An ELOVL2-Based Epigenetic Clock for Forensic Age Prediction: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Search
2.2. Exclusion and Inclusion Criteria
2.3. Statistical Analysis
3. Results
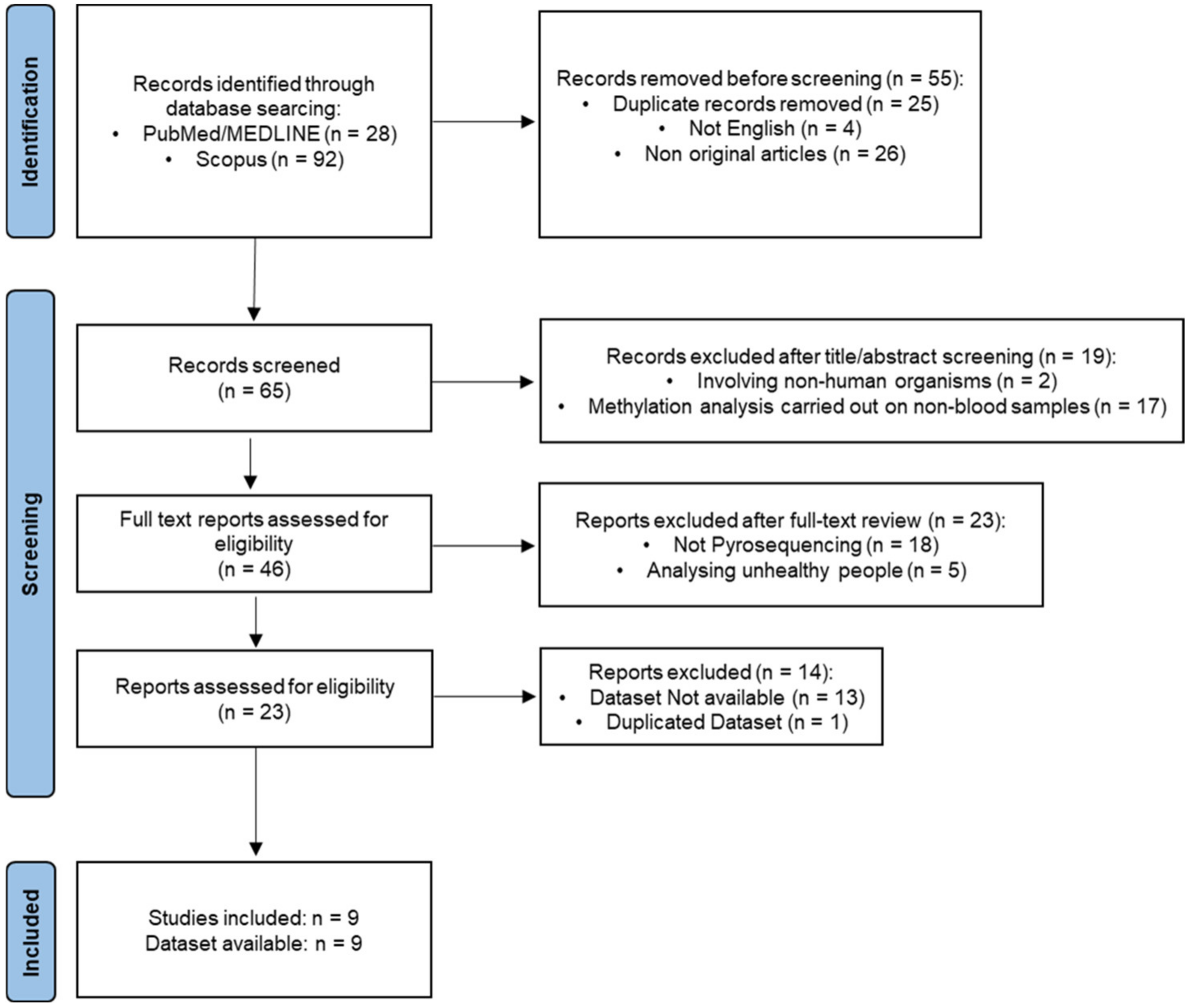
3.1. Study Selection
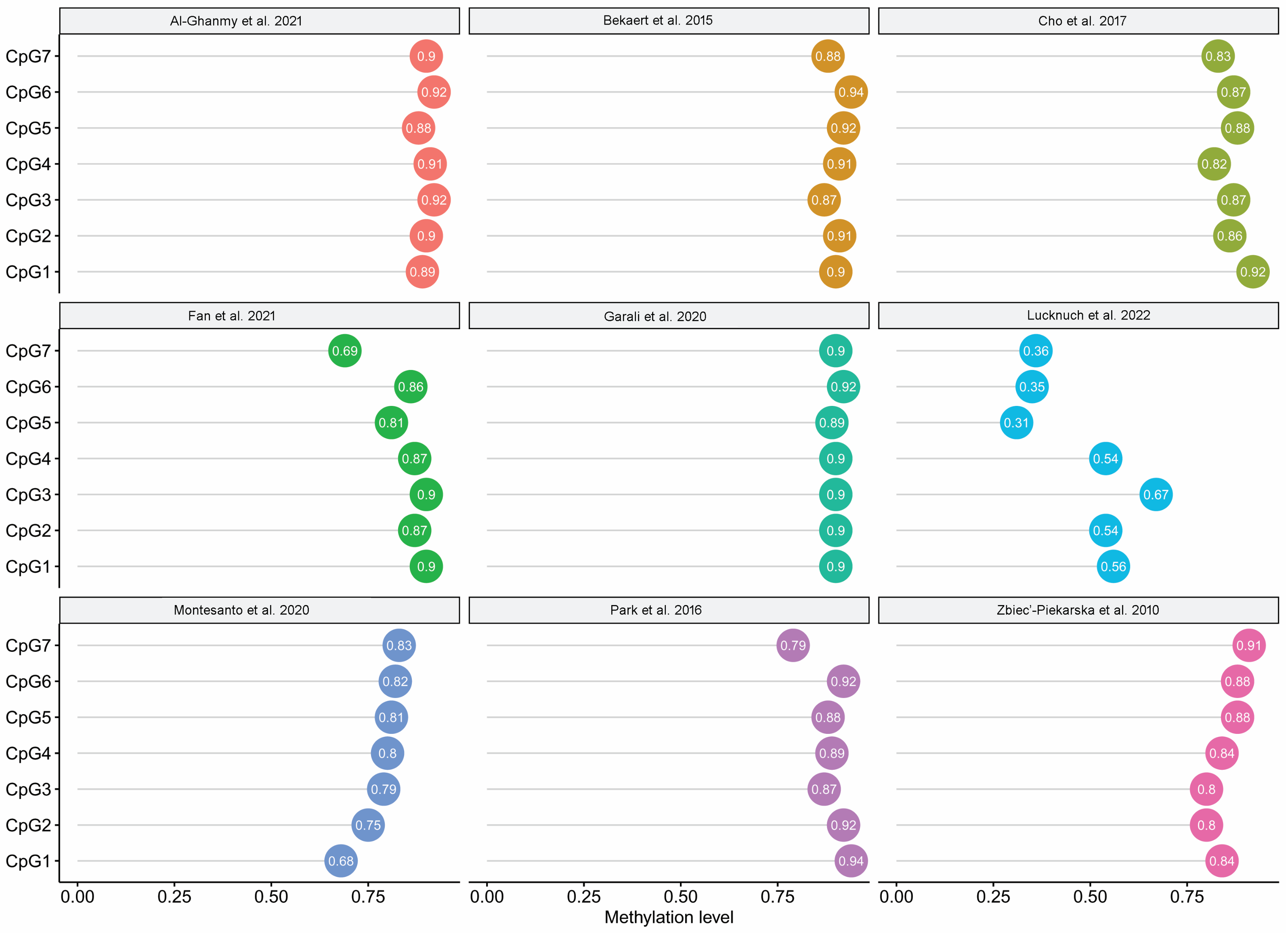
3.2. PCA Results
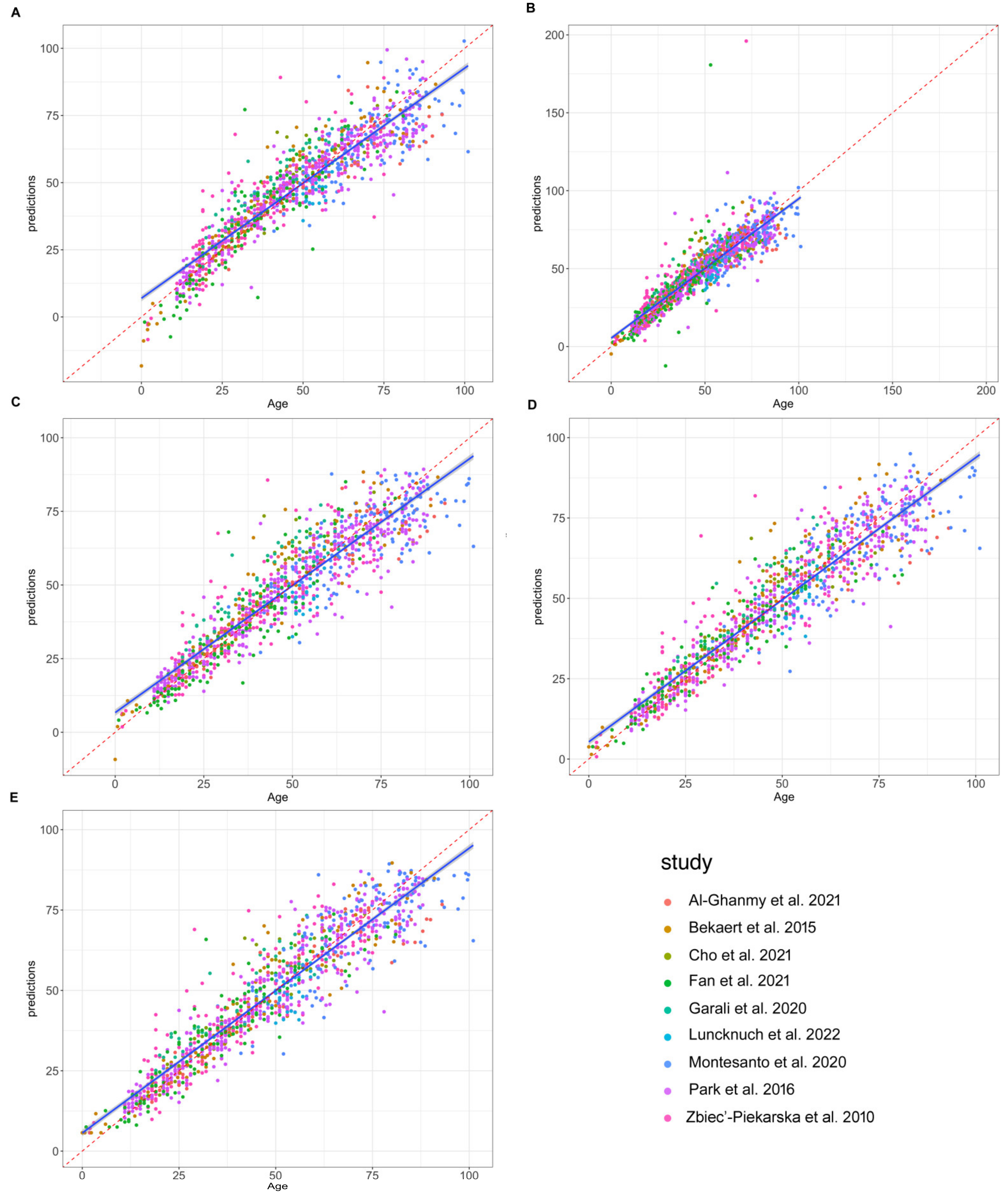
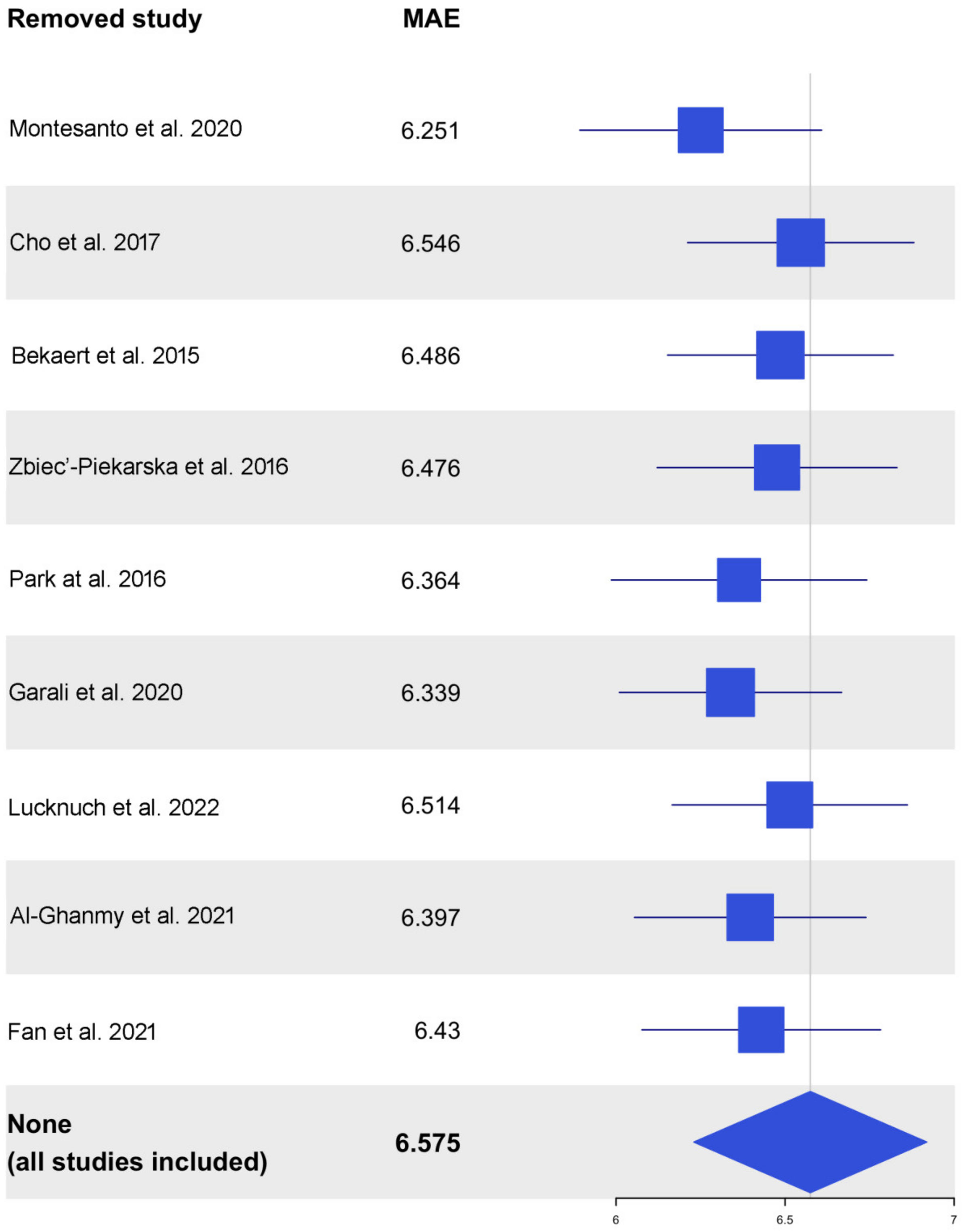
3.3. ELOVL2 Methylation-Based Prediction Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Correia Dias, H.; Cunha, E.; Corte Real, F.; Manco, L. Challenges and (Un)Certainties for DNAm Age Estimation in Future. Forensic Sci. 2022, 2, 44. [Google Scholar] [CrossRef]
- Bell, C.G.; Lowe, R.; Adams, P.D.; Baccarelli, A.A.; Beck, S.; Bell, J.T.; Christensen, B.C.; Gladyshev, V.N.; Heijmans, B.T.; Horvath, S.; et al. DNA methylation aging clocks: Challenges and recommendations. Genome Biol. 2019, 20, 249. [Google Scholar] [CrossRef] [PubMed]
- Cunha, E.; Baccino, E.; Martrille, L.; Ramsthaler, F.; Prieto, J.; Schuliar, Y.; Lynnerup, N.; Cattaneo, C. The problem of aging human remains and living individuals: A review. Forensic Sci. Int. 2009, 193, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- Zaguia, A.; Pandey, D.; Painuly, S.; Pal, S.K.; Garg, V.K.; Goel, N. DNA Methylation Biomarkers-Based Human Age Prediction Using Machine Learning. Comput. Intell. Neurosci. 2022, 2022, 8393498. [Google Scholar] [CrossRef]
- Fan, H.; Xie, Q.; Zhang, Z.; Wang, J.; Chen, X.; Qiu, P. Chronological Age Prediction: Developmental Evaluation of DNA Methylation-Based Machine Learning Models. Front. Bioeng. Biotechnol. 2021, 9, 819991. [Google Scholar] [CrossRef]
- Xu, C.; Qu, H.; Wang, G.; Xie, B.; Shi, Y.; Yang, Y.; Zhao, Z.; Hu, L.; Fang, X.; Yan, J.; et al. A novel strategy for forensic age prediction by DNA methylation and support vector regression model. Sci. Rep. 2015, 5, 17788. [Google Scholar] [CrossRef]
- Thong, Z.; Tan, J.Y.Y.; Loo, E.S.; Phua, Y.W.; Chan, X.L.S.; Syn, C.K. Artificial neural network, predictor variables and sensitivity threshold for DNA methylation-based age prediction using blood samples. Sci. Rep. 2021, 11, 1744. [Google Scholar] [CrossRef]
- Li, X.; Li, W.; Xu, Y. Human Age Prediction Based on DNA Methylation Using a Gradient Boosting Regressor. Genes 2018, 9, 424. [Google Scholar] [CrossRef]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Gori, D.; Giuliani, C.; Mari, D.; Di Blasio, A.M.; Gentilini, D.; Vitale, G.; Collino, S.; et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 2012, 11, 1132–1134. [Google Scholar] [CrossRef]
- Daunay, A.; Baudrin, L.G.; Deleuze, J.F.; How-Kit, A. Evaluation of six blood-based age prediction models using DNA methylation analysis by pyrosequencing. Sci. Rep. 2019, 9, 8862. [Google Scholar] [CrossRef]
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Parys-Proszek, A.; Makowska, Ż.; Pałeczka, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci. Int. Genet. 2015, 17, 173–179. [Google Scholar] [CrossRef]
- Garali, I.; Sahbatou, M.; Daunay, A.; Baudrin, L.G.; Renault, V.; Bouyacoub, Y.; Deleuze, J.F.; How-Kit, A. Improvements and inter-laboratory implementation and optimization of blood-based single-locus age prediction models using DNA methylation of the ELOVL2 promoter. Sci. Rep. 2020, 10, 15652. [Google Scholar] [CrossRef]
- Manco, L.; Dias, H.C. DNA methylation analysis of ELOVL2 gene using droplet digital PCR for age estimation purposes. Forensic Sci. Int. 2022, 333, 111206. [Google Scholar] [CrossRef]
- Slieker, R.C.; Relton, C.L.; Gaunt, T.R.; Slagboom, P.E.; Heijmans, B.T. Age-related DNA methylation changes are tissue-specific with ELOVL2 promoter methylation as exception. Epigenetics Chromatin 2018, 11, 25. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Al-Ghanmy, H.S.G.; Al-Rashedi, N.A.M.; Ayied, A.Y. Age estimation by DNA methylation levels in Iraqi subjects. Gene Rep. 2021, 23, 101022. [Google Scholar] [CrossRef]
- Bekaert, B.; Kamalandua, A.; Zapico, S.C.; Van de Voorde, W.; Decorte, R. Improved age determination of blood and teeth samples using a selected set of DNA methylation markers. Epigenetics 2015, 10, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jung, S.E.; Hong, S.R.; Lee, E.H.; Lee, J.H.; Lee, S.D.; Lee, H.Y. Independent validation of DNA-based approaches for age prediction in blood. Forensic Sci. Int. Genet. 2017, 29, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Lucknuch, T.; Praihirunkit, P. Evaluation of age-associated DNA methylation markers in colorectal cancer of Thai population. Forensic Sci. Int. Rep. 2022, 5, 100265. [Google Scholar] [CrossRef]
- Montesanto, A.; D’Aquila, P.; Lagani, V.; Paparazzo, E.; Geracitano, S.; Formentini, L.; Giacconi, R.; Cardelli, M.; Provinciali, M.; Bellizzi, D.; et al. A New Robust Epigenetic Model for Forensic Age Prediction. J. Forensic Sci. 2020, 65, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Park, J.L.; Kim, J.H.; Seo, E.; Bae, D.H.; Kim, S.Y.; Lee, H.C.; Woo, K.M.; Kim, Y.S. Identification and evaluation of age-correlated DNA methylation markers for forensic use. Forensic Sci. Int. Genet. 2016, 23, 64–70. [Google Scholar] [CrossRef]
- Patsopoulos, N.A.; Evangelou, E.; Ioannidis, J.P. Sensitivity of between-study heterogeneity in meta-analysis: Proposed metrics and empirical evaluation. Int. J. Epidemiol. 2008, 37, 1148–1157. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.J. Learning with Kernels: Support vector Machines, Regularization, Optimization, and beyond; Adaptive computation and machine learning; MIT Press: Cambridge, MA, USA, 2002; p. xviii. 626p. [Google Scholar]
- Freire-Aradas, A.; Phillips, C.; Mosquera-Miguel, A.; Girón-Santamaría, L.; Gómez-Tato, A.; de Cal, M.C.; Álvarez-Dios, J.; Ansede-Bermejo, J.; Torres-Español, M.; Schneider, P.; et al. Development of a methylation marker set for forensic age estimation using analysis of public methylation data and the Agena Bioscience EpiTYPER system. Forensic Sci. Int. Genet. 2016, 24, 65–74. [Google Scholar] [CrossRef]
- Zolotarenko, A.D.; Chekalin, E.V.; Bruskin, S.A. Modern Molecular Genetic Methods for Age Estimation in Forensics. Russ. J. Genet. 2019, 55, 1460–1471. [Google Scholar] [CrossRef]
- Goel, N.; Karir, P.; Garg, V.K. Role of DNA methylation in human age prediction. Mech. Ageing Dev. 2017, 166, 33–41. [Google Scholar] [CrossRef]
- Ghemrawi, M.; Tejero, N.F.; Duncan, G.; McCord, B. Pyrosequencing: Current forensic methodology and future applications—A review. Electrophoresis 2022, 44, 298–312. [Google Scholar] [CrossRef]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.-H.; Erbel, R.; Mühleisen, T.W.; et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef]
- Fleckhaus, J.; Schneider, P.M. Novel multiplex strategy for DNA methylation-based age prediction from small amounts of DNA via Pyrosequencing. Forensic Sci. Int. Genet. 2020, 44, 102189. [Google Scholar] [CrossRef]
- Vidaki, A.; Ballard, D.; Aliferi, A.; Miller, T.H.; Barron, L.P.; Court, D.S. DNA methylation-based forensic age prediction using artificial neural networks and next generation sequencing. Forensic Sci. Int. Genet. 2017, 28, 225–236. [Google Scholar] [CrossRef]
- Naue, J.; Hoefsloot, H.C.; Mook, O.R.; Rijlaarsdam-Hoekstra, L.; van der Zwalm, M.C.; Henneman, P.; Kloosterman, A.D.; Verschure, P.J. Chronological age prediction based on DNA methylation: Massive parallel sequencing and random forest regression. Forensic Sci. Int. Genet. 2017, 31, 19–28. [Google Scholar] [CrossRef]
- Naue, J.; Sänger, T.; Hoefsloot, H.C.; Lutz-Bonengel, S.; Kloosterman, A.D.; Verschure, P.J. Proof of concept study of age-dependent DNA methylation markers across different tissues by massive parallel sequencing. Forensic Sci. Int. Genet. 2018, 36, 152–159. [Google Scholar] [CrossRef]
- Aliferi, A.; Ballard, D.; Gallidabino, M.; Thurtle, H.; Barron, L.; Court, D.S. DNA methylation-based age prediction using massively parallel sequencing data and multiple machine learning models. Forensic Sci. Int. Genet. 2018, 37, 215–226. [Google Scholar] [CrossRef]
- Dias, H.C.; Cordeiro, C.; Pereira, J.; Pinto, C.; Real, F.C.; Cunha, E.; Manco, L. DNA methylation age estimation in blood samples of living and deceased individuals using a multiplex SNaPshot assay. Forensic Sci. Int. 2020, 311, 110267. [Google Scholar] [CrossRef]
- Hong, S.R.; Shin, K.J.; Jung, S.E.; Lee, E.H.; Lee, H.Y. Platform-independent models for age prediction using DNA methylation data. Forensic Sci. Int. Genet. 2019, 38, 39–47. [Google Scholar] [CrossRef]
- Freire-Aradas, A.; Phillips, C.; Girón-Santamaría, L.; Miguel, A.M.; Gómez-Tato, A.; de Cal, M.C.; Alvarez-Dios, J.A.; Lareu, M.V. Tracking age-correlated DNA methylation markers in the young. Forensic Sci. Int. Genet. 2018, 36, 50–59. [Google Scholar] [CrossRef]
- Gensous, N.; Sala, C.; Pirazzini, C.; Ravaioli, F.; Milazzo, M.; Kwiatkowska, K.M.; Marasco, E.; De Fanti, S.; Giuliani, C.; Pellegrini, C.; et al. A Targeted Epigenetic Clock for the Prediction of Biological Age. Cells 2022, 11, 4044. [Google Scholar] [CrossRef]
- Freire-Aradas, A.; Pośpiech, E.; Aliferi, A.; Girón-Santamaría, L.; Mosquera-Miguel, A.; Pisarek, A.; Ambroa-Conde, A.; Phillips, C.; De Cal, M.A.C.; Gómez-Tato, A.; et al. A Comparison of Forensic Age Prediction Models Using Data From Four DNA Methylation Technologies. Front. Genet. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Carratto, T.M.T.; Moraes, V.M.S.; Recalde, T.S.F.; de Oliveira, M.L.G.; Mendes-Junior, C.T. Applications of massively parallel sequencing in forensic genetics. Genet. Mol. Biol. 2022, 45, e20220077. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Müller, P.; Roewer, L.; Willuweit, S.; Budowle, B.; Parson, W. European survey on forensic applications of massively parallel sequencing. Forensic Sci. Int. Genet. 2017, 29, e23–e25. [Google Scholar] [CrossRef] [PubMed]
- Bruijns, B.; Tiggelaar, R.M.; Gardeniers, J. Massively parallel sequencing techniques for forensics: A review. Electrophoresis 2018, 39, 2642–2654. [Google Scholar] [CrossRef]
- Heidegger, A.; Xavier, C.; Niederstätter, H.; de la Puente, M.; Pośpiech, E.; Pisarek, A.; Kayser, M.; Branicki, W.; Parson, W. Development and optimization of the VISAGE basic prototype tool for forensic age estimation. Forensic Sci. Int. Genet. 2020, 48, 102322. [Google Scholar] [CrossRef]
- Freire-Aradas, A.; Girón-Santamaría, L.; Mosquera-Miguel, A.; Ambroa-Conde, A.; Phillips, C.; de Cal, M.C.; Gómez-Tato, A.; Álvarez-Dios, J.; Pospiech, E.; Aliferi, A.; et al. A common epigenetic clock from childhood to old age. Forensic Sci. Int. Genet. 2022, 60, 102743. [Google Scholar] [CrossRef]
- Fransquet, P.D.; Wrigglesworth, J.; Woods, R.L.; Ernst, M.E.; Ryan, J. The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin. Epigenetics 2019, 11, 62. [Google Scholar] [CrossRef]
- Maulani, C.; Auerkari, E.I. Age estimation using DNA methylation technique in forensics: A systematic review. Egypt. J. Forensic Sci. 2020, 10, 38. [Google Scholar] [CrossRef]
- Becker, J.; Böhme, P.; Reckert, A.; Eickhoff, S.B.; Koop, B.E.; Blum, J.; Gündüz, T.; Takayama, M.; Wagner, W.; Ritz-Timme, S. Evidence for differences in DNA methylation between Germans and Japanese. Int. J. Legal Med. 2022, 136, 405–413. [Google Scholar] [CrossRef]
- Márquez-Ruiz, A.B.; González-Herrera, L.; Luna, J.D.; Valenzuela, A. DNA methylation levels and telomere length in human teeth: Usefulness for age estimation. Int. J. Legal Med. 2020, 134, 451–459. [Google Scholar] [CrossRef]
- Fokias, K.; Dierckx, L.; Van de Voorde, W.; Bekaert, B. Age determination through DNA methylation patterns of fingernails and toenails. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Makowska, Ż.; Spas, A.; Parys-Proszek, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci. Int. Genet. 2015, 14, 161–167. [Google Scholar] [CrossRef]
- Pfeifer, M.; Bajanowski, T.; Helmus, J.; Poetsch, M. Inter-laboratory adaption of age estimation models by DNA methylation analysis-problems and solutions. Int. J. Leg. Med. 2020, 134, 953–961. [Google Scholar] [CrossRef]
- Goncearenco, A.; LaBarre, B.A.; Petrykowska, H.M.; Jaratlerdsiri, W.; Bornman, M.S.R.; Turner, S.D.; Hayes, V.M.; Elnitski, L. DNA methylation profiles unique to Kalahari KhoeSan individuals. Epigenetics 2020, 16, 537–553. [Google Scholar] [CrossRef]
- Jonkman, T.H.; Dekkers, K.F.; Slieker, R.C.; Grant, C.D.; Ikram, M.A.; van Greevenbroek, M.M.J.; Franke, L.; Veldink, J.H.; Boomsma, D.I.; Slagboom, P.E.; et al. Functional genomics analysis identifies T and NK cell activation as a driver of epigenetic clock progression. Genome Biol. 2022, 23, 24. [Google Scholar] [CrossRef]
- Paparazzo, E.; Geracitano, S.; Lagani, V.; Bartolomeo, D.; Aceto, M.A.; D’Aquila, P.; Citrigno, L.; Bellizzi, D.; Passarino, G.; Montesanto, A. A Blood-Based Molecular Clock for Biological Age Estimation. Cells 2023, 12, 32. [Google Scholar] [CrossRef]
- Daunay, A.; Hardy, L.M.; Bouyacoub, Y.; Sahbatou, M.; Touvier, M.; Blanché, H.; Deleuze, J.F.; How-Kit, A. Centenarians consistently present a younger epigenetic age than their chronological age with four epigenetic clocks based on a small number of CpG sites. Aging 2022, 14, 7718–7733. [Google Scholar] [CrossRef]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sánchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS One 2011, 6, e14821. [Google Scholar] [CrossRef]
- Ogata, A.; Kondo, M.; Yoshikawa, M.; Okano, M.; Tsutsumi, T.; Aboshi, H. Dental age estimation based on DNA methylation using real-time methylation-specific PCR. Forensic Sci. Int. 2022, 340, 111445. [Google Scholar] [CrossRef]
- Kondo, M.; Aboshi, H.; Yoshikawa, M.; Ogata, A.; Murayama, R.; Takei, M.; Aizawa, S. A newly developed age estimation method based on CpG methylation of teeth-derived DNA using real-time methylation-specific PCR. J. Oral Sci. 2020, 63, 54–58. [Google Scholar] [CrossRef]
- Fitzgerald, K.N.; Hodges, R.; Hanes, D.; Stack, E.; Cheishvili, D.; Szyf, M.; Henkel, J.; Twedt, M.W.; Giannapoulou, D.; Herdell, J.; et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: A pilot randomized clinical trial. Aging 2021, 13, 9419–9432. [Google Scholar] [CrossRef] [PubMed]
- Galow, A.M. and S. Peleg. How to Slow down the Ticking Clock: Age-Associated Epigenetic Alterations and Related Interventions to Extend Life Span. Cells 2022, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Fiorito, G.; Caini, S.; Palli, D.; Bendinelli, B.; Saieva, C.; Ermini, I.; Valentini, V.; Assedi, M.; Rizzolo, P.; Ambrogetti, D.; et al. DNA methylation-based biomarkers of aging were slowed down in a two-year diet and physical activity intervention trial: The DAMA study. Aging Cell 2021, 20, e13439. [Google Scholar] [CrossRef] [PubMed]
- Fahy, G.M.; Brooke, R.T.; Watson, J.P.; Good, Z.; Vasanawala, S.S.; Maecker, H.; Leipold, M.D.; Lin, D.T.S.; Kobor, M.S.; Horvath, S. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell 2019, 18, e13028. [Google Scholar] [CrossRef]

| PubMed/MEDLINE | Scopus |
|---|---|
| ((((ELOVL fatty acid elongase 2[All Fields]) OR ELOVL2[All Fields]))) AND (((Age[Title/Abstract]) OR aging[Title/Abstract]) OR ageing[Title/Abstract]) AND (pyrosequencing[All Fields]) | ((ALL (ELOVL AND fatty AND acid AND elongase AND 2) OR ALL (ELOVL2))) AND ((TITLE-ABS-KEY (age) OR TITLE-ABS-KEY (aging) OR TITLE-ABS-KEY (ageing))) AND (ALL (pyrosequencing)) |
| Study | Year of Publication | Population | Age Range (Years) | Sample Size | Reference |
|---|---|---|---|---|---|
| Al-Ghanmy et al. | 2021 | Iraqi | 18–93 | 92 | [20] |
| Bekaert et al. | 2015 | Belgium | 0–91 | 206 | [21] |
| Cho et al. | 2017 | Korean | 20–74 | 100 | [22] |
| Fan et al. | 2021 | Chinese | 1–81 | 240 | [8] |
| Garali et al. | 2020 | French | 19–65 | 100 | [16] |
| Lucknuch et al. | 2022 | Thailand | 5–60 | 52 * | [23] |
| Montesanto et al. | 2020 | Italian | 20–89 | 323 ** | [24] |
| Park et al. | 2016 | Korean | 1–100 | 765 | [25] |
| Zbieć-Piekarska et al. | 2015 | Polish | 2–75 | 420 | [15] |
| MLR | MQR | SVM | GBR | PC | |
|---|---|---|---|---|---|
| MLR | 6.575 | 0.1174 | <0.001 | <0.001 | 0.0118 |
| MQR | 0.256 (−0.083, 0.558) | 6.319 | <0.001 | <0.001 | 0.8826 |
| SVM | 0.93 (0.708, 1.151) | 0.674 (0.351, 1.042) | 5.645 | 0.4295 | <0.001 |
| GBR | 0.989 (0.766, 1.213) | 0.733 (0.385, 1.131) | 0.059 (−0.087, 0.206) | 5.586 | <0.001 |
| PC | 0.226 (0.048, 0.404) | −0.029 (−0.376, 0.367) | −0.703 (−0.908, −0.492) | −0.762 (−0.971, −0.554) | 6.348 |
| Holdout Study | Best Model | Best MAE | MAE Garali SVM 6,7 | MAE Garali GBM 6,7 * |
|---|---|---|---|---|
| Montesanto et al. [24] | SVM | 7.77 | 7.380 | 7.976 |
| Cho et al. [22] | GBR | 5.214 | 13.666 | 13.293 |
| Bekaert et al. [21] | SVM | 5.984 | 4.478 | 4.355 |
| Zbieć-Piekarska et al. [15] | SVM | 6.136 | 5.399 | 4.398 |
| Park et al. [25] | SVM | 7.303 | 5.871 | 4.924 |
| Garali et al. [16] | GBR | 8.572 | 4.173 | 5.430 |
| Lucknuch et al. [23] | GBR | 7.892 | 27.301 | 27.865 |
| Al-Ghanmy et al. [20] | SVM | 9.218 | 24.452 | 23.468 |
| Fan et al. [8] | SVM | 7.367 | 13.125 | 15.275 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paparazzo, E.; Lagani, V.; Geracitano, S.; Citrigno, L.; Aceto, M.A.; Malvaso, A.; Bruno, F.; Passarino, G.; Montesanto, A. An ELOVL2-Based Epigenetic Clock for Forensic Age Prediction: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 2254. https://doi.org/10.3390/ijms24032254
Paparazzo E, Lagani V, Geracitano S, Citrigno L, Aceto MA, Malvaso A, Bruno F, Passarino G, Montesanto A. An ELOVL2-Based Epigenetic Clock for Forensic Age Prediction: A Systematic Review. International Journal of Molecular Sciences. 2023; 24(3):2254. https://doi.org/10.3390/ijms24032254
Chicago/Turabian StylePaparazzo, Ersilia, Vincenzo Lagani, Silvana Geracitano, Luigi Citrigno, Mirella Aurora Aceto, Antonio Malvaso, Francesco Bruno, Giuseppe Passarino, and Alberto Montesanto. 2023. "An ELOVL2-Based Epigenetic Clock for Forensic Age Prediction: A Systematic Review" International Journal of Molecular Sciences 24, no. 3: 2254. https://doi.org/10.3390/ijms24032254
APA StylePaparazzo, E., Lagani, V., Geracitano, S., Citrigno, L., Aceto, M. A., Malvaso, A., Bruno, F., Passarino, G., & Montesanto, A. (2023). An ELOVL2-Based Epigenetic Clock for Forensic Age Prediction: A Systematic Review. International Journal of Molecular Sciences, 24(3), 2254. https://doi.org/10.3390/ijms24032254







