Carbonyl Cyanide 3-Chloro Phenyl Hydrazone (CCCP) Restores the Colistin Sensitivity in Brucella intermedia
Abstract
1. Introduction
2. Results
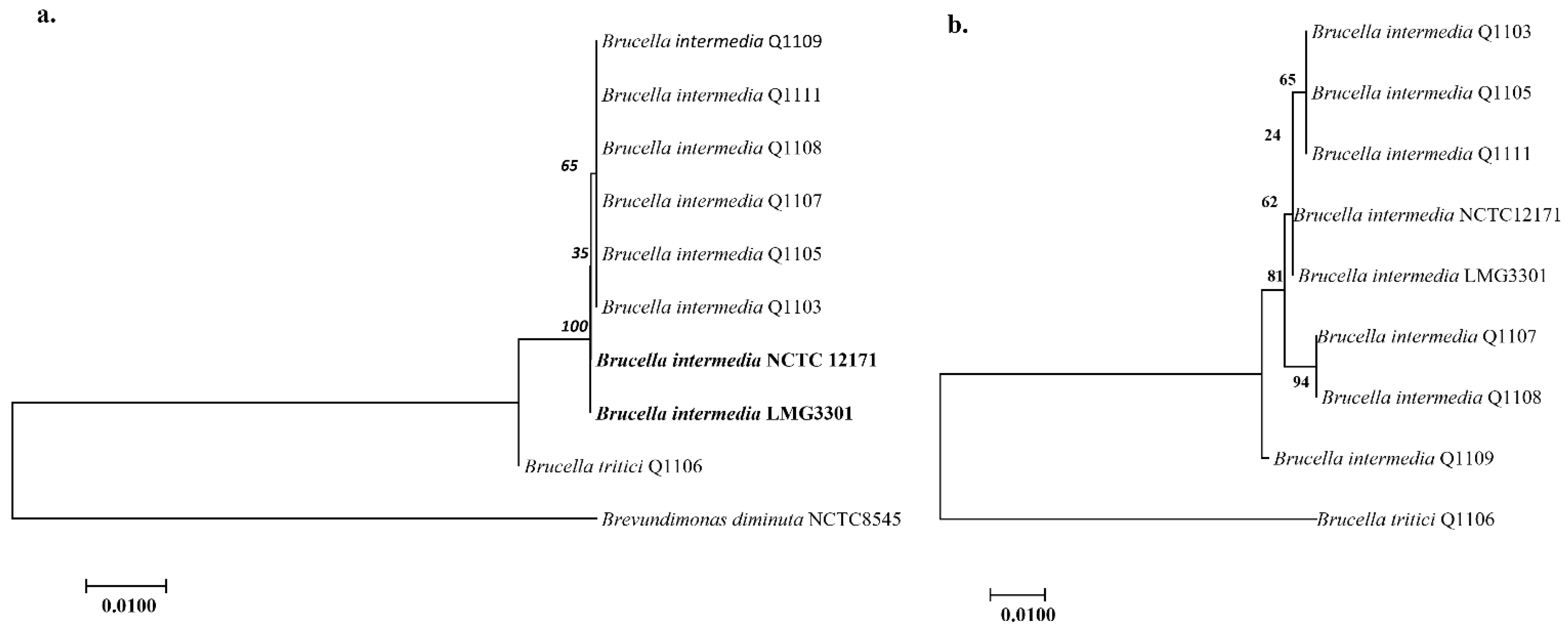
2.1. Bacterial Isolation and Identification
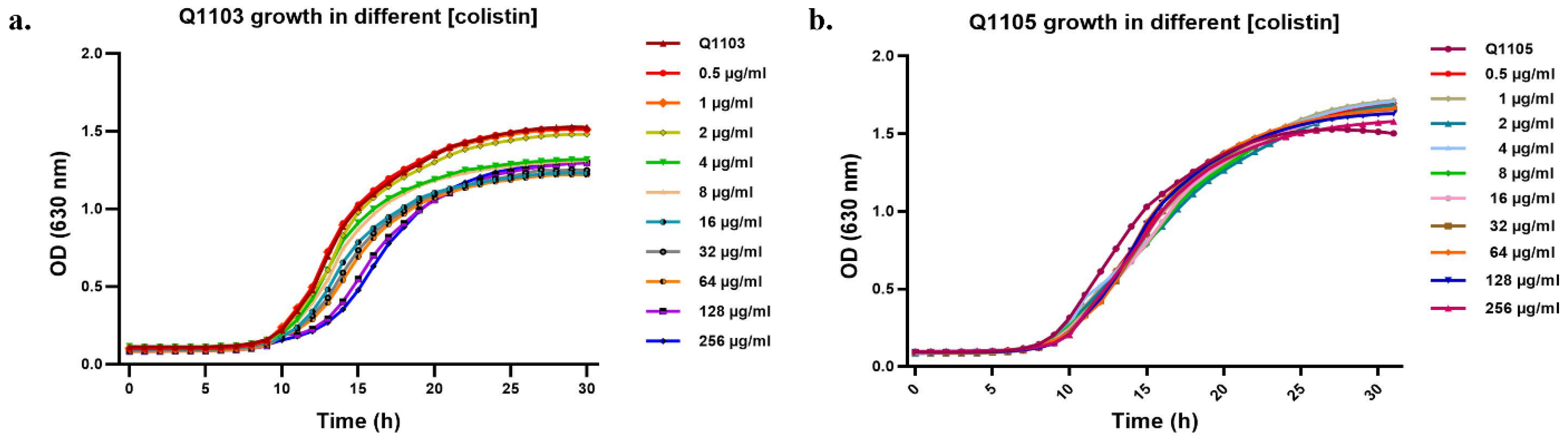
2.2. CO, CCCP and CO/CCCP MIC Determination
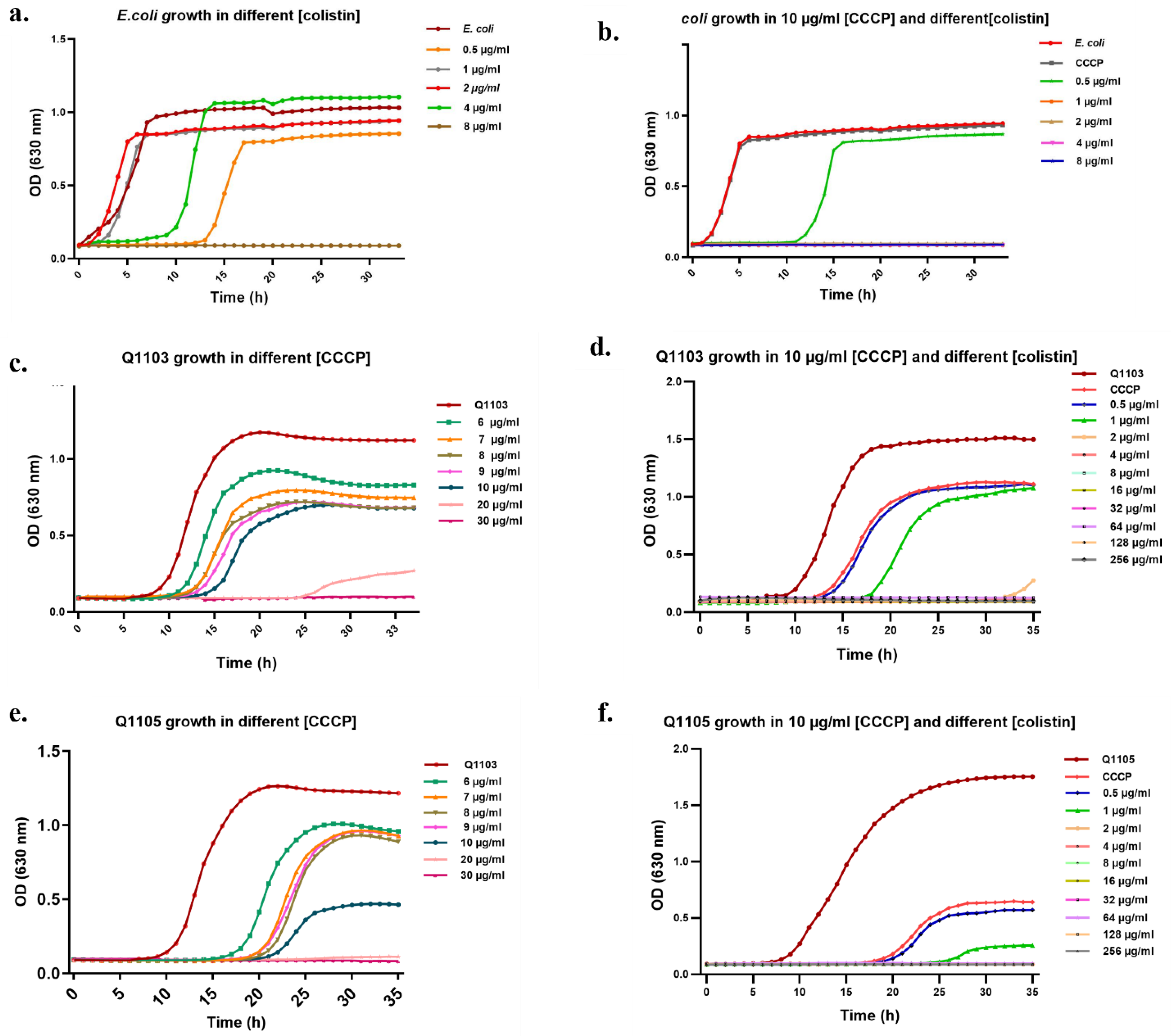
2.3. Synergistic Effect of CCCP-CO Inhibits Bacterial Growth
2.4. Bactericidal Effect of CCCP–CO
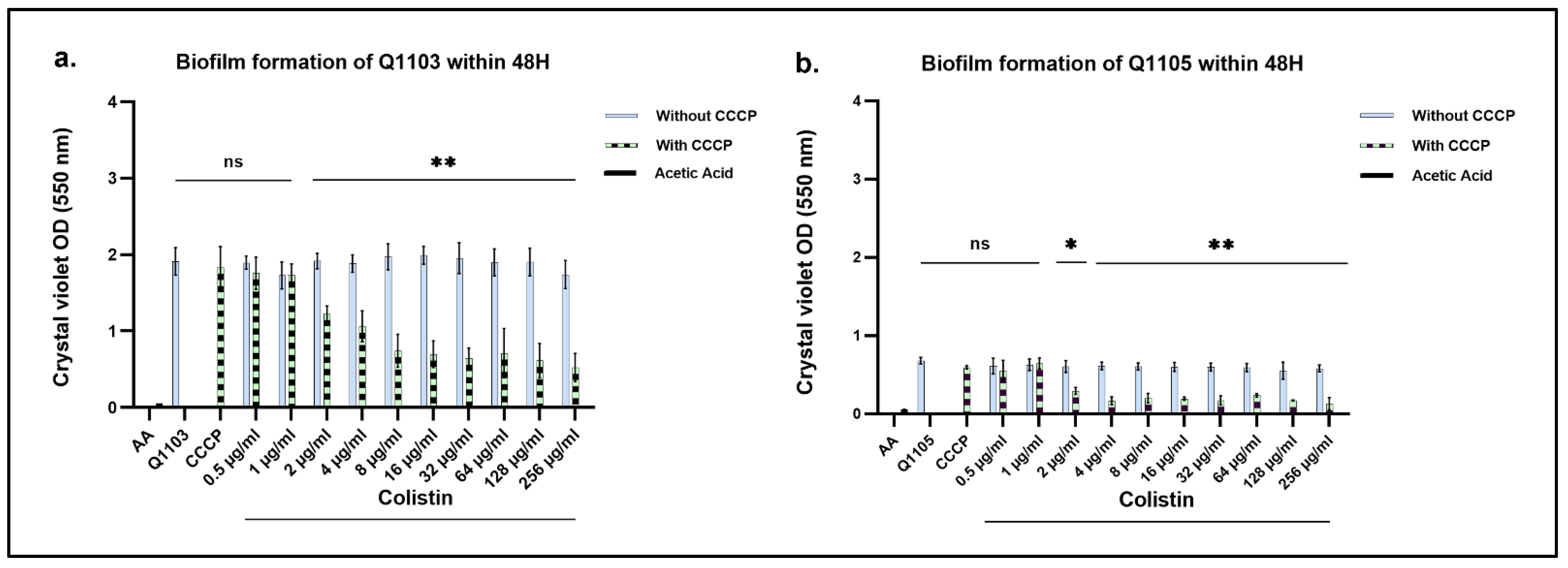
2.5. CO-Treatment with CO/CCCP Inhibits Biofilm Formation of B. intermedia
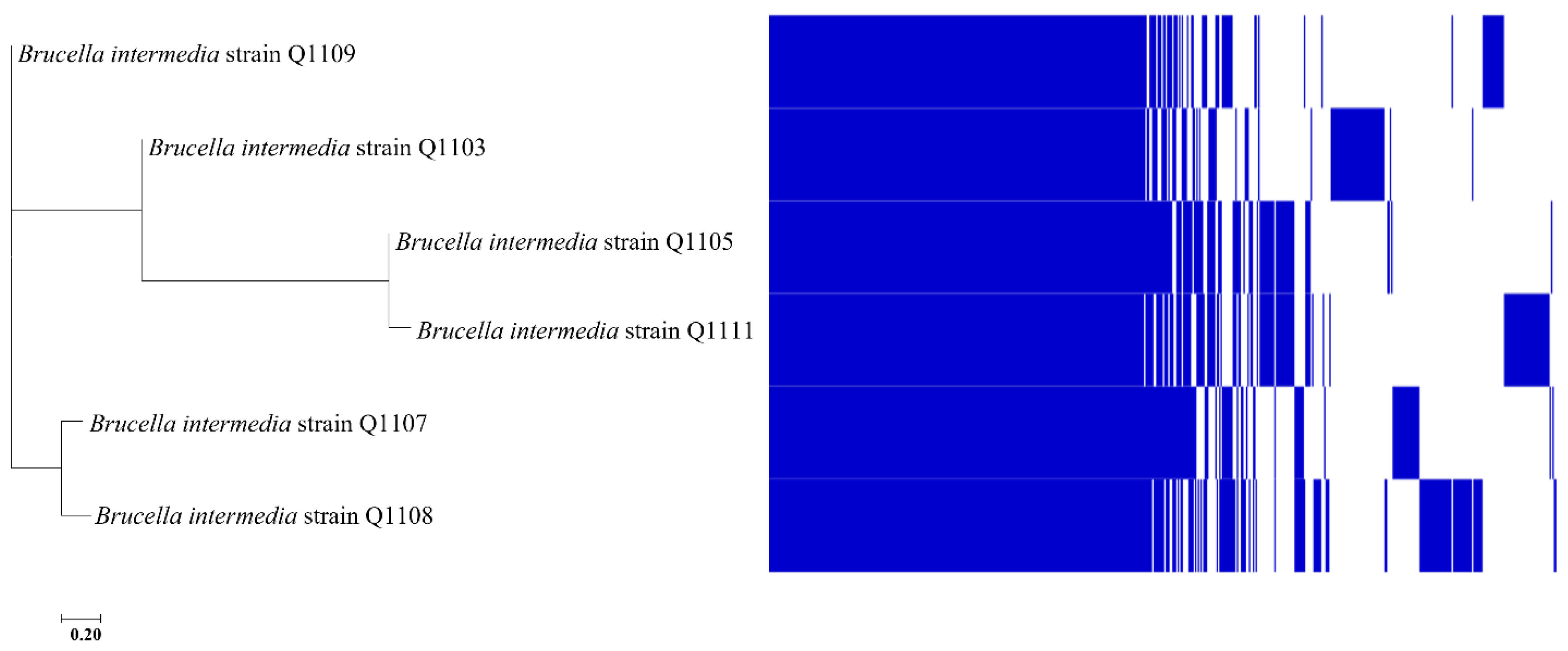
2.6. Pangenomic Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolation and Culture Conditions
4.2. Genome Sequencing, Annotation and Phylogenetic Analyses
4.3. Chemicals and Stock Solution
4.4. Minimal Inhibitory Concentration (MICs) Determination
4.5. MIC Fold Change and Synergy Index
4.6. Effect of CCCP on CO MIC on Agar Plates
4.7. Crystal Blue Assay
4.8. Growth Curves
4.9. Time-Kill Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Storm, D.R.; Rosenthal, K.S.; Swanson, P.E. Polymyxin and Related Peptide Antibiotics. Annu. Rev. Biochem. 1977, 46, 723–763. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.; Giani, T.; Raffone, M.; Arena, F.; Garcia-Fernandez, A.; Pollini, S.; Collective Network EuSCAPE-Italy; Grundmann, H.; Pantosti, A.; Rossolini, G.M. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: A rapidly evolving problem in Italy, November 2013 to April 2014. Eurosurveillance 2014, 19, 20939. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Cai, Y.; Qin, W.; Lin, J.; Qiu, J. Polymyxin E Induces Rapid Paenibacillus polymyxa Death by Damaging Cell Membrane while Ca2+ Can Protect Cells from Damage. PLoS ONE 2015, 10, e0135198. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. In Polymyxin Antibiotics: From Laboratory Bench to Bedside; Li, J., Nation, R.L., Kaye, K.S., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2019; Volume 1145, pp. 55–71. ISBN 978-3-030-16371-6. [Google Scholar]
- Baron, S.; Hadjadj, L.; Rolain, J.-M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef]
- Fernández, L.; Gooderham, W.J.; Bains, M.; McPhee, J.B.; Wiegand, I.; Hancock, R.E.W. Adaptive Resistance to the “Last Hope” Antibiotics Polymyxin B and Colistin in Pseudomonas aeruginosa Is Mediated by the Novel Two-Component Regulatory System ParR-ParS. Antimicrob. Agents Chemother. 2010, 54, 3372–3382. [Google Scholar] [CrossRef]
- Ito-Kagawa, M.; Koyama, Y. Selective cleavage of a peptide antibiotic, colistin by colistinase. J. Antibiot. 1980, 33, 1551–1555. [Google Scholar] [CrossRef]
- Yin, J.; Wang, G.; Cheng, D.; Fu, J.; Qiu, J.; Yu, Z. Inactivation of Polymyxin by Hydrolytic Mechanism. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Hamel, M.; Rolain, J.-M.; Baron, S. The History of Colistin Resistance Mechanisms in Bacteria: Progress and Challenges. Microorganisms 2021, 9, 442. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Campos, M.A.; Vargas, M.A.; Regueiro, V.; Llompart, C.M.; Albertí, S.; Bengoechea, J.A. Capsule Polysaccharide Mediates Bacterial Resistance to Antimicrobial Peptides. Infect. Immun. 2004, 72, 7107–7114. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-F.; Lin, Y.-Y.; Lan, C.-Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 2017, 55, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Puja, H.; Bolard, A.; Noguès, A.; Plésiat, P.; Jeannot, K. The Efflux Pump MexXY/OprM Contributes to the Tolerance and Acquired Resistance of Pseudomonas aeruginosa to Colistin. Antimicrob. Agents Chemother. 2020, 64, e02033-19. [Google Scholar] [CrossRef] [PubMed]
- Aujoulat, F.; Romano-Bertrand, S.; Masnou, A.; Marchandin, H.; Jumas-Bilak, E. Niches, Population Structure and Genome Reduction in Ochrobactrum intermedium: Clues to Technology-Driven Emergence of Pathogens. PLoS ONE 2014, 9, e83376. [Google Scholar] [CrossRef]
- Teyssier, C.; Jumas-Bilak, E.; Marchandin, H.; Jean-Pierre, H.; Jeannot, J.; Dusart, G.; Foulongne, V.; de Buochberg, M.S. Identification d’espèce et épidémiologie moléculaire des bactéries du genre Ochrobactrum. Pathol. Biol. 2003, 51, 5–12. [Google Scholar] [CrossRef]
- Teyssier, C.; Jumas-Bilak, E.; Counil, F.; Masnou, A.; Aleyrangues, L.; Chiron, R.; Marchandin, H. Ochrobactrum and Agrobacterium spp.: Emerging opportunistic pathogens in cystic fibrosis patients? J. Cyst. Fibros. 2009, 8, S47. [Google Scholar] [CrossRef]
- Apisarnthanarak, A.; Kiratisin, P.; Mundy, L.M. Evaluation of Ochrobactrum intermedium bacteremia in a patient with bladder cancer. Diagn. Microbiol. Infect. Dis. 2005, 53, 153–155. [Google Scholar] [CrossRef]
- Vaidya, S.A.; Citron, D.M.; Fine, M.B.; Murakami, G.; Goldstein, E.J.C. Pelvic Abscess Due to Ochrobactrum anthropi in an Immunocompetent Host: Case Report and Review of the Literature. J. Clin. Microbiol. 2007, 45, 1672. [Google Scholar] [CrossRef]
- Dharne, M.S.; Misra, S.P.; Misra, V.; Dwivedi, M.; Patole, M.S.; Shouche, Y.S. Isolation of Urease-Positive Ochrobactrum Intermedium in the Stomach of a Non-Ulcer Dyspeptic Patient from North India. J. Microbiol. Immunol. Infect. 2008, 41, 183–186. [Google Scholar]
- Bharucha, T.; Sharma, D.; Sharma, H.; Kandil, H.; Collier, S. Ochromobactrum intermedium: An emerging opportunistic pathogen—Case of recurrent bacteraemia associated with infective endocarditis in a haemodialysis patient. New Microbes New Infect. 2016, 15, 14–15. [Google Scholar] [CrossRef][Green Version]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, ume 12, 965–975. [Google Scholar] [CrossRef]
- Olaitan, A.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Ko, K.S. Effect of carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on killing Acinetobacter baumannii by colistin. J. Microbiol. 2015, 53, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Li, Y.; Guan, J.; Zhao, J.; Cui, J.; Wang, R.; Liu, Y. Effects of Efflux Pump Inhibitors on Colistin Resistance in Multidrug-Resistant Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2016, 60, 3215–3218. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Wei, X.; Wan, X.; Ding, Z.; Ding, Y.; Liu, J. The role and relationship with efflux pump of biofilm formation in Klebsiella pneumoniae. Microb. Pathog. 2020, 147, 104244. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Teng, T.; Zhang, Z.; Shang, Y.; Xiao, H.; Jiang, G.; Wang, F.; Jia, J.; Dong, L.; Zhao, L.; et al. Carbonyl Cyanide 3-Chlorophenylhydrazone (CCCP) Exhibits Direct Antibacterial Activity Against Mycobacterium abscessus. Infect. Drug Resist. 2021, ume 14, 1199–1208. [Google Scholar] [CrossRef]
- A Baron, S.; Rolain, J.-M. Efflux pump inhibitor CCCP to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and Gram-negative bacteria. J. Antimicrob. Chemother. 2018, 73, 1862–1871. [Google Scholar] [CrossRef]
- Wang, Z.; Bie, P.; Cheng, J.; Lu, L.; Cui, B.; Wu, Q. The ABC transporter YejABEF is required for resistance to antimicrobial peptides and the virulence of Brucella melitensis. Sci. Rep. 2016, 6, 31876. [Google Scholar] [CrossRef]
- Cheah, S.-E.; Johnson, M.; Zhu, Y.; Tsuji, B.T.; Forrest, A.; Bulitta, J.; Boyce, J.; Nation, R.L.; Li, J. Polymyxin Resistance in Acinetobacter baumannii: Genetic Mutations and Transcriptomic Changes in Response to Clinically Relevant Dosage Regimens. Sci. Rep. 2016, 6, 26233. [Google Scholar] [CrossRef]
- Fehlner-Gardiner, C.C.; A Valvano, M. Cloning and characterization of the Burkholderia vietnamiensis norM gene encoding a multi-drug efflux protein. FEMS Microbiol. Lett. 2002, 215, 279–283. [Google Scholar] [CrossRef][Green Version]
- Bialer, M.G.; Ruiz-Ranwez, V.; Sycz, G.; Estein, S.M.; Russo, D.M.; Altabe, S.; Sieira, R.; Zorreguieta, A. MapB, the Brucella Suis TamB Homologue, Is Involved in Cell Envelope Biogenesis, Cell Division and Virulence. Sci. Rep. 2019, 9, 2158. [Google Scholar] [CrossRef] [PubMed]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global Colistin Use: A Review of the Emergence of Resistant Enterobacterales and the Impact on Their Genetic Basis. FEMS Microbiol. Rev. 2021, 46, fuab049. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J.; Amoako, D.G. Carbonyl Cyanide M-Chlorophenylhydrazine (CCCP) Reverses Resistance to Colistin, but Not to Carbapenems and Tigecycline in Multidrug-Resistant Enterobacteriaceae. Front. Microbiol. 2017, 8, 228. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Prevention of Drug Access to Bacterial Targets: Permeability Barriers and Active Efflux. Science 1994, 264, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Montville, T.J.; Bruno, M.E.C. Evidence That Dissipation of Proton Motive Force Is a Common Mechanism of Action for Bacteriocins and Other Antimicrobial Proteins. Int. J. Food Microbiol. 1994, 24, 53–74. [Google Scholar] [CrossRef] [PubMed]
- Le, D.; Krasnopeeva, E.; Sinjab, F.; Pilizota, T.; Kim, M. Active Efflux Leads to Heterogeneous Dissipation of Proton Motive Force by Protonophores in Bacteria. mBio 2021, 12, e00676-21. [Google Scholar] [CrossRef] [PubMed]
- Borges-Walmsley, M.I.; Beauchamp, J.; Kelly, S.M.; Jumel, K.; Candlish, D.; Harding, S.E.; Price, N.C.; Walmsley, A.R. Identification of Oligomerization and Drug-Binding Domains of the Membrane Fusion Protein EmrA. J. Biol. Chem. 2003, 278, 12903–12912. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Gil-Ramírez, Y.; Conde-Álvarez, R.; Palacios-Chaves, L.; Zúñiga-Ripa, A.; Grilló, M.-J.; Arce-Gorvel, V.; Hanniffy, S.; Moriyón, I.; Iriarte, M. The Identification of WadB, a New Glycosyltransferase Gene, Confirms the Branched Structure and the Role in Virulence of the Lipopolysaccharide Core of Brucella Abortus. Microb. Pathog. 2014, 73, 53–59. [Google Scholar] [CrossRef]
- Matson, J.S.; Yoo, H.J.; Hakansson, K.; DiRita, V.J. Polymyxin B Resistance in El Tor Vibrio Cholerae Requires Lipid Acylation Catalyzed by MsbB. J. Bacteriol. 2010, 192, 2044–2052. [Google Scholar] [CrossRef]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef] [PubMed]
- Liautard, J.; Ouahrani-Bettache, S.; Jubier-Maurin, V.; Lafont, V.; Köhler, S.; Liautard, J.-P. Identification and Isolation of Brucella Suis Virulence Genes Involved in Resistance to the Human Innate Immune System. Infect. Immun. 2007, 75, 5167–5174. [Google Scholar] [CrossRef] [PubMed]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A Next Generation Sequencing Platform for Genomic Analysis. In Disease Gene Identification: Methods and Protocols; DiStefano, J.K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 223–232. ISBN 978-1-4939-7471-9. [Google Scholar]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An Interactive Viewer for Bacterial Population Genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; André, C.; Poirel, L.; Dubois, V. Evaluation of Three Broth Microdilution Systems to Determine Colistin Susceptibility of Gram-Negative Bacilli. J. Antimicrob. Chemother. 2018, 73, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Lamers, R.P.; Cavallari, J.F.; Burrows, L.L. The Efflux Inhibitor Phenylalanine-Arginine Beta-Naphthylamide (PAβN) Permeabilizes the Outer Membrane of Gram-Negative Bacteria. PLoS ONE 2013, 8, e60666. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Terminology Relating to Methods for the Determination of Susceptibility of Bacteria to Antimicrobial Agents. Clin. Microbiol. Infect. 2000, 6, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef]


| Microdilution Method | ||||||
|---|---|---|---|---|---|---|
| MIC (µg/mL) | FIC Index | |||||
| Strains | CO | CCCP | CO/CCCP | MIC Fold Change | Value | Interpretation |
| E. coli | >4–8 | >30 | <1 | 4–8 | 0.1–0.2 | Synergism |
| Q1103 | >512 | <30 | <2–4 | 128–256 | 0.0 | Synergism |
| Q1105 | >512 | <20 | <1 | 512 | 0.0 | Synergism |
| Q1107 | >512 | <20 | <2 | 256 | 0.0 | Synergism |
| Q1108 | >512 | <15 | <0.5 | 512 | 0.0 | Synergism |
| Q1109 | >512 | <20 | <1 | 512 | 0.0 | Synergism |
| Q1111 | >512 | <15 | <1 | 512 | 0.0 | Synergism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zoaiter, M.; Zeaiter, Z.; Mediannikov, O.; Sokhna, C.; Fournier, P.-E. Carbonyl Cyanide 3-Chloro Phenyl Hydrazone (CCCP) Restores the Colistin Sensitivity in Brucella intermedia. Int. J. Mol. Sci. 2023, 24, 2106. https://doi.org/10.3390/ijms24032106
Zoaiter M, Zeaiter Z, Mediannikov O, Sokhna C, Fournier P-E. Carbonyl Cyanide 3-Chloro Phenyl Hydrazone (CCCP) Restores the Colistin Sensitivity in Brucella intermedia. International Journal of Molecular Sciences. 2023; 24(3):2106. https://doi.org/10.3390/ijms24032106
Chicago/Turabian StyleZoaiter, Malak, Zaher Zeaiter, Oleg Mediannikov, Cheikh Sokhna, and Pierre-Edouard Fournier. 2023. "Carbonyl Cyanide 3-Chloro Phenyl Hydrazone (CCCP) Restores the Colistin Sensitivity in Brucella intermedia" International Journal of Molecular Sciences 24, no. 3: 2106. https://doi.org/10.3390/ijms24032106
APA StyleZoaiter, M., Zeaiter, Z., Mediannikov, O., Sokhna, C., & Fournier, P.-E. (2023). Carbonyl Cyanide 3-Chloro Phenyl Hydrazone (CCCP) Restores the Colistin Sensitivity in Brucella intermedia. International Journal of Molecular Sciences, 24(3), 2106. https://doi.org/10.3390/ijms24032106






