury. Cell Death Dis. 2020, 11, 929. [Google Scholar] [CrossRef]
- Ding, H.; Luo, Y.; Hu, K.; Liu, P.; Xiong, M. Linc00467 Promotes Lung Adenocarcinoma Proliferation via Sponging MiR-20b-5p to Activate CCND1 Expression. Onco. Targets Ther. 2019, 12, 6733–6743. [Google Scholar] [CrossRef]
- Yang, H.; Lin, J.; Jiang, J.; Ji, J.; Wang, C.; Zhang, J. MiR-20b-5p Functions as Tumor Suppressor MicroRNA by Targeting CyclinD1 in Colon Cancer. Cell Cycle 2020, 19, 2939–2954. [Google Scholar] [CrossRef]
- Tang, D.; Yang, Z.; Long, F.; Luo, L.; Yang, B.; Zhu, R.; Sang, X.; Cao, G.; Wang, K. Long Noncoding RNA MALAT1 Mediates Stem Cell-like Properties in Human Colorectal Cancer Cells by Regulating MiR-20b-5p/Oct4 Axis. J. Cell. Physiol. 2019, 234, 20816–20828. [Google Scholar] [CrossRef]
- Zhou, L.; Liang, X.; Zhang, L.; Yang, L.; Nagao, N.; Wu, H.; Liu, C.; Lin, S.; Cai, G.; Liu, J. MiR-27a-3p Functions as an Oncogene in Gastric Cancer by Targeting BTG2. Oncotarget 2016, 7, 51943–51954. [Google Scholar] [CrossRef]
- Zheng, F.; Wang, M.; Li, Y.; Huang, C.; Tao, D.; Xie, F.; Zhang, H.; Sun, J.; Zhang, C.; Gu, C.; et al. CircNR3C1 Inhibits Proliferation of Bladder Cancer Cells by Sponging MiR-27a-3p and Downregulating Cyclin D1 Expression. Cancer Lett. 2019, 460, 139–151. [Google Scholar] [CrossRef]
- Yang, X.; Chen, J.; Liao, Y.; Huang, L.; Wen, C.; Lin, M.; Li, W.; Zhu, Y.; Wu, X.; Iwamoto, A.; et al. MiR-27b-3p Promotes Migration and Invasion in Colorectal Cancer Cells by Targeting HOXA10. Biosci. Rep. 2019, 39, BSR20191087. [Google Scholar] [CrossRef]
- Li, H.-L.; Wei, J.-F.; Fan, L.-Y.; Wang, S.-H.; Zhu, L.; Li, T.-P.; Lin, G.; Sun, Y.; Sun, Z.-J.; Ding, J.; et al. MiR-302 Regulates Pluripotency, Teratoma Formation and Differentiation in Stem Cells via an AKT1/OCT4-Dependent Manner. Cell Death Dis. 2016, 7, e2078. [Google Scholar] [CrossRef]
- Liang, J.; Tang, J.; Shi, H.; Li, H.; Zhen, T.; Duan, J.; Kang, L.; Zhang, F.; Dong, Y.; Han, A. MiR-27a-3p Targeting RXRα Promotes Colorectal Cancer Progression by Activating Wnt/β-Catenin Pathway. Oncotarget 2017, 8, 82991–83008. [Google Scholar] [CrossRef]
- Shen, S.; Sun, Q.; Liang, Z.; Cui, X.; Ren, X.; Chen, H.; Zhang, X.; Zhou, Y. A Prognostic Model of Triple-Negative Breast Cancer Based on MiR-27b-3p and Node Status. PLoS ONE 2014, 9, e100664. [Google Scholar] [CrossRef]
- Tao, J.; Zhi, X.; Zhang, X.; Fu, M.; Huang, H.; Fan, Y.; Guan, W.; Zou, C. MiR-27b-3p Suppresses Cell Proliferation through Targeting Receptor Tyrosine Kinase like Orphan Receptor 1 in Gastric Cancer. J. Exp. Clin. Cancer Res. 2015, 34, 139. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, T.; Cao, Y.-W.; Ding, X.-Q. Antitumor Effect of MiR-27b-3p on Lung Cancer Cells via Targeting Fzd7. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4113–4123. [Google Scholar]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L.; et al. MiR-27b-3p Inhibits Proliferation and Potentially Reverses Multi-Chemoresistance by Targeting CBLB/GRB2 in Breast Cancer Cells. Cell Death Dis. 2018, 9, 188. [Google Scholar] [CrossRef]
- Mueller, T.; Luetzkendorf, J.; Nerger, K.; Schmoll, H.-J.; Mueller, L.P. Analysis of OCT4 Expression in an Extended Panel of Human Tumor Cell Lines from Multiple Entities and in Human Mesenchymal Stem Cells. Cell. Mol. Life Sci. 2009, 66, 495–503. [Google Scholar] [CrossRef]
- Mueller, T.; Hantsch, C.; Volkmer, I.; Staege, M.S. Differentiation-Dependent Regulation of Human Endogenous Retrovirus K Sequences and Neighboring Genes in Germ Cell Tumor Cells. Front. Microbiol. 2018, 9, 1253. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Feng, J.; Cui, X.; Huang, W.; Li, Y.; Su, F.; Liu, Q.; Zhu, J.; Lv, X.; et al. Lin28 Induces Epithelial-to-Mesenchymal Transition and Stemness via Downregulation of Let-7a in Breast Cancer Cells. PLoS ONE 2013, 8, e83083. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Miki, Y.; Masuda, M.; Hata, S.; Shibahara, Y.; Hirakawa, H.; Suzuki, T.; Sasano, H. LIN28: A Regulator of Tumor-Suppressing Activity of Let-7 MicroRNA in Human Breast Cancer. J. Steroid Biochem. Mol. Biol. 2012, 131, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Chang, H.-M.; Piskounova, E.; Gregory, R.I. Lin28-Mediated Control of Let-7 MicroRNA Expression by Alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef]
- De Martino, M.; Esposito, F.; Pellecchia, S.; Cortez Cardoso Penha, R.; Botti, G.; Fusco, A.; Chieffi, P. HMGA1-Regulating MicroRNAs Let-7a and MiR-26a Are Downregulated in Human Seminomas. Int. J. Mol. Sci. 2020, 21, 3014. [Google Scholar] [CrossRef]
- Batool, A.; Wang, Y.-Q.; Hao, X.-X.; Chen, S.-R.; Liu, Y.-X. A MiR-125b/CSF1-CX3CL1/Tumor-Associated Macrophage Recruitment Axis Controls Testicular Germ Cell Tumor Growth. Cell Death Dis. 2018, 9, 962. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Fan, H.; Zhang, Z.; Li, N. MiR-125b-5p Inhibits Breast Cancer Cell Proliferation, Migration and Invasion by Targeting KIAA1522. Biochem. Biophys. Res. Commun. 2018, 504, 277–282. [Google Scholar] [CrossRef]
- Yang, D.; Zhan, M.; Chen, T.; Chen, W.; Zhang, Y.; Xu, S.; Yan, J.; Huang, Q.; Wang, J. MiR-125b-5p Enhances Chemotherapy Sensitivity to Cisplatin by down-Regulating Bcl2 in Gallbladder Cancer. Sci. Rep. 2017, 7, 43109. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Q.; Wang, Y. MiR-125b-5p Suppresses the Bladder Cancer Progression via Targeting HK2 and Suppressing PI3K/AKT Pathway. Hum. Cell 2020, 33, 185–194. [Google Scholar] [CrossRef]
- D’Angelo, E.; Fassan, M.; Maretto, I.; Pucciarelli, S.; Zanon, C.; Digito, M.; Rugge, M.; Nitti, D.; Agostini, M. Serum MiR-125b Is a Non-Invasive Predictive Biomarker of the Pre-Operative Chemoradiotherapy Responsiveness in Patients with Rectal Adenocarcinoma. Oncotarget 2016, 7, 28647–28657. [Google Scholar] [CrossRef]
- Peng, B.; Theng, P.Y.; Le, M.T.N. Essential Functions of MiR-125b in Cancer. Cell Prolif. 2021, 54, e12913. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, W.; Wang, Y.; Zou, T.; Zhang, B.; Xu, Y.; Pang, T.; Hu, Q.; Chen, M.; Wang, L.; et al. Hypoxia-Induced MiR-214 Expression Promotes Tumour Cell Proliferation and Migration by Enhancing the Warburg Effect in Gastric Carcinoma Cells. Cancer Lett. 2018, 414, 44–56. [Google Scholar] [CrossRef]
- Dettori, D.; Orso, F.; Penna, E.; Baruffaldi, D.; Brundu, S.; Maione, F.; Turco, E.; Giraudo, E.; Taverna, D. Therapeutic Silencing of MiR-214 Inhibits Tumor Progression in Multiple Mouse Models. Mol. Ther. 2018, 26, 2008–2018. [Google Scholar] [CrossRef]
- Hsieh, T.-H.; Liu, Y.-R.; Chang, T.-Y.; Liang, M.-L.; Chen, H.-H.; Wang, H.-W.; Yen, Y.; Wong, T.-T. Global DNA Methylation Analysis Reveals MiR-214-3p Contributes to Cisplatin Resistance in Pediatric Intracranial Nongerminomatous Malignant Germ Cell Tumors. Neuro. Oncol. 2018, 20, 519–530. [Google Scholar] [CrossRef]
- Liu, F.; Lou, K.; Zhao, X.; Zhang, J.; Chen, W.; Qian, Y.; Zhao, Y.; Zhu, Y.; Zhang, Y. MiR-214 Regulates Papillary Thyroid Carcinoma Cell proliferation and Metastasis by Targeting PSMD10. Int. J. Mol. Med. 2018, 42, 3027–3036. [Google Scholar] [CrossRef]
- Chen, X.; Du, J.; Jiang, R.; Li, L. MicroRNA-214 Inhibits the Proliferation and Invasion of Lung Carcinoma Cells by Targeting JAK1. Am. J. Transl. Res. 2018, 10, 1164–1171. [Google Scholar]
- Cheung, H.H.; Lee, T.L.; Davis, A.J.; Taft, D.H.; Rennert, O.M.; Chan, W.Y. Genome-Wide DNA Methylation Profiling Reveals Novel Epigenetically Regulated Genes and Non-Coding RNAs in Human Testicular Cancer. Br. J. Cancer 2010, 102, 419–427. [Google Scholar] [CrossRef]
- Gu, S.; Cheung, H.H.; Lee, T.L.; Lu, G.; Poon, W.S.; Chan, W.Y. Molecular Mechanisms of Regulation and Action of MicroRNA-199a in Testicular Germ Cell Tumor and Glioblastomas. PLoS ONE 2013, 8, e83980. [Google Scholar] [CrossRef]
- Cheung, H.-H.; Davis, A.J.; Lee, T.-L.; Pang, A.L.; Nagrani, S.; Rennert, O.M.; Chan, W.-Y. Methylation of an Intronic Region Regulates MiR-199a in Testicular Tumor Malignancy. Oncogene 2011, 30, 3404–3415. [Google Scholar] [CrossRef]
- Lee, Y.-B.; Bantounas, I.; Lee, D.-Y.; Phylactou, L.; Caldwell, M.A.; Uney, J.B. Twist-1 Regulates the MiR-199a/214 Cluster during Development. Nucleic Acids Res. 2009, 37, 123–128. [Google Scholar] [CrossRef]
- Sakurai, K.; Furukawa, C.; Haraguchi, T.; Inada, K.; Shiogama, K.; Tagawa, T.; Fujita, S.; Ueno, Y.; Ogata, A.; Ito, M.; et al. MicroRNAs MiR-199a-5p and -3p Target the Brm Subunit of SWI/SNF to Generate a Double-Negative Feedback Loop in a Variety of Human Cancers. Cancer Res. 2011, 71, 1680–1689. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-F.; Suen, Y.-K.; Gu, S.; Li, L.; Chan, W.-Y. A MiR-199a/MiR-214 Self-Regulatory Network via PSMD10, TP53 and DNMT1 in Testicular Germ Cell Tumor. Sci. Rep. 2014, 4, 6413. [Google Scholar] [CrossRef]
- Chen, J.; Shin, V.Y.; Siu, M.T.; Ho, J.C.W.; Cheuk, I.; Kwong, A. MiR-199a-5p Confers Tumor-Suppressive Role in Triple-Negative Breast Cancer. BMC Cancer 2016, 16, 887. [Google Scholar] [CrossRef] [PubMed]
- Beyer, J.; Albers, P.; Altena, R.; Aparicio, J.; Bokemeyer, C.; Busch, J.; Cathomas, R.; Cavallin-Stahl, E.; Clarke, N.W.; Claßen, J.; et al. Maintaining Success, Reducing Treatment Burden, Focusing on Survivorship: Highlights from the Third European Consensus Conference on Diagnosis and Treatment of Germ-Cell Cancer. Ann. Oncol. 2013, 24, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Albers, P.; Albrecht, W.; Algaba, F.; Bokemeyer, C.; Cohn-Cedermark, G.; Fizazi, K.; Horwich, A.; Laguna, M.P.; Nicolai, N.; Oldenburg, J. Guidelines on Testicular Cancer: 2015 Update. Eur. Urol. 2015, 68, 1054–1068. [Google Scholar] [CrossRef]
- Boormans, J.L.; Mayor de Castro, J.; Marconi, L.; Yuan, Y.; Laguna Pes, M.P.; Bokemeyer, C.; Nicolai, N.; Algaba, F.; Oldenburg, J.; Albers, P. Testicular Tumour Size and Rete Testis Invasion as Prognostic Factors for the Risk of Relapse of Clinical Stage I Seminoma Testis Patients Under Surveillance: A Systematic Review by the Testicular Cancer Guidelines Panel. Eur. Urol. 2018, 73, 394–405. [Google Scholar] [CrossRef]
- Tandstad, T.; Kollmannsberger, C.K.; Roth, B.J.; Jeldres, C.; Gillessen, S.; Fizazi, K.; Daneshmand, S.; Lowrance, W.T.; Hanna, N.H.; Albany, C.; et al. Practice Makes Perfect: The Rest of the Story in Testicular Cancer as a Model Curable Neoplasm. J. Clin. Oncol. 2017, 35, 3525–3528. [Google Scholar] [CrossRef]
- Doherty, A.P.; Bower, M.; Christmas, T.J. The Role of Tumour Markers in the Diagnosis and Treatment of Testicular Germ Cell Cancers. Br. J. Urol. 1997, 79, 247–252. [Google Scholar] [CrossRef]
- Eyben, F.E. von Laboratory Markers and Germ Cell Tumors. Crit. Rev. Clin. Lab. Sci. 2003, 40, 377–427. [Google Scholar] [CrossRef]
- Montgomery, J.; Weizer, A.; Filson, C.; Milose, J. Khaled Hafez Role of Biochemical Markers in Testicular Cancer: Diagnosis, Staging, and Surveillance. Open Access J. Urol. 2011, 2012, 1–8. [Google Scholar] [CrossRef]
- Sequeira, J.P.; Lobo, J.; Constâncio, V.; Brito-Rocha, T.; Carvalho-Maia, C.; Braga, I.; Maurício, J.; Henrique, R.; Jerónimo, C. DigiMir Test: Establishing a Novel Pipeline for MiR-371a Quantification Using Droplet Digital PCR in Liquid Biopsies from Testicular Germ Cell Tumor Patients. Front. Oncol. 2022, 12, 876732. [Google Scholar] [CrossRef]
- Lobo, J.; Costa, A.L.; Vilela-Salgueiro, B.; Rodrigues, Â.; Guimarães, R.; Cantante, M.; Lopes, P.; Antunes, L.; Jerónimo, C.; Henrique, R. Testicular Germ Cell Tumors: Revisiting a Series in Light of the New WHO Classification and AJCC Staging Systems, Focusing on Challenges for Pathologists. Hum. Pathol. 2018, 82, 113–124. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of Functional MicroRNA Targets by Integrative Modeling of MicroRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of MicroRNA Targeting Efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Garcia, D.M.; Baek, D.; Shin, C.; Bell, G.W.; Grimson, A.; Bartel, D.P. Weak Seed-Pairing Stability and High Target-Site Abundance Decrease the Proficiency of Lsy-6 and Other MicroRNAs. Nat. Struct. Mol. Biol. 2011, 18, 1139–1146. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER Version 16: A Revised Family Classification, Tree-Based Classification Tool, Enhancer Regions and Extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for Large-Scale Genome and Gene Function Analysis with the PANTHER Classification System (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Thomas, P.D.; Kejariwal, A.; Guo, N.; Mi, H.; Campbell, M.J.; Muruganujan, A.; Lazareva-Ulitsky, B. Applications for Protein Sequence-Function Evolution Data: MRNA/Protein Expression Analysis and Coding SNP Scoring Tools. Nucleic Acids Res. 2006, 34, W645–W650. [Google Scholar] [CrossRef]
- Mi, H.; Thomas, P. PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar]

| miRNA | TargetScanHuman Predicted Targets [n (Conserved Sites; Poorly Conserved Sites)] | miRDB Predicted Targets (n) | Overlap of the Same Target Genes (n) |
|---|---|---|---|
| miR-218-5p | 1102 (1262; 453) | 1084 | 686 |
| miR-31-5p | 477 (510; 228) | 595 | 221 |
| miR-199a-5p | 634 (723; 287) | 562 | 326 |
| miR-125b-5p | 931 (1038; 452) | 925 | 569 |
| miR-27b-3p | 1421 (1613; 762) | 1497 | 895 |
| let-7a-5p | 1207 (1386; 243) | 990 | 755 |
| miR-517a-3p | 557 | 58 | 54 |
| miRNAs | Forward (5′-′3) | Reverse (5′-′3) |
|---|---|---|
| miR-218-5p | CGCAGTTGTGCTTGATCTAAC | CAGGTCCAGTTTTTTTTTTTTTTTACAT |
| miR-31-5p | GCAGAGGCAAGATGCTG | TCCAGTTTTTTTTTTTTTTTAGCTAT |
| miR-199a-5p | GCAGCCCAGTGTTCAGA | CCAGTTTTTTTTTTTTTTTGAACA |
| miR-214-3p | GCAGACAGCAGGCACA | AGTTTTTTTTTTTTTTTACTGCCT |
| miR-125b-5p | GCAGTCCCTGAGACCCTAA | TCCAGTTTTTTTTTTTTTTTTCACA |
| miR-27b-3p | GCAGTTCACAGTGGCTAAG | CCAGTTTTTTTTTTTTTTTGCAG |
| miR-320a-5p | GCGCAGGCCTTCTCTT | TCCAGTTTTTTTTTTTTTTTGGAA |
| miR-324-5p | GCAGCGCATCCCCTA | CAGTTTTTTTTTTTTTTTCACCAAT |
| miR-374b-5p | CAGCGCAGATATAATACAACCT | GGTCCAGTTTTTTTTTTTTTTTCACTT |
| miR-378a-3p | GCAGACTGGACTTGGAGT | CCAGTTTTTTTTTTTTTTTGCCTT |
| miR-20b-5p | CGCAGCAAAGTGCTCATA | CCAGTTTTTTTTTTTTTTTCTACCT |
| miR-30e-3p | CGCAGCTTTCAGTCGGA | TCCAGTTTTTTTTTTTTTTTGCTGT |
| let-7a-5p | CGCAGTGAGGTAGTAGGT | GTCCAGTTTTTTTTTTTTTTTAACTATA |
| miR-375-5p | GGCGACGAGCCCCTC | CAGTTTTTTTTTTTTTTTGGTTTGT |
| miR-517a-3p | GCAGATCGTGCATCCCTTTA | GGTCCAGTTTTTTTTTTTTTTTACAC |
| Housekeepers | ||
| SNORD44 | CAGCCTGGATGATGATAAGCAAAT | GGTCCAGTTTTTTTTTTTTTTTAGTCAGTT |
| SNORD38B | GCAGCTCAGTGATGAAAACTTTGT | AGGTCCAGTTTTTTTTTTTTTTTTCTCCT |
| SNORD66 | GCAGTTCCTCTGATGACTTCCT | GTCCAGTTTTTTTTTTTTTTTTTCCTCAGA |
| miR-191-5p | GCAGCAACGGAATCCCAA | CCAGTTTTTTTTTTTTTTTCAGCT |
| miR-93-5p | CGCAGCAAAGTGCTGTT | TCCAGTTTTTTTTTTTTTTTCTACCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roška, J.; Lobo, J.; Ivovič, D.; Wachsmannová, L.; Mueller, T.; Henrique, R.; Jerónimo, C.; Chovanec, M.; Jurkovičová, D. Integrated Microarray-Based Data Analysis of miRNA Expression Profiles: Identification of Novel Biomarkers of Cisplatin-Resistance in Testicular Germ Cell Tumours. Int. J. Mol. Sci. 2023, 24, 2495. https://doi.org/10.3390/ijms24032495
Roška J, Lobo J, Ivovič D, Wachsmannová L, Mueller T, Henrique R, Jerónimo C, Chovanec M, Jurkovičová D. Integrated Microarray-Based Data Analysis of miRNA Expression Profiles: Identification of Novel Biomarkers of Cisplatin-Resistance in Testicular Germ Cell Tumours. International Journal of Molecular Sciences. 2023; 24(3):2495. https://doi.org/10.3390/ijms24032495
Chicago/Turabian StyleRoška, Jan, João Lobo, Danica Ivovič, Lenka Wachsmannová, Thomas Mueller, Rui Henrique, Carmen Jerónimo, Miroslav Chovanec, and Dana Jurkovičová. 2023. "Integrated Microarray-Based Data Analysis of miRNA Expression Profiles: Identification of Novel Biomarkers of Cisplatin-Resistance in Testicular Germ Cell Tumours" International Journal of Molecular Sciences 24, no. 3: 2495. https://doi.org/10.3390/ijms24032495
APA StyleRoška, J., Lobo, J., Ivovič, D., Wachsmannová, L., Mueller, T., Henrique, R., Jerónimo, C., Chovanec, M., & Jurkovičová, D. (2023). Integrated Microarray-Based Data Analysis of miRNA Expression Profiles: Identification of Novel Biomarkers of Cisplatin-Resistance in Testicular Germ Cell Tumours. International Journal of Molecular Sciences, 24(3), 2495. https://doi.org/10.3390/ijms24032495












