Complexes of Cationic Pyridylphenylene Dendrimers with Anionic Liposomes: The Role of Dendrimer Composition in Membrane Structural Changes
Abstract
1. Introduction
2. Results and Discussion
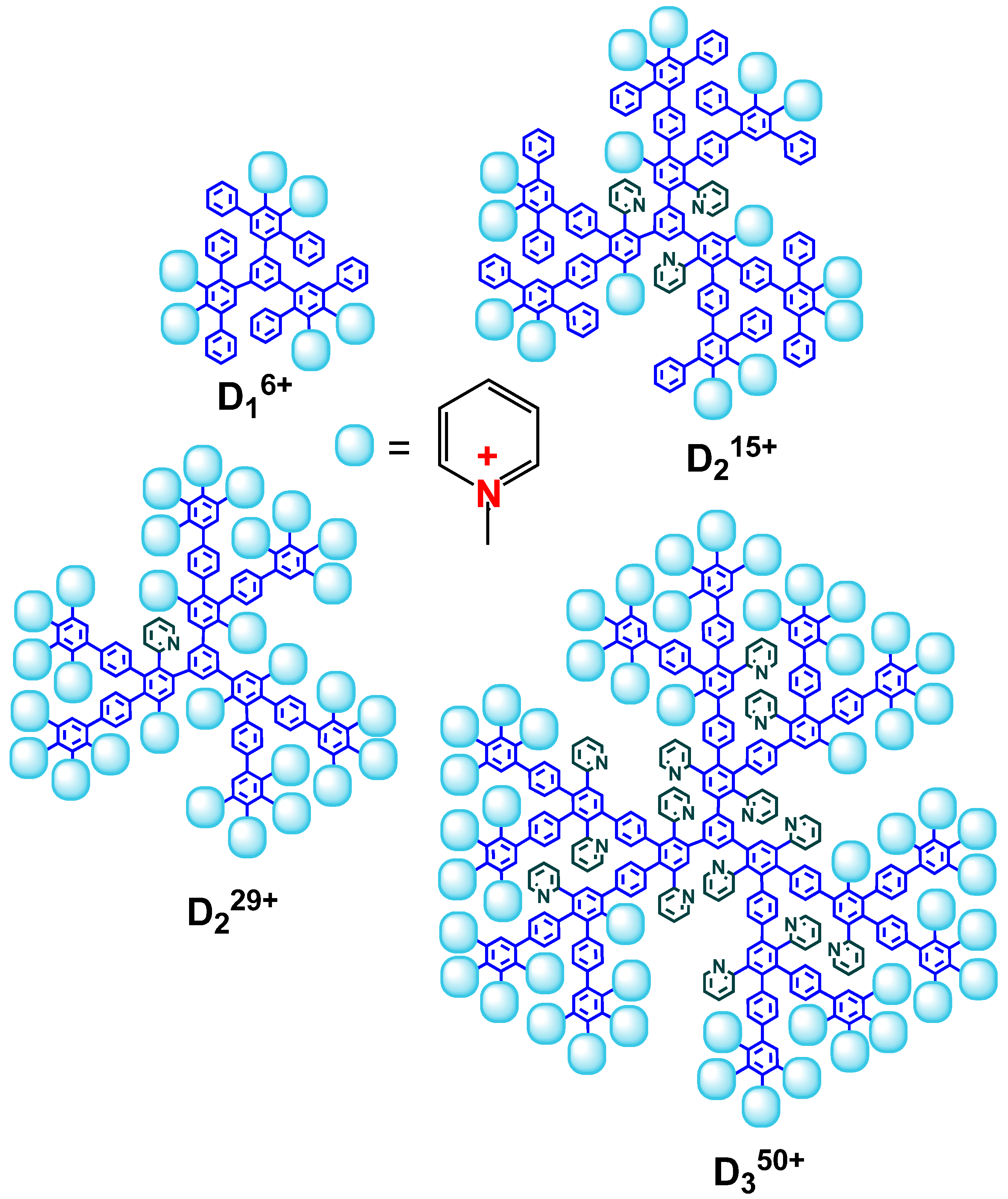
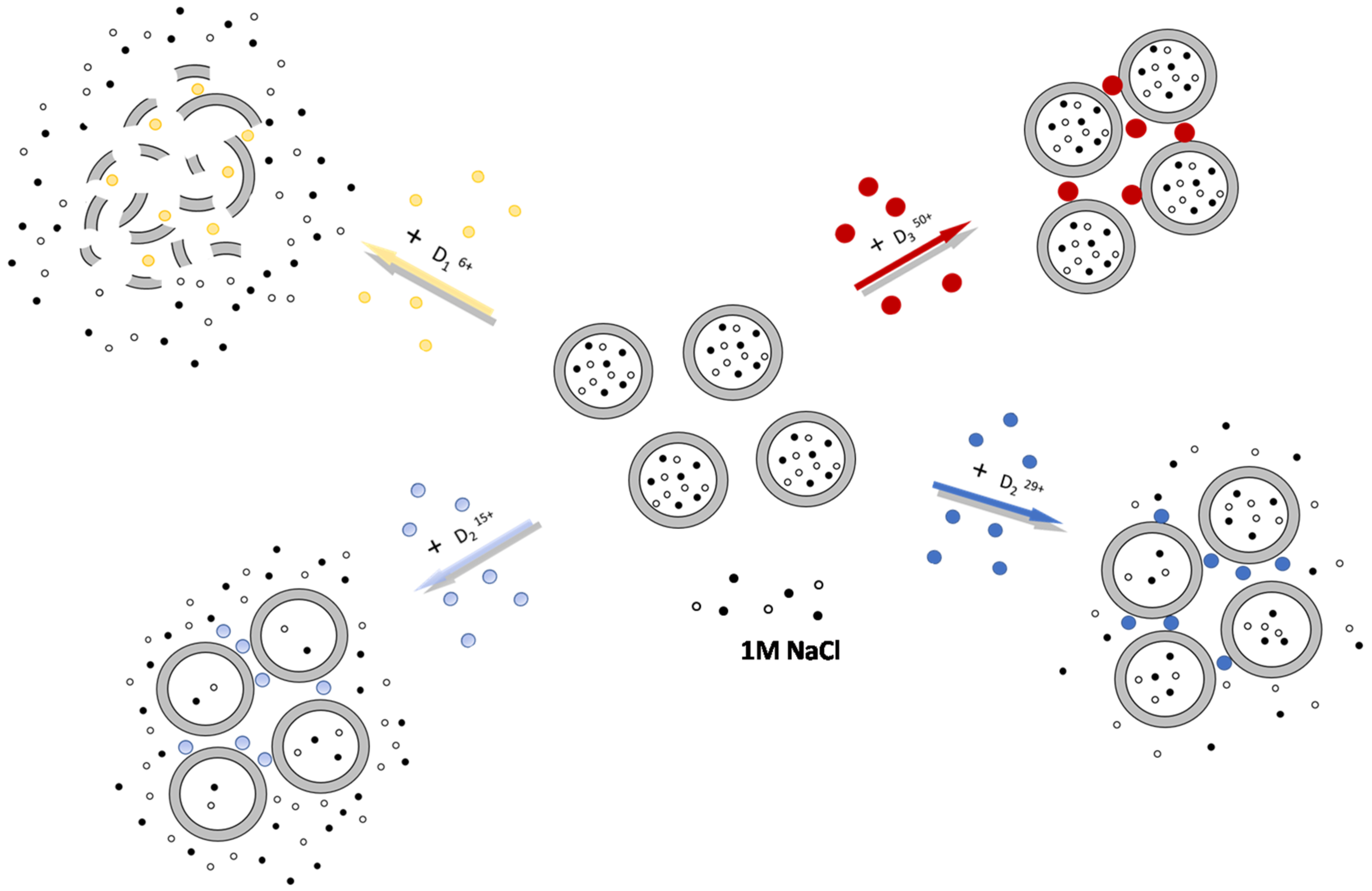
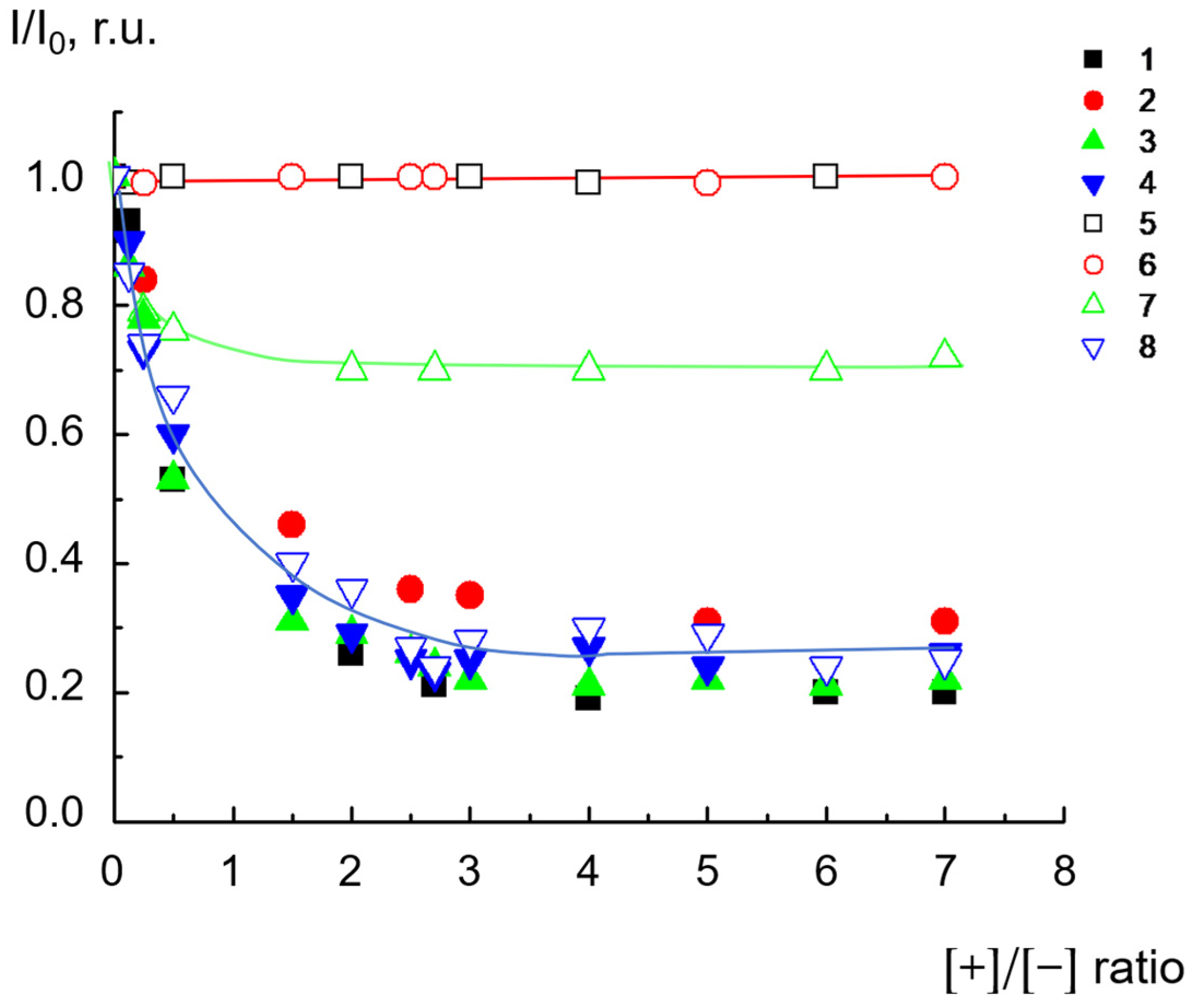
2.1. Complexes of Dendrimers with Anionic CL/Chol/DOPC liposomes
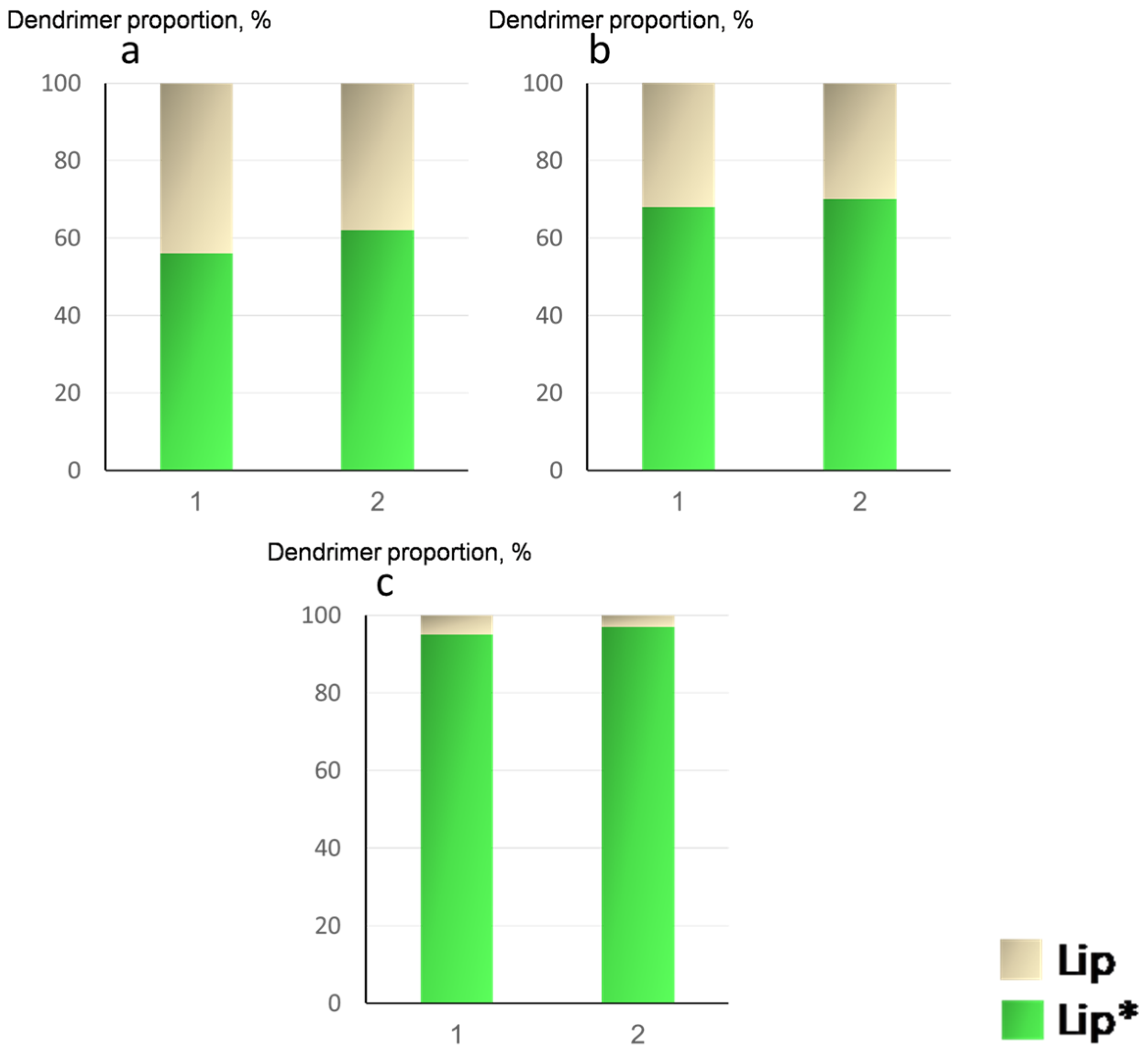
2.2. Integrity of Lipid Bilayer upon Complexation with Dendrimers
2.3. Reversibility of Interactions
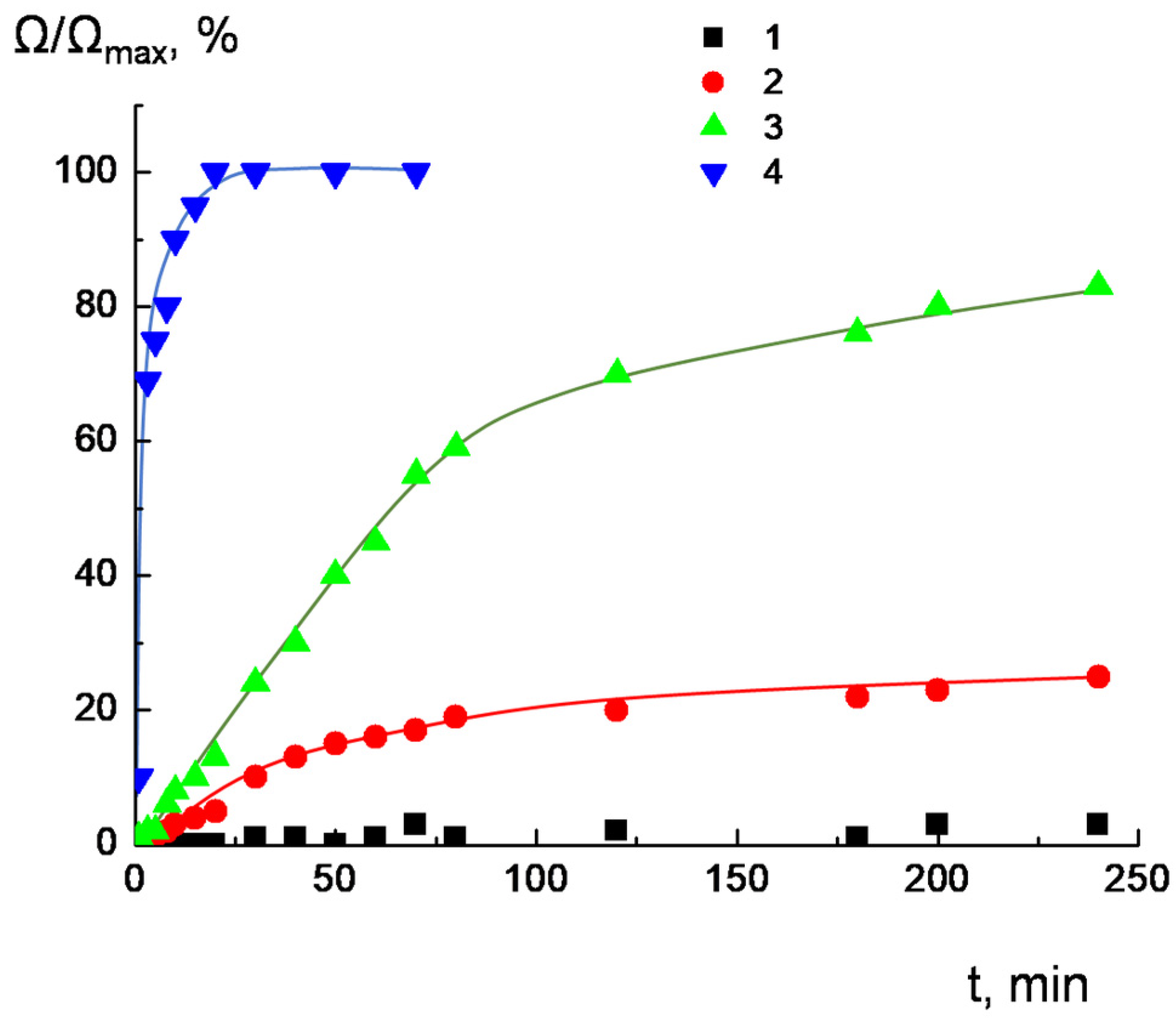
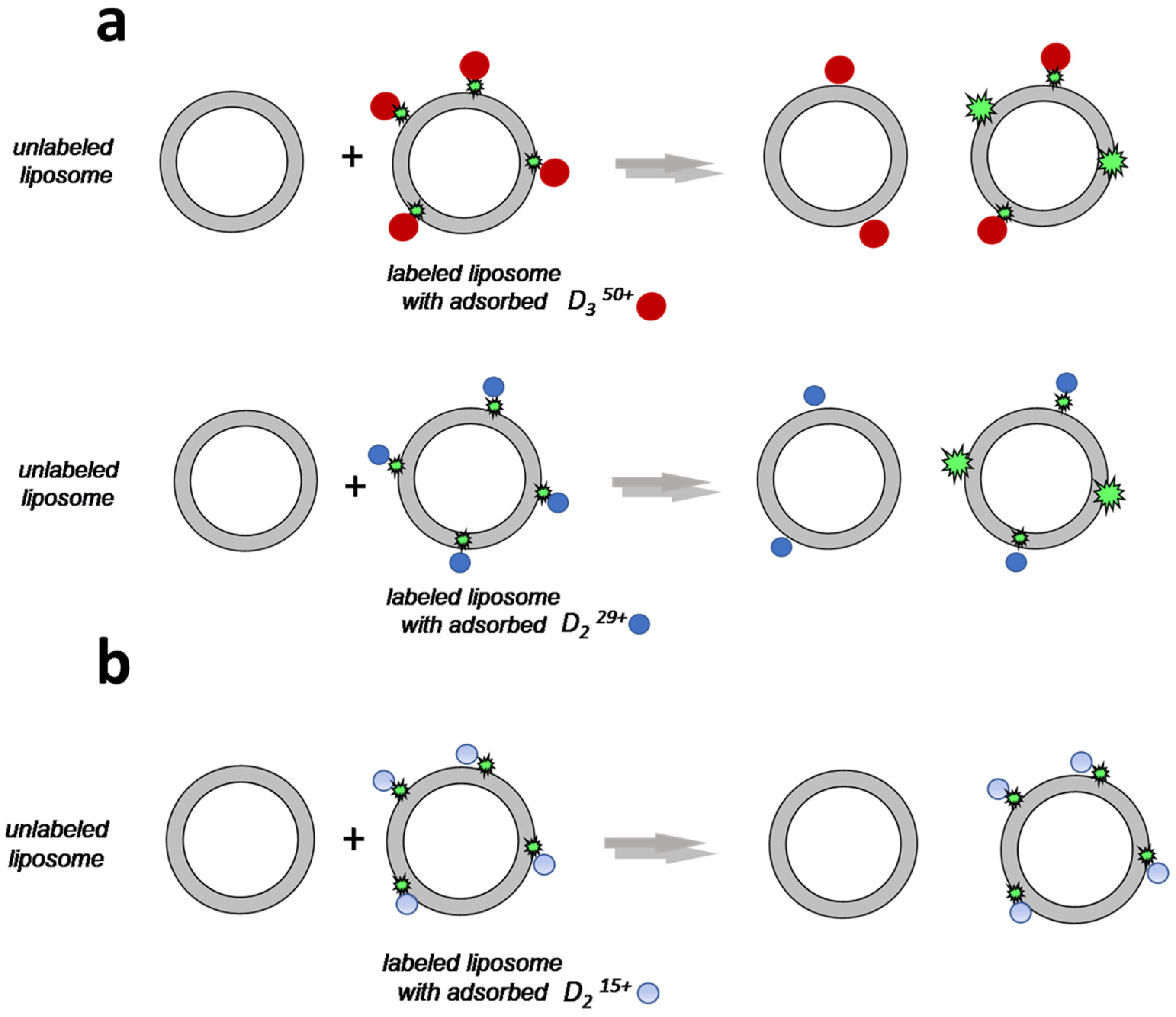
2.4. Migration of Dendrimers between Liposomes
3. Materials and Methods
3.1. Dendrimer Synthesis and Characterization
3.2. Chemicals
3.3. Synthesis of Anionic Liposomes
3.4. Preparation of Dendrimer–Liposome Complexes
3.5. Degree of Binding of Dendrimers with Anionic Liposomes
3.6. Dendrimer Migration Experiment
3.7. Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Araújo, R.V.D.; Santos, S.D.S.; Igne Ferreira, E.; Giarolla, J. New advances in general biomedical applications of PAMAM dendrimers. Molecules 2018, 23, 2849. [Google Scholar] [CrossRef] [PubMed]
- Pedziwiatr-Werbicka, E.; Milowska, K.; Dzmitruk, V.; Ionov, M.; Shcharbin, D.; Bryszewska, M. Dendrimers and hyperbranched structures for biomedical applications. Eur. Polym. J. 2019, 119, 61–73. [Google Scholar] [CrossRef]
- Huang, K.W.; Hsu, F.F.; Qiu, J.T.; Chern, G.J.; Lee, Y.A.; Chang, C.C.; Huang, Y.T.; Sung, Y.C.; Chiang, C.C.; Huang, R.L.; et al. Highly efficient and tumor-selective nanoparticles for dual-targeted immunogene therapy against cancer. Sci. Adv. 2020, 6, eaax5032. [Google Scholar] [CrossRef] [PubMed]
- Chis, A.A.; Dobrea, C.; Morgovan, C.; Arseniu, A.M.; Rus, L.L.; Butuca, A.; Juncan, A.M.; Totan, M.; Vonica-Tincu, A.L.; Cormos, G.; et al. Applications and limitations of dendrimers in biomedicine. Molecules 2020, 25, 3982. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, C.; Pang, Z. Dendrimer-based drug delivery systems for brain targeting. Biomolecule 2019, 9, 790. [Google Scholar] [CrossRef] [PubMed]
- Contin, M.; Garcia, C.; Dobrecky, C.; Lucangioli, S.; D’Accorso, N. Advances in drug delivery, gene delivery and therapeutic agents based on dendritic materials. Future Med. Chem. 2019, 11, 1791–1810. [Google Scholar] [CrossRef]
- Chauhan, A.S. Dendrimers for drug delivery. Molecules 2018, 23, 938. [Google Scholar] [CrossRef]
- El-Beyrouthy, J.; Freeman, E. Characterizing the Structure and Interactions of Model Lipid Membranes Using Electrophysiology. Membranes 2021, 11, 319. [Google Scholar] [CrossRef]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef]
- Yaroslavov, A.A.; Efimova, A.A.; Pyatnikova, D.A. Complexes of a cationic polymer with oxidized anionic liposomes: Composition, structure, and properties. Polym. Sci. Ser. B 2017, 59, 577–585. [Google Scholar] [CrossRef]
- Wilkosz, N.; Jamróz, D.; Kopec, W.; Nakai, K.; Yusa, S.I.; Wytrwal-Sarna, M.; Bednar, J.; Nowakowska, M.; Kepczynski, M. Effect of polycation structure on interaction with lipid membranes. J. Phys. Chem. B 2017, 121, 7318–7326. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Dong, X.; Chen, X. Polymer-Modified Liposomes for Drug Delivery: From Fundamentals to Applications. Pharmaceutics 2022, 14, 778. [Google Scholar] [CrossRef] [PubMed]
- Billard, A.; Pourchet, L.; Malaise, S.; Alcouffe, P.; Montembault, A.; Ladavière, C. Liposome-loaded chitosan physical hydrogel: Toward a promising delayed-release biosystem. Carbohydr. Polym. 2015, 115, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Yaroslavov, A.A.; Efimova, A.A.; Krasnikov, E.A.; Trosheva, K.S.; Popov, A.S.; Melik-Nubarov, N.S.; Krivtsov, G.G. Chitosan-based multi-liposomal complexes: Synthesis, biodegradability and cytotoxicity. Int. J. Biol. Macromol. 2021, 177, 455–462. [Google Scholar] [CrossRef]
- Kopec, W.; Żak, A.; Jamróz, D.; Nakahata, R.; Yusa, S.I.; Gapsys, V.; Kepczynski, M. Polycation–anionic lipid membrane interactions. Langmuir 2020, 36, 12435–12450. [Google Scholar] [CrossRef]
- Efimova, A.A.; Kostenko, S.N.; Orlov, V.N.; Yaroslavov, A.A. Effect of cholesterol on the phase state and permeability of mixed liposomes composed of anionic diphosphatidylglycerol and zwitterionic dipalmitoylphosphatidylcholine. Mendeleev Commun. 2016, 26, 99–100. [Google Scholar] [CrossRef]
- Porcar, L.; Gerelli, Y. On the lipid flip-flop and phase transition coupling. Soft Matter 2020, 16, 7696–7703. [Google Scholar] [CrossRef]
- Yaroslavov, A.A.; Efimova, A.A.; Sybachin, A.V.; Izumrudov, V.A.; Samoshin, V.V.; Potemkin, I.I. Stability of anionic liposome-cationic polymer complexes in water-salt media. Colloid J. 2011, 73, 430–435. [Google Scholar] [CrossRef]
- Awasthi, N.; Kopec, W.; Wilkosz, N.; Jamróz, D.; Hub, J.S.; Zatorska, M.; Petka, R.; Nowakowska, M.; Kepczynski, M. Molecular mechanism of polycation-induced pore formation in biomembranes. ACS Biomater. Sci. Eng. 2018, 5, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Efimova, A.A.; Abramova, T.A.; Popov, A.S. Complexes of Negatively Charged Liposomes with Chitosan: Effect of Phase State of the Lipid Bilayer. Russ. J. Gen. Chem. 2021, 91, 2079–2085. [Google Scholar] [CrossRef]
- Kepczynski, M.; Jamróz, D.; Wytrwal, M.; Bednar, J.; Rzad, E.; Nowakowska, M. Interactions of a hydrophobically modified polycation with zwitterionic lipid membranes. Langmuir 2012, 28, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Yaroslavov, A.A.; Rakhnyanskaya, A.A.; Yaroslavova, E.G.; Efimova, A.A.; Menger, F.M. Polyelectrolyte-coated liposomes: Stabilization of the interfacial complexes. Adv. Colloid Interface Sci. 2008, 142, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Yaroslavov, A.A.; Sitnikova, T.A.; Rakhnyanskaya, A.A.; Yaroslavova, E.G.; Sybachin, A.V.; Melik-Nubarov, N.S.; Khomutov, G.B. Variable and low-toxic polyampholytes: Complexation with biological membranes. Colloid Polym. Sci. 2017, 295, 1405–1417. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Efimova, A.A.; Lentin, I.I.; Le-Deygen, I.M.; Izumrudov, V.A. Interaction of ionenes with lipid membrane: Unusual impact of charge density. Langmuir 2020, 36, 14717–14727. [Google Scholar] [CrossRef]
- Wrobel, D.; Appelhans, D.; Signorelli, M.; Wiesner, B.; Fessas, D.; Scheler, U.; Voit, B.; Maly, J. Interaction study between maltose-modified PPI dendrimers and lipidic model membranes. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1490–1501. [Google Scholar] [CrossRef]
- Lombardo, D.; Calandra, P.; Bellocco, E.; Laganà, G.; Barreca, D.; Magazù, S.; Wanderlingh, M.A.; Kiselev, M.A. Effect of anionic and cationic polyamidoamine (PAMAM) dendrimers on a model lipid membrane. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 2769–2777. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.P.; Lyng, F.M.; Garcia, A.; Davoren, M.; Byrne, H.J. Mechanistic studies of in vitro cytotoxicity of poly(amidoamine) dendrimers in mammalian cells. Toxicol. Appl. Pharmacol. 2010, 248, 259–268. [Google Scholar] [CrossRef]
- Janaszewska, A.; Lazniewska, J.; Trzepiński, P.; Marcinkowska, M.; Klajnert-Maculewicz, B. Cytotoxicity of dendrimers. Biomolecules 2019, 9, 330. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, S.A.; Stroylova, Y.Y.; Shifrina, Z.B.; Muronetz, V.I. Disruption of amyloid prion protein aggregates by cationic pyridylphenylene dendrimers. Macromol. Biosci. 2016, 16, 266–275. [Google Scholar] [CrossRef]
- Stroylova, Y.; Sorokina, S.; Stroylov, V.; Melnikova, A.; Gaillard, C.; Shifrina, Z.; Haertlé, T.; Muronetz, V.I. Spontaneous formation of nanofilms under interaction of 4th generation pyrydylphenylene dendrimer with proteins. Polymer 2018, 137, 186–194. [Google Scholar] [CrossRef]
- Sorokina, S.; Semenyuk, P.; Stroylova, Y.; Muronetz, V.; Shifrina, Z. Complexes between cationic pyridylphenylene dendrimers and ovine prion protein: Do hydrophobic interactions matter? RSC Adv. 2017, 7, 16565–16574. [Google Scholar] [CrossRef]
- Shifrina, Z.B.; Kuchkina, N.V.; Rutkevich, P.N.; Vlasik, T.N.; Sushko, A.D.; Izumrudov, V.A. Water-soluble cationic aromatic dendrimers and their complexation with DNA. Macromolecules 2009, 42, 9548–9560. [Google Scholar] [CrossRef]
- Sorokina, S.A.; Stroylova, Y.Y.; Tishina, S.A.; Shifrina, Z.B.; Muronetz, V.I. Promising anti-amyloid behavior of cationic pyridylphenylene dendrimers: Role of structural features and mechanism of action. Eur. Polym. J. 2019, 116, 20–29. [Google Scholar] [CrossRef]
- Trosheva, K.S.; Sorokina, S.A.; Efimova, A.A.; Semenyuk, P.I.; Berkovich, A.K.; Yaroslavov, A.A.; Shifrina, Z.B. Interaction of multicomponent anionic liposomes with cationic pyridylphenylene dendrimer: Does the complex behavior depend on the liposome composition? Biochim. Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183761. [Google Scholar] [CrossRef]
- Pinkwart, K.; Schneider, F.; Lukoseviciute, M.; Sauka-Spengler, T.; Lyman, E.; Eggeling, C.; Sezgin, E. Nanoscale dynamics of cholesterol in the cell membrane. J. Biol. Chem. 2019, 294, 12599–12609. [Google Scholar] [CrossRef] [PubMed]
- Doole, F.T.; Kumarage, T.; Ashkar, R.; Brown, M.F. Cholesterol Stiffening of Lipid Membranes. J. Membr. Biol. 2022, 255, 385–405. [Google Scholar] [CrossRef]
- Wrobel, D.; Kłys, A.; Ionov, M.; Vitovic, P.; Waczulikowa, I.; Hianik, T.; Gomez-Ramirez, R.; de la Mata, J.; Klajnert, B.; Bryszewska, M. Cationic carbosilane dendrimers–lipid membrane interactions. Chem. Phys. Lipids 2012, 165, 401–407. [Google Scholar] [CrossRef]
- Peng, S.F.; Yang, M.J.; Su, C.J.; Chen, H.L.; Lee, P.W.; Wei, M.C.; Sung, H.W. Effects of incorporation of poly (γ-glutamic acid) in chitosan/DNA complex nanoparticles on cellular uptake and transfection efficiency. Biomaterials 2009, 30, 1797–1808. [Google Scholar] [CrossRef]
- Kabanov, V.A.; Yaroslavov, A.A.; Sukhishvili, S.A.; Polynsky, A.S.; Chechik, O.S. Recognition of target colloid species by conjugates of a linear polyelectrolyte with a vector protein. FEBS Lett. 1990, 275, 231–234. [Google Scholar] [CrossRef]
- Davydov, D.A.; Yaroslavova, E.G.; Rakhnyanskaya, A.A.; Efimova, A.A.; Ermakov, Y.A.; Menger, F.M.; Yaroslavov, A.A. Polymer migration among phospholipid liposomes. Langmuir 2009, 25, 13528–13533. [Google Scholar] [CrossRef]
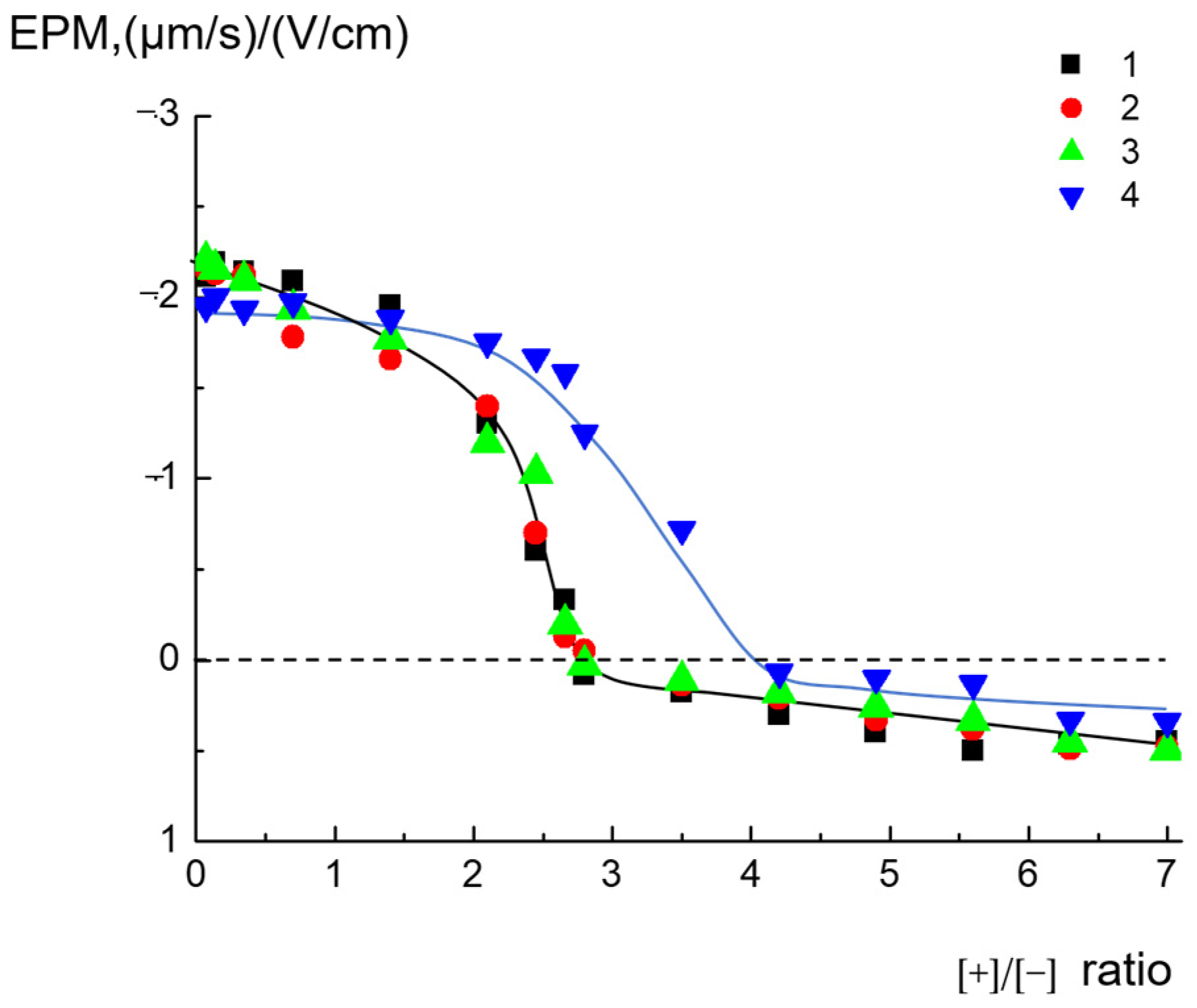
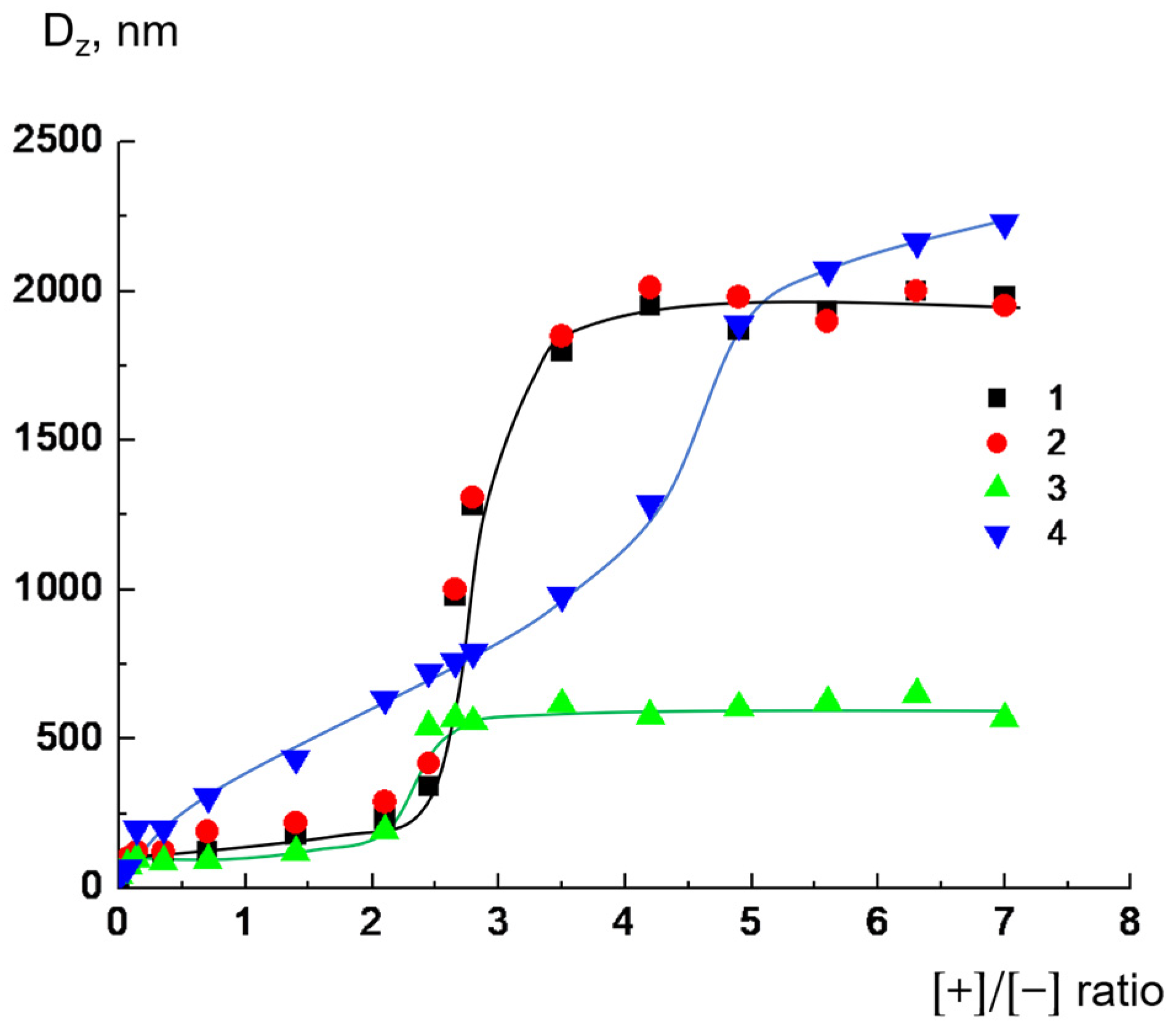
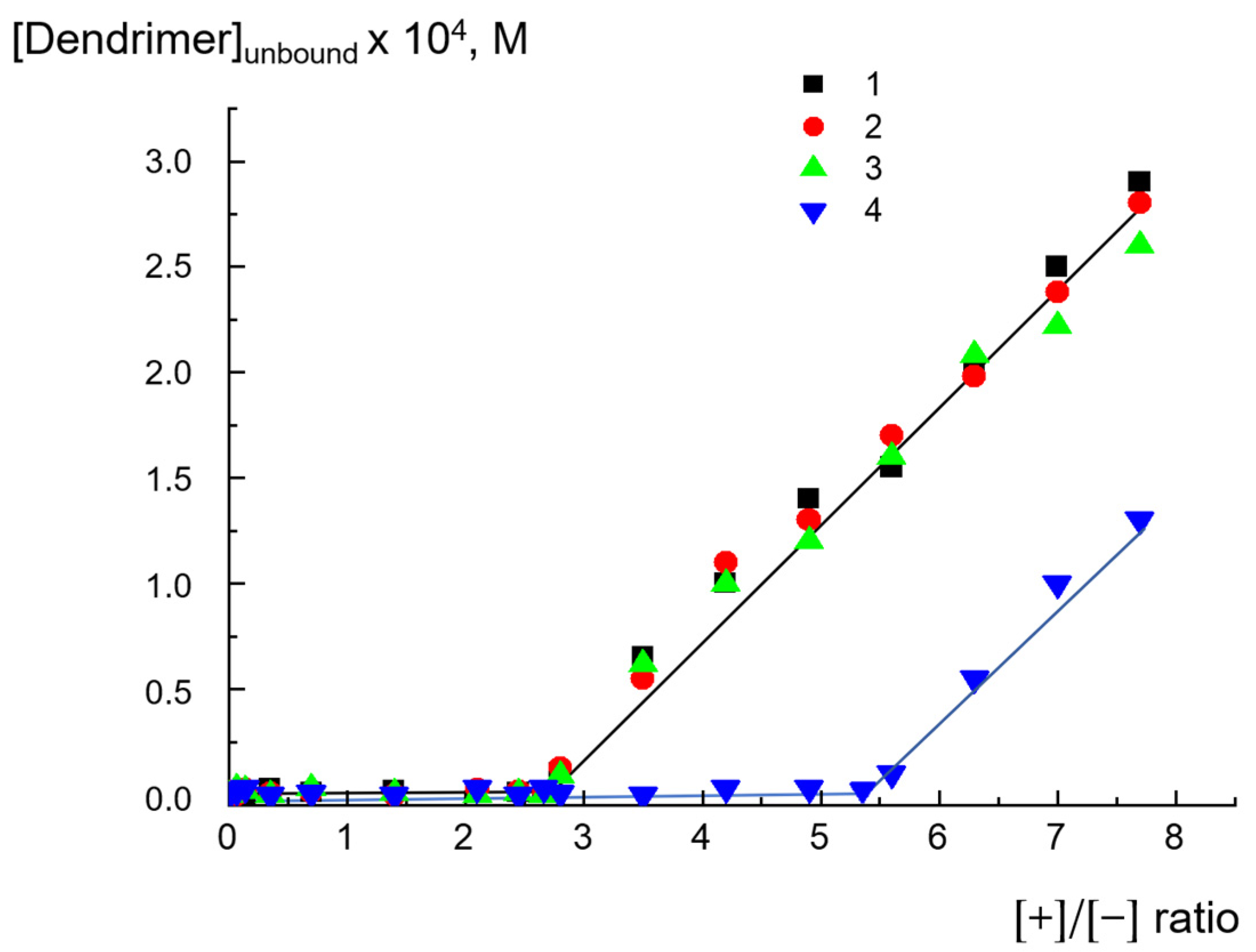
| Total Lipid Concentration, mg/mL | ||||||
|---|---|---|---|---|---|---|
| 2 | 1 | 0.8 | 0.6 | 0.4 | 0.2 | |
| EPM, (μm/s)/(V/cm) | −2.35 ± 0.14 | −2.45 ± 0.15 | −2.38 ± 0.10 | −2.41 ± 0.13 | −2.42 ± 0.12 | −2.43 ± 0.09 |
| Hydrodynamic diameter, nm | 45 ± 5 | 42 ± 4 | 39 ± 2 | 44 ± 2 | 34 ± 4 | 43 ± 7 |
| Dendrimer | Alkylation Degree, % | Number of Ionogenic Groups | Molecular Weight, Da | Hydrodynamic Diameter, nm |
|---|---|---|---|---|
| D16+ | 100 | 6 | 1981 | 3.0 |
| D215+ | 83 | 15 | 5142 | 5.0 |
| D229+ | 96 | 29 | 7190 | 4.5 |
| D350+ | 78 | 50 | 14,440 | 5 |
| Lipid | Formula | Molar Content in Liposomes |
|---|---|---|
| CL |  | 0.1 * |
| DOPC | 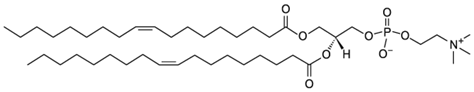 | 0.9 |
| Chol | 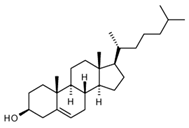 | 0.1 |
| DOPE-CF | 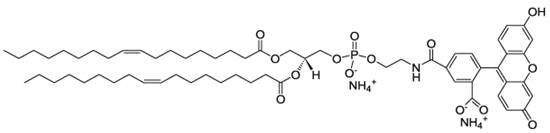 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efimova, A.A.; Sorokina, S.A.; Trosheva, K.S.; Yaroslavov, A.A.; Shifrina, Z.B. Complexes of Cationic Pyridylphenylene Dendrimers with Anionic Liposomes: The Role of Dendrimer Composition in Membrane Structural Changes. Int. J. Mol. Sci. 2023, 24, 2225. https://doi.org/10.3390/ijms24032225
Efimova AA, Sorokina SA, Trosheva KS, Yaroslavov AA, Shifrina ZB. Complexes of Cationic Pyridylphenylene Dendrimers with Anionic Liposomes: The Role of Dendrimer Composition in Membrane Structural Changes. International Journal of Molecular Sciences. 2023; 24(3):2225. https://doi.org/10.3390/ijms24032225
Chicago/Turabian StyleEfimova, Anna A., Svetlana A. Sorokina, Kseniya S. Trosheva, Alexander A. Yaroslavov, and Zinaida B. Shifrina. 2023. "Complexes of Cationic Pyridylphenylene Dendrimers with Anionic Liposomes: The Role of Dendrimer Composition in Membrane Structural Changes" International Journal of Molecular Sciences 24, no. 3: 2225. https://doi.org/10.3390/ijms24032225
APA StyleEfimova, A. A., Sorokina, S. A., Trosheva, K. S., Yaroslavov, A. A., & Shifrina, Z. B. (2023). Complexes of Cationic Pyridylphenylene Dendrimers with Anionic Liposomes: The Role of Dendrimer Composition in Membrane Structural Changes. International Journal of Molecular Sciences, 24(3), 2225. https://doi.org/10.3390/ijms24032225






