Wheat Long Noncoding RNAs from Organelle and Nuclear Genomes Carry Conserved microRNA Precursors Which May Together Comprise Intricate Networks in Insect Responses
Abstract
1. Introduction
2. Results
2.1. Different Wheat Genotypes May Contain Several Lineage-Specific lncRNAs
2.2. A Subset of Wheat lncRNAs Contain Potential Precursors for miRNAs
2.3. Genomic Loci Associated with Insect Resistance Contain Putative lncRNAs
2.4. Some lncRNAs May Derive from Organellar Genomes
3. Discussion
4. Materials and Methods
4.1. RNA-Sequencing Data and Transcriptome Assembly
4.2. In Silico Identification of Long Noncoding RNAs
4.3. In Silico Identification of microRNAs and Prediction of Targets
4.4. Comparison of the Putative lncRNAs to the Organellar Genomes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Liu, S.; Nie, X.; Weining, S.; Wu, L. Conservation Analysis of Long Non-Coding RNAs in Plants. Sci. China Life Sci. 2017, 61, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Brant, E.J.; Budak, H. Plant Small Non-Coding RNAs and Their Roles in Biotic Stresses. Front. Plant Sci. 2018, 9, 1038. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long Non-Coding RNA in Plants in the Era of Reference Sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Martínez, H.N.; Recillas-Targa, F. Emerging Functions of LncRNA Loci beyond the Transcript Itself. Int. J. Mol. Sci. 2022, 23, 6258. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, A.T.; Blevins, T.; Swiezewski, S. Long Noncoding RNAs in Plants. Annu. Rev. Plant Biol. 2021, 72, 245–271. [Google Scholar] [CrossRef]
- Zhao, Z.; Zang, S.; Zou, W.; Pan, Y.B.; Yao, W.; You, C.; Que, Y. Long Non-Coding RNAs: New Players in Plants. Int. J. Mol. Sci. 2022, 23, 9301. [Google Scholar] [CrossRef]
- Holley, C.L.; Topkara, V.K. An Introduction to Small Non-Coding RNAs: MiRNA and SnoRNA. Cardiovasc. Drugs Ther. 2011, 25, 151–159. [Google Scholar] [CrossRef]
- Hung, T.; Chang, H.Y. Long Noncoding RNA in Genome Regulation: Prospects and Mechanisms. RNA Biol. 2011, 7, 582–585. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Biyiklioglu, S.; Budak, H. Assembly and Annotation of Transcriptome Provided Evidence of MiRNA Mobility between Wheat and Wheat Stem Sawfly. Front. Plant Sci. 2017, 8, 1653. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Lu, P.; Xu, Y.; Li, Z.; Yu, S.; Liu, J.; Wang, H.; Chua, N.H.; Cao, P. PLncDB V2.0: A Comprehensive Encyclopedia of Plant Long Noncoding RNAs. Nucleic Acids Res. 2021, 49, D1489–D1495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An Updated Database Dedicated to Long Non-Coding RNA Annotation in Both Animals and Plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Taneja, M.; Tyagi, S.; Singh, K.; Upadhyay, S.K. Survey of High Throughput RNA-Seq Data Reveals Potential Roles for LncRNAs during Development and Stress Response in Bread Wheat. Front. Plant Sci. 2017, 8, 1019. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, K.; Yu, R.; Zhou, B.; Huang, P.; Cao, Z.; Zhou, Y.; Wang, J. From “Dark Matter” to “Star”: Insight Into the Regulation Mechanisms of Plant Functional Long Non-Coding RNAs. Front. Plant Sci. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Chen, J.; Zhong, Y.; Qi, X. LncRNA TCONS_00021861 Is Functionally Associated with Drought Tolerance in Rice (Oryza Sativa L.) via Competing Endogenous RNA Regulation. BMC Plant Biol. 2021, 21, 410. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Nong, W.; Zhao, S.; Lin, X.; Xie, Y.; Cheung, M.Y.; Xiao, Z.; Wong, A.Y.P.; Chan, T.F.; Hui, J.H.L.; et al. Differential MicroRNA Expression, MicroRNA Arm Switching, and MicroRNA:Long Noncoding RNA Interaction in Response to Salinity Stress in Soybean. BMC Genom. 2022, 23, 65. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, H.; Hu, W.; Ji, W. The Emerging Role of Long Non-Coding RNAs in Plant Defense against Fungal Stress. Int. J. Mol. Sci. 2020, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Biyiklioglu, S.; Alptekin, B.; Akpinar, B.A.; Varella, A.C.; Hofland, M.L.; Weaver, D.K.; Bothner, B.; Budak, H. A Large-Scale Multiomics Analysis of Wheat Stem Solidness and the Wheat Stem Sawfly Feeding Response, and Syntenic Associations in Barley, Brachypodium, and Rice. Funct. Integr. Genom. 2018, 18, 241–259. [Google Scholar] [CrossRef]
- Muslu, T.; Akpinar, B.A.; Biyiklioglu-Kaya, S.; Yuce, M.; Budak, H. Comparative Analysis of Coding and Non-Coding Features within Insect Tolerance Loci in Wheat with Their Homologs in Cereal Genomes. Int. J. Mol. Sci. 2021, 22, 22. [Google Scholar] [CrossRef]
- Nilsen, K.T.; Walkowiak, S.; Xiang, D.; Gao, P.; Quilichini, T.D.; Willick, I.R.; Byrns, B.; N’Diaye, A.; Ens, J.; Wiebe, K.; et al. Copy Number Variation of TdDof Controls Solid-Stemmed Architecture in Wheat. Proc. Natl. Acad. Sci. USA 2020, 117, 28708–28718. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple Wheat Genomes Reveal Global Variation in Modern Breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Shi, W.; Xu, M.; Liu, J.; Zhang, F. Genome-Wide Identification and Characterization of Long Non-Coding RNA in Wheat Roots in Response to Ca2+ Channel Blocker. Front. Plant Sci. 2018, 9, 244. [Google Scholar] [CrossRef]
- Madhawan, A.; Sharma, A.; Bhandawat, A.; Rahim, M.S.; Kumar, P.; Mishra, A.; Parveen, A.; Sharma, H.; Verma, S.K.; Roy, J. Identification and Characterization of Long Non-Coding RNAs Regulating Resistant Starch Biosynthesis in Bread Wheat (Triticum Aestivum L.). Genomics 2020, 112, 3065–3074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.; Liu, H.; Fu, Y.; Cheng, Y.; Zhang, L.; Shi, W.; Zhang, Y.; Chen, J. Transcriptomic and Metabolomic Analysis of Wheat Kernels in Response to the Feeding of Orange Wheat Blossom Midges (Sitodiplosis Mosellana) in the Field. J. Agric. Food Chem. 2022, 70, 1477–1493. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H.M. Long Non-Coding RNAs: Emerging Players Regulating Plant Abiotic Stress Response and Adaptation. BMC Plant Biol. 2020, 20, 1–20. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z.; Budak, H. Mirmachine: A One-Stop Shop for Plant Mirna Annotation. J. Vis. Exp. 2021, 2021, 1–10. [Google Scholar] [CrossRef]
- Ke, L.; Zhou, Z.; Xu, X.W.; Wang, X.; Liu, Y.; Xu, Y.; Huang, Y.; Wang, S.; Deng, X.; Chen, L.L.; et al. Evolutionary Dynamics of LincRNA Transcription in Nine Citrus Species. Plant J. 2019, 98, 912–927. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 176, 407–431. [Google Scholar] [CrossRef]
- Budak, H.; Akpinar, A. Dehydration Stress-Responsive Mirna in Brachypodium Distachyon: Evident by Genome-Wide Screening of Micrornas Expression. Omi. A J. Integr. Biol. 2011, 15, 791–799. [Google Scholar] [CrossRef]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Guo, F.; Xu, Q.; Cang, J. LncRNA Improves Cold Resistance of Winter Wheat by Interacting with MiR398. Funct. Plant Biol. 2020, 47, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Megha, S.; Basu, U.; Rahman, M.H.; Kav, N.N.V. Genome Wide Identification and Functional Prediction of Long Non-Coding RNAs Responsive to Sclerotinia Sclerotiorum Infection in Brassica Napus. PLoS ONE 2016, 11, 1–19. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Hong, Y.H.; Liu, Y.R.; Cui, J.; Luan, Y.S. Function Identification of MiR394 in Tomato Resistance to Phytophthora Infestans. Plant Cell Rep. 2021, 40, 1831–1844. [Google Scholar] [CrossRef]
- Sun, L.; Sun, G.; Shi, C.; Sun, D. Transcriptome Analysis Reveals New MicroRNAs-Mediated Pathway Involved in Anther Development in Male Sterile Wheat. BMC Genom. 2018, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-Wide Changes in MicroRNA Expression during Short and Prolonged Heat Stress and Recovery in Contrasting Rice Cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef]
- Sobhani Najafabadi, A.; Naghavi, M.R. Mining Ferula Gummosa Transcriptome to Identify MiRNAs Involved in the Regulation and Biosynthesis of Terpenes. Gene 2018, 645, 41–47. [Google Scholar] [CrossRef]
- López-Galiano, M.J.; González-Hernández, A.I.; Crespo-Salvador, O.; Rausell, C.; Real, M.D.; Escamilla, M.; Camañes, G.; García-Agustín, P.; González-Bosch, C.; García-Robles, I. Epigenetic Regulation of the Expression of WRKY75 Transcription Factor in Response to Biotic and Abiotic Stresses in Solanaceae Plants. Plant Cell Rep. 2018, 37, 167–176. [Google Scholar] [CrossRef]
- Calderon, R.H.; Strand, Å. How Retrograde Signaling Is Intertwined with the Evolution of Photosynthetic Eukaryotes. Curr. Opin. Plant Biol. 2021, 63, 102093. [Google Scholar] [CrossRef]
- Zhelyazkova, P.; Sharma, C.M.; Forstner, K.U.; Liere, K.; Vogel, J.; Borner, T. The Primary Transcriptome of Barley Chloroplasts: Numerous Noncoding RNAs and the Dominating Role of the Plastid-Encoded RNA Polymerase. Plant Cell 2012, 24, 123–136. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum Wheat Genome Highlights Past Domestication Signatures and Future Improvement Targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 1–19. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool Using an Alignment-Free Logistic Regression Model. Nucleic Acids Res. 2013, 41, 1–7. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Guo, G.; Ni, Z.; Sunkar, R.; Du, J.; Zhu, J.K.; Sun, Q. Cloning and Characterization of MicroRNAs from Wheat (Triticum Aestivum L.). Genome Biol. 2007, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Jian, C.; Lv, J.; Yan, Y.; Chi, Q.; Li, Z.; Wang, Q.; Zhang, J. Identification and Characterization of MicroRNAs in the Flag Leaf and Developing Seed of Wheat ( Triticum Aestivum L.). BMC Genom. 2014, 15, 289. [Google Scholar] [CrossRef]
- Wei, B.; Cai, T.; Zhang, R.; Li, A.; Huo, N.; Li, S.; Gu, Y.Q.; Vogel, J.; Jia, J.; Qi, Y.; et al. Novel MicroRNAs Uncovered by Deep Sequencing of Small RNA Transcriptomes in Bread Wheat (Triticum Aestivum L.) and Brachypodium Distachyon (L.) Beauv. Funct. Integr. Genom. 2009, 9, 499–511. [Google Scholar] [CrossRef]
- Chu, Z.; Chen, J.; Xu, H.; Dong, Z.; Chen, F.; Cui, D. Identification and Comparative Analysis of MicroRNA in Wheat (Triticum Aestivum L.) Callus Derived from Mature and Immature Embryos during In Vitro Culture. Front. Plant Sci. 2016, 7, 1302. [Google Scholar] [CrossRef]
- Jin, X.; Jia, L.; Wang, Y.; Li, B.; Sun, D.; Chen, X. Identification of Fusarium Graminearum-Responsive MiRNAs and Their Targets in Wheat by SRNA Sequencing and Degradome Analysis. Funct. Integr. Genom. 2020, 20, 51–61. [Google Scholar] [CrossRef]
- Li, Y.F.; Wei, K.; Wang, M.; Wang, L.; Cui, J.; Zhang, D.; Guo, J.; Zhao, M.; Zheng, Y. Identification and Temporal Expression Analysis of Conserved and Novel MicroRNAs in the Leaves of Winter Wheat Grown in the Field. Front. Genet. 2019, 10, 779. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Integrated Analysis of Small RNA, Transcriptome, and Degradome Sequencing Reveals the Water-Deficit and Heat Stress Response Network in Durum Wheat. Int. J. Mol. Sci. 2021, 21, 6017. [Google Scholar] [CrossRef]
- Ragupathy, R.; Ravichandran, S.; Mahdi, M.S.R.; Huang, D.; Reimer, E.; Domaratzki, M.; Cloutier, S. Deep Sequencing of Wheat SRNA Transcriptome Reveals Distinct Temporal Expression Pattern of MiRNAs in Response to Heat, Light and UV. Sci. Rep. 2016, 6, 39373. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Able, A.J.; Able, J.A. Multi-Omics Analysis of Small RNA, Transcriptome, and Degradome in T. Turgidum—Regulatory Networks of Grain Development and Abiotic Stress Response. Int. J. Mol. Sci. 2020, 21, 7772. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Liu, H.; Wang, K.; Liu, L.; Wang, S.; Zhao, Y.; Yin, J. Development-Associated MicroRNAs in Grains of Wheat ( Triticum Aestivum L.). BMC Plant Biol. 2013, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Noyszewski, A.K.; Ghavami, F.; Alnemer, L.M.; Soltani, A.; Gu, Y.Q.; Huo, N.; Meinhardt, S.; Kianian, P.M.A.; Kianian, S.F. Accelerated Evolution of the Mitochondrial Genome in an Alloplasmic Line of Durum Wheat. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef] [PubMed]
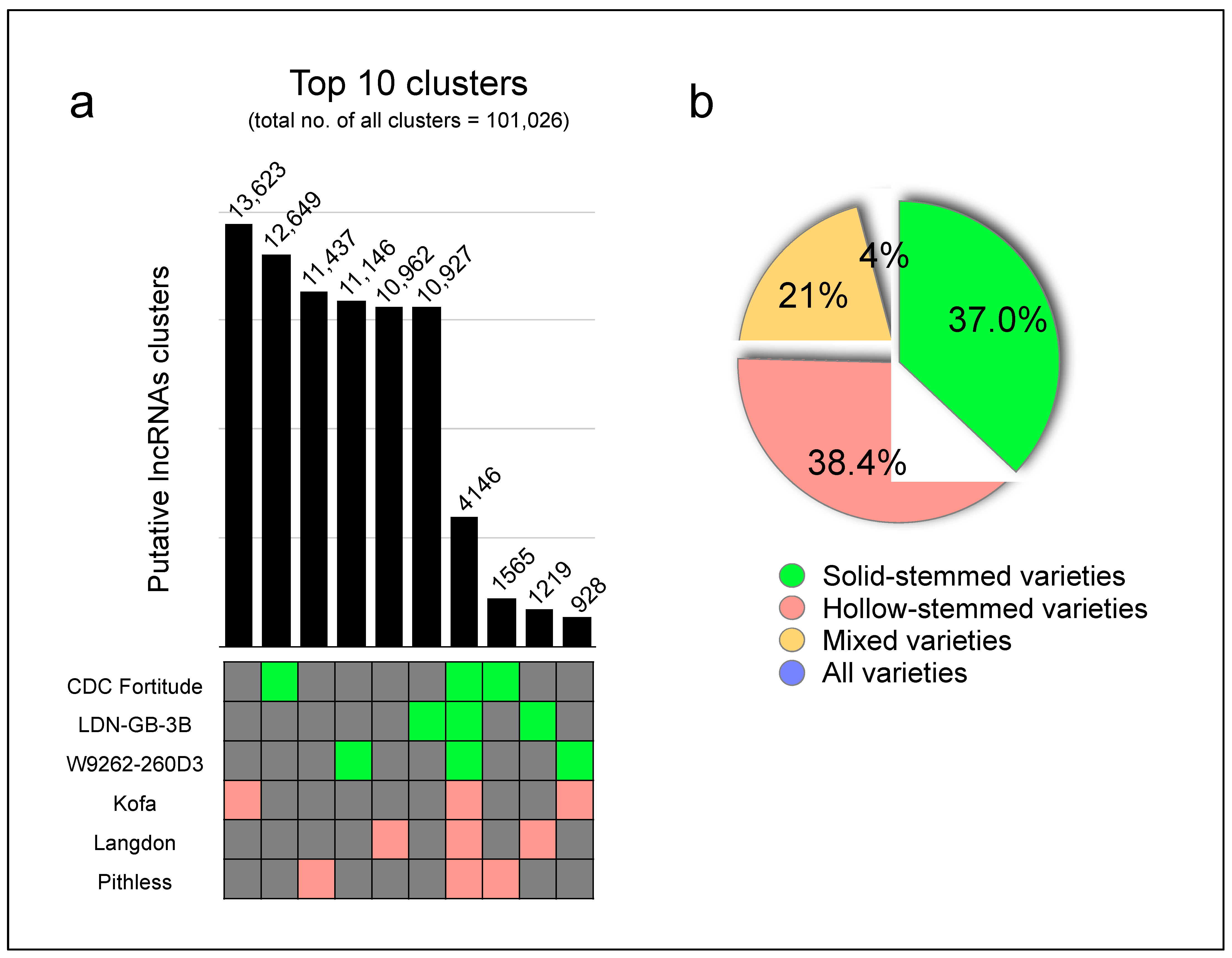
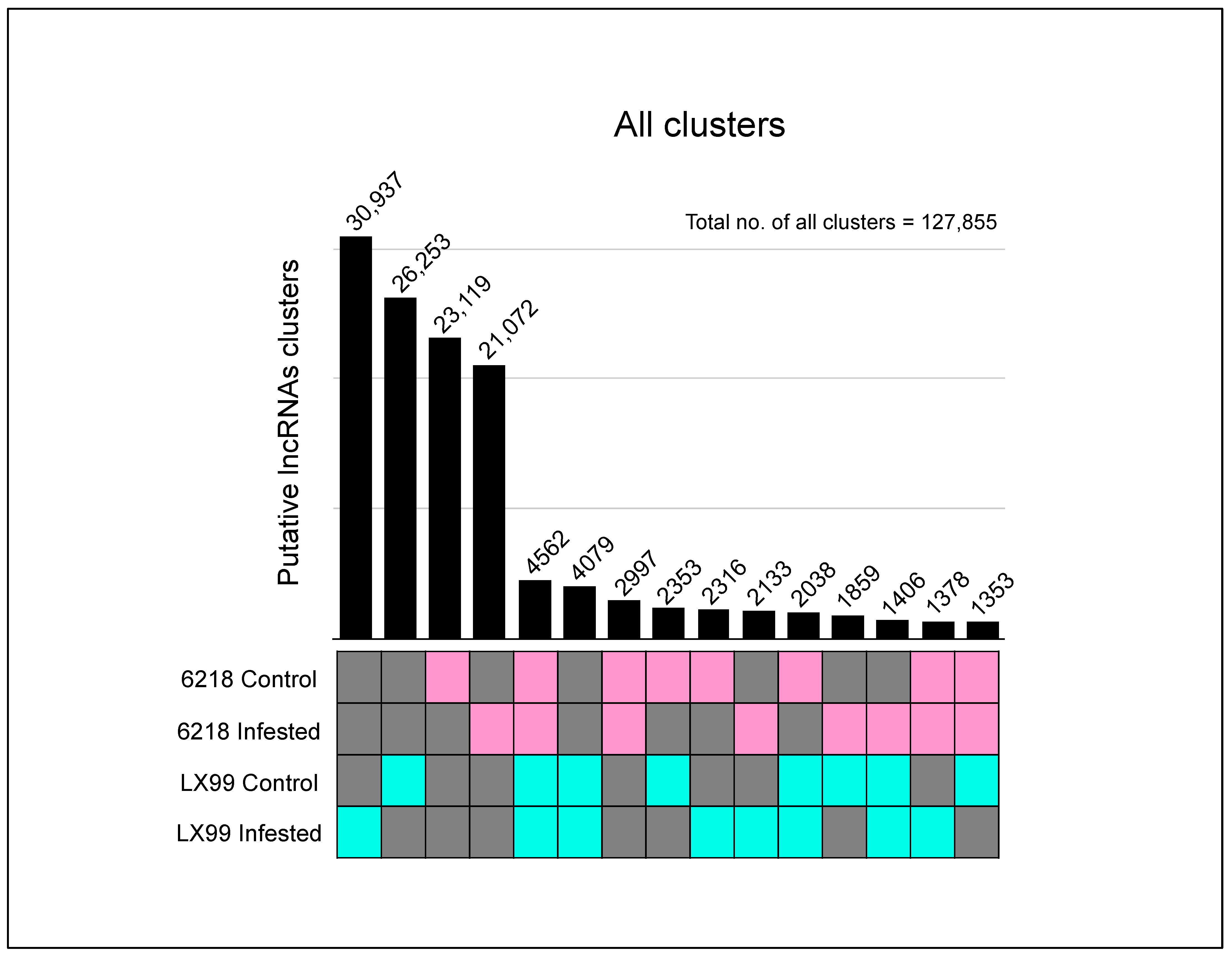
| Variety/ Sample | Total Number of Assembled Transcripts | Number of Transcripts Remaining after Removal of Other Noncoding RNA Types (>200 nt) | Number of Transcripts with Potential ORFs < 100 aa | Number of Transcripts with No Coding Potential—CPAT | Number of Transcripts with No Coding Potential—CPC2 | Number of Transcripts with No Coding Potential—BOTH (Hence Retained) | Number of Transcripts Not Matching Coding Sequences | Number of Transcripts Partially Matching Coding Sequences |
|---|---|---|---|---|---|---|---|---|
| Triticum durum (AABB) varieties | ||||||||
| CDC Fortitude | 173,462 | 168,753 | 57,278 | 55,589 | 54,847 | 53,519 | 38,356 | 6075 |
| LDN-GB-3B | 161,633 | 157,632 | 51,472 | 49,875 | 49,351 | 48,063 | 34,122 | 5443 |
| W9262-260D3 | 160,015 | 155,787 | 51,128 | 49,523 | 48,932 | 47,673 | 33,832 | 5646 |
| Kofa | 170,654 | 166,414 | 55,698 | 53,985 | 53,398 | 52,032 | 37,210 | 5897 |
| Langdon | 162,270 | 158,438 | 51,900 | 50,241 | 49,553 | 48,263 | 33,946 | 5796 |
| Pithless | 165,947 | 161,925 | 53,462 | 51,825 | 51,259 | 49,955 | 35,386 | 5722 |
| Triticum aestivum (AABBDD) varieties | ||||||||
| 6218 Control | 214,161 | 209,624 | 72,672 | 69,717 | 69,387 | 67,398 | 47,146 | 5874 |
| 6218 Infested | 202,040 | 197,970 | 66,991 | 64,270 | 64,118 | 62,225 | 42,427 | 5461 |
| LX99 Control | 210,529 | 206,501 | 77,489 | 74,829 | 74,596 | 72,724 | 51,560 | 5188 |
| LX99 Infested | 219,575 | 215,276 | 85,389 | 82,452 | 82,164 | 80,156 | 57,166 | 5640 |
| Variety/ Sample | No. of lncRNAs from Insect Resistance Loci | LncRNAs from Insect Resistance Potentially Targeted by miRNAs | Targeting miRNA Families Predicted from lncRNAs | Targeting Known Wheat miRNA Families | |
|---|---|---|---|---|---|
| Triticum durum (AABB) varieties | |||||
| CDC Fortitude | 177 | STRG.49702.1 | miR1118, miR1436, miR5174 | miR1118 | |
| STRG.49256.1 | - | Unknown family | |||
| LDN-GB-3B | 164 | STRG.46147.1 | miR1118, miR1436 | miR1118 | |
| STRG.46190.1 | miR1130_miR1122 | miR1130 | |||
| STRG.45094.1 | - | Unknown family | |||
| W9262-260D3 | 148 | STRG.46057.1 | miR1118, miR1130, miR1436, miR5174, | miR1118, miR1138 | |
| STRG.46179.1 | miR5174, miR5181 | - | |||
| Kofa | 211 | STRG.49467.1 | miR395 | - | |
| Langdon | 180 | STRG.49026.1 | miR169 | - | |
| STRG.45799.1 | miR169 | miR6197 | |||
| Pithless | 159 | STRG.47553.1 | miR1439 | - | |
| Triticum aestivum (AABBDD) varieties | |||||
| 6218 Control | 109 | STRG.29249.1 | - | miR9673 | |
| 6218 Infested | 97 | STRG.26323.1 | miR1128 | - | |
| STRG.26415.1 | mR6197 | - | |||
| LX99 Control | 108 | STRG.30297.1 | miR1127 | miR1127 | |
| LX99 Infested | 145 | STRG.31964.1 | miR1127 | miR1127 | |
| STRG.31924.1 | miR5174 | miR1133 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akpinar, B.A.; Muslu, T.; Reddy, G.V.P.; Dogramaci, M.; Budak, H. Wheat Long Noncoding RNAs from Organelle and Nuclear Genomes Carry Conserved microRNA Precursors Which May Together Comprise Intricate Networks in Insect Responses. Int. J. Mol. Sci. 2023, 24, 2226. https://doi.org/10.3390/ijms24032226
Akpinar BA, Muslu T, Reddy GVP, Dogramaci M, Budak H. Wheat Long Noncoding RNAs from Organelle and Nuclear Genomes Carry Conserved microRNA Precursors Which May Together Comprise Intricate Networks in Insect Responses. International Journal of Molecular Sciences. 2023; 24(3):2226. https://doi.org/10.3390/ijms24032226
Chicago/Turabian StyleAkpinar, Bala Ani, Tugdem Muslu, Gadi V. P. Reddy, Munevver Dogramaci, and Hikmet Budak. 2023. "Wheat Long Noncoding RNAs from Organelle and Nuclear Genomes Carry Conserved microRNA Precursors Which May Together Comprise Intricate Networks in Insect Responses" International Journal of Molecular Sciences 24, no. 3: 2226. https://doi.org/10.3390/ijms24032226
APA StyleAkpinar, B. A., Muslu, T., Reddy, G. V. P., Dogramaci, M., & Budak, H. (2023). Wheat Long Noncoding RNAs from Organelle and Nuclear Genomes Carry Conserved microRNA Precursors Which May Together Comprise Intricate Networks in Insect Responses. International Journal of Molecular Sciences, 24(3), 2226. https://doi.org/10.3390/ijms24032226









