The Intertwined Role of 8-oxodG and G4 in Transcription Regulation
Abstract
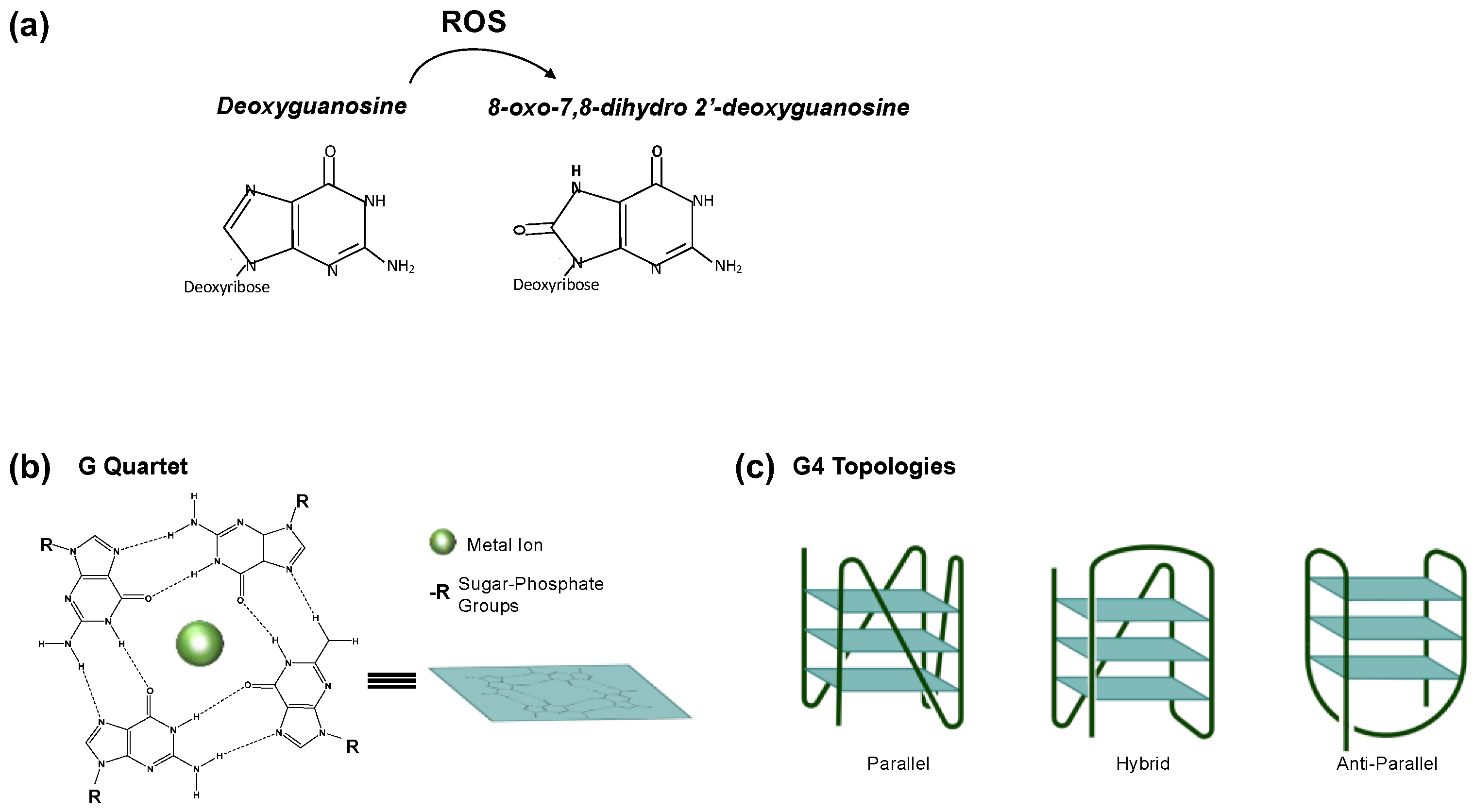
1. Introduction
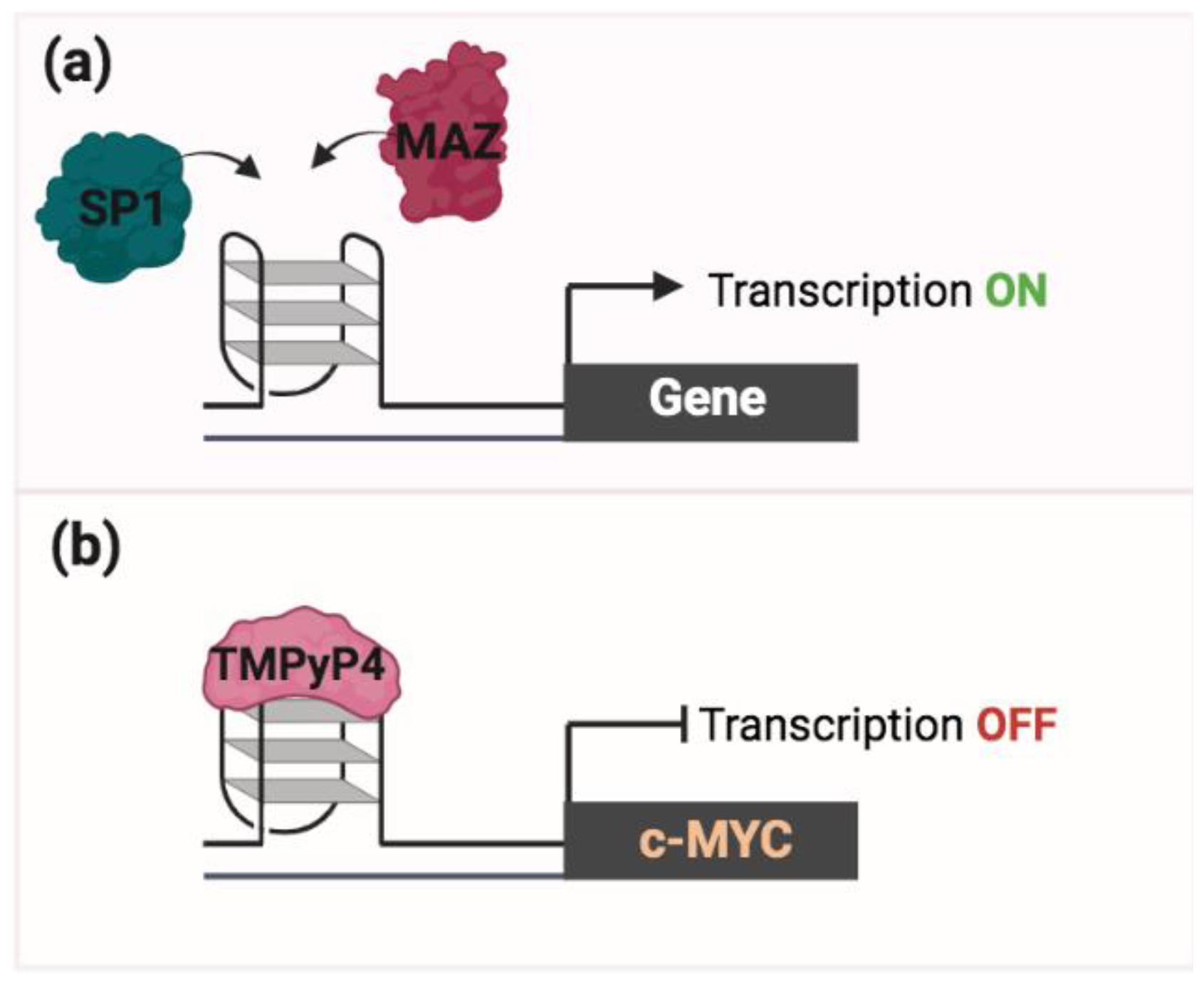
2. PQS/G4 in Transcription Regulation
3. G4 as a Pharmacological Target
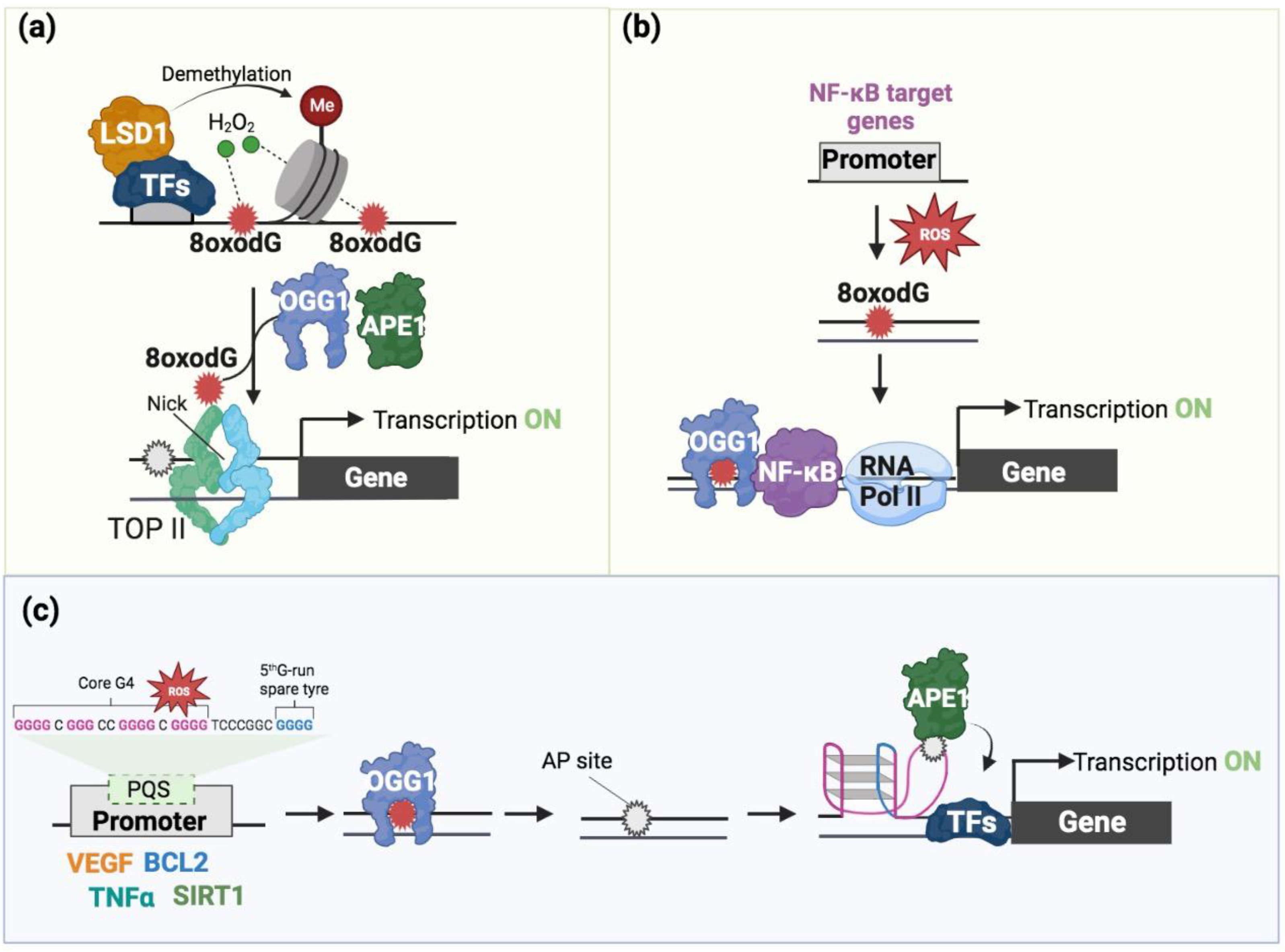
4. 8-oxodG in Transcription Regulation
5. The Interplay of 8-oxodG and G4 in Transcription
6. The Potential Interplay of 8-oxodG and G4 in Chromatin Organization
7. 8-oxodG and BER’s Proteins as a Pharmacological Target
8. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gorini, F.; Scala, G.; Cooke, M.S.; Majello, B.; Amente, S. Towards a Comprehensive View of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine: Highlighting the Intertwined Roles of DNA Damage and Epigenetics in Genomic Instability. DNA Repair 2021, 97, 103027. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Baik, M.H.; Silverman, J.S.; Yang, I.V.; Ropp, P.A.; Szalai, V.A.; Yang, W.; Thorp, H.H. Using Density Functional Theory to Design DNA Base Analogues with Low Oxidation Potentials. J. Phys. Chem. B 2001, 105, 6437–6444. [Google Scholar] [CrossRef]
- van Loon, B.; Markkanen, E.; Hübscher, U. Oxygen as a Friend and Enemy: How to Combat the Mutational Potential of 8-Oxo-Guanine. DNA Repair 2010, 9, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA Damage: Mechanisms, Mutation, and Disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA Damage and Disease: Induction, Repair and Significance. Mutat. Res.-Rev. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef]
- de Rosa, M.; Johnson, S.A.; Opresko, P.L. Roles for the 8-Oxoguanine DNA Repair System in Protecting Telomeres From Oxidative Stress. Front. Cell Dev. Biol. 2021, 9, 758402. [Google Scholar] [CrossRef]
- Giorgio, M.; Dellino, G.I.; Gambino, V.; Roda, N.; Pelicci, P.G. On the Epigenetic Role of Guanosine Oxidation. Redox Biol. 2020, 29, 101398. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. 8-Oxo-7,8-Dihydroguanine, Friend and Foe: Epigenetic-like Regulator versus Initiator of Mutagenesis. DNA Repair 2017, 56, 75–83. [Google Scholar] [CrossRef]
- Wang, R.; Hao, W.; Pan, L.; Boldogh, I.; Ba, X. The Roles of Base Excision Repair Enzyme OGG1 in Gene Expression. Cell. Mol. Life Sci. 2018, 75, 3741–3750. [Google Scholar] [CrossRef]
- Ding, Y.; Fleming, A.M.; Burrows, C.J. Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-Dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017, 139, 2569–2572. [Google Scholar] [CrossRef] [PubMed]
- Amente, S.; Di Palo, G.; Scala, G.; Castrignanò, T.; Gorini, F.; Cocozza, S.; Moresano, A.; Pucci, P.; Ma, B.; Stepanov, I.; et al. Genome-Wide Mapping of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine Reveals Accumulation of Oxidatively-Generated Damage at DNA Replication Origins within Transcribed Long Genes of Mammalian Cells. Nucleic Acids Res. 2019, 47, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhu, J.; Manage, S.A.H.; Burrows, C.J. Human NEIL3 Gene Expression Regulated by Epigenetic-Like Oxidative DNA Modification. J. Am. Chem. Soc. 2019, 141, 11036–11049. [Google Scholar] [CrossRef]
- Redstone, S.C.J.; Fleming, A.M.; Burrows, C.J. Oxidative Modification of the Potential G-Quadruplex Sequence in the PCNA Gene Promoter Can Turn on Transcription. Chem. Res. Toxicol. 2019, 32, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Gorini, F.; Scala, G.; Di Palo, G.; Dellino, G.I.; Cocozza, S.; Pelicci, P.G.; Lania, L.; Majello, B.; Amente, S. The Genomic Landscape of 8-OxodG Reveals Enrichment at Specific Inherently Fragile Promoters. Nucleic Acids Res. 2020, 48, 4309–4324. [Google Scholar] [CrossRef]
- An, J.; Yin, M.; Yin, J.; Wu, S.; Selby, C.P.; Yang, Y.; Sancar, A.; Xu, G.-L.; Qian, M.; Hu, J. Genome-Wide Analysis of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine at Single-Nucleotide Resolution Unveils Reduced Occurrence of Oxidative Damage at G-Quadruplex Sites. Nucleic Acids Res. 2021, 49, 12252–12267. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Oxidative Stress-Mediated Epigenetic Regulation by G-Quadruplexes. NAR Cancer 2021, 3, 1–16. [Google Scholar] [CrossRef]
- Huppert, J.L. Structure, Location and Interactions of G-Quadruplexes. FEBS J. 2010, 277, 3452–3458. [Google Scholar] [CrossRef]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making Sense of G-Quadruplex and i-Motif Functions in Oncogene Promoters. FEBS J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Visser, J.A.; Zhu, J.; Burrows, C.J. Human DNA Repair Genes Possess Potential G-Quadruplex Sequences in Their Promoters and 5′-Untranslated Regions. Biochemistry 2018, 57, 991–1002. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of Parallel Four-Stranded Complexes by Guanine-Rich Motifs in DNA and Its Implications for Meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Schiavone, D.; Guilbaud, G.; Murat, P.; Papadopoulou, C.; Sarkies, P.; Prioleau, M.-N.; Balasubramanian, S.; Sale, J.E. Determinants of G Quadruplex-Induced Epigenetic Instability in REV1-Deficient Cells. EMBO J. 2014, 33, 2507–2520. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Miglietta, G.; Russo, M.; Capranico, G. G-Quadruplex-R-Loop Interactions and the Mechanism of Anticancer G-Quadruplex Binders. Nucleic Acids Res. 2020, 48, 11942–11957. [Google Scholar] [CrossRef]
- Miglietta, G.; Marinello, J.; Russo, M.; Capranico, G. Ligands Stimulating Antitumour Immunity as the next G-Quadruplex Challenge. Mol. Cancer 2022, 21, 180. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Raguseo, F.; Nuccio, S.P.; Liano, D.; Di Antonio, M. DNA G-Quadruplex Structures: More than Simple Roadblocks to Transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [Google Scholar] [CrossRef]
- Kotsantis, P.; Segura-Bayona, S.; Margalef, P.; Marzec, P.; Ruis, P.; Hewitt, G.; Bellelli, R.; Patel, H.; Goldstone, R.; Poetsch, A.R.; et al. RTEL1 Regulates G4/R-Loops to Avert Replication-Transcription Collisions. Cell Rep. 2020, 33, 108546. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-Quadruplex Formation within the Promoter of the KRAS Proto-Oncogene and Its Effect on Transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef]
- Cogoi, S.; Shchekotikhin, A.E.; Xodo, L.E. HRAS Is Silenced by Two Neighboring G-Quadruplexes and Activated by MAZ, a Zinc-Finger Transcription Factor with DNA Unfolding Property. Nucleic Acids Res. 2014, 42, 8379–8388. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Xodo, L.E. G4 DNA in Ras Genes and Its Potential in Cancer Therapy. Biochim. Biophys. Acta-Gene Regul. Mech. 2016, 1859, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.W.; Phang, T.; Edwards, M.G.; Geraci, M.W.; Gillespie, M.N. Promoter G-Quadruplex Sequences Are Targets for Base Oxidation and Strand Cleavage during Hypoxia-Induced Transcription. Free Radic. Biol. Med. 2012, 53, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Raiber, E.A.; Kranaster, R.; Lam, E.; Nikan, M.; Balasubramanian, S. A Non-Canonical DNA Structure Is a Binding Motif for the Transcription Factor SP1 in Vitro. Nucleic Acids Res. 2012, 40, 1499–1508. [Google Scholar] [CrossRef]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Moreno, H.D.; Schotta, G.; Richter, S.N. Promoter G-Quadruplexes and Transcription Factors Cooperate to Shape the Cell Type-Specific Transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-Quadruplex Structures Mark Human Regulatory Chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c-MYC Transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-Throughput Sequencing of DNA G-Quadruplex Structures in the Human Genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole Genome Experimental Maps of DNA G-Quadruplexes in Multiple Species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef]
- Tu, J.; Duan, M.; Liu, W.; Lu, N.; Zhou, Y.; Sun, X.; Lu, Z. Direct Genome-Wide Identification of G-Quadruplex Structures by Whole-Genome Resequencing. Nat. Commun. 2021, 12, 6014. [Google Scholar] [CrossRef]
- Zheng, K.; Zhang, J.; He, Y.; Gong, J.; Wen, C.; Chen, J.; Hao, Y.; Zhao, Y.; Tan, Z. Detection of Genomic G-Quadruplexes in Living Cells Using a Small Artificial Protein. Nucleic Acids Res. 2020, 48, 11706–11720. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Shao, R.; Yung, P.Y.K.; Elsässer, S.J. Genome-Wide Mapping of G-Quadruplex Structures with CUT&Tag. Nucleic Acids Res. 2022, 50, e13. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Lan, L. The DNA Secondary Structures at Telomeres and Genome Instability. Cell Biosci. 2020, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M. Mechanisms of DNA Replication and Repair: Insights from the Study of G-Quadruplexes. Molecules 2019, 24, 3439. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M. G-Quadruplexes at Telomeres: Friend or Foe? Molecules 2020, 25, 3686. [Google Scholar] [CrossRef] [PubMed]
- Lerner, L.K.; Sale, J.E. Replication of G Quadruplex DNA. Genes 2019, 10, 95. [Google Scholar] [CrossRef]
- Carvalho, J.; Mergny, J.-L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-Quadruplex, Friend or Foe: The Role of the G-Quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Yang, Q.-F.; Lin, X.; Chen, D.; Wang, Z.-Y.; Chen, B.; Han, H.-Y.; Chen, H.-D.; Cai, K.-C.; Li, Q.; et al. G4LDB 2.2: A Database for Discovering and Studying G-Quadruplex and i-Motif Ligands. Nucleic Acids Res. 2022, 50, D150–D160. [Google Scholar] [CrossRef]
- Micco, M.; Collie, G.W.; Dale, A.G.; Ohnmacht, S.A.; Pazitna, I.; Gunaratnam, M.; Reszka, A.P.; Neidle, S. Structure-Based Design and Evaluation of Naphthalene Diimide G-Quadruplex Ligands as Telomere Targeting Agents in Pancreatic Cancer Cells. J. Med. Chem. 2013, 56, 2959–2974. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Vankayalapati, H.; Shin-Ya, K.; Wierzba, K.; Hurley, L.H. Telomestatin, a Potent Telomerase Inhibitor That Interacts Quite Specifically with the Human Telomeric Intramolecular g-Quadruplex. J. Am. Chem. Soc. 2002, 124, 2098–2099. [Google Scholar] [CrossRef] [PubMed]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-Quadruplex-Interactive Molecule BRACO-19 Inhibits Tumor Growth, Consistent with Telomere Targeting and Interference with Telomerase Function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Izbicka, E.; Wheelhouse, R.T.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of Cationic Porphyrins as G-Quadruplex Interactive Agents in Human Tumor Cells. Cancer Res. 1999, 59, 639–644. [Google Scholar]
- Gowan, S.M.; Heald, R.; Stevens, M.F.; Kelland, L.R. Potent Inhibition of Telomerase by Small-Molecule Pentacyclic Acridines Capable of Interacting with G-Quadruplexes. Mol. Pharmacol. 2001, 60, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Müller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.-F.; Balasubramanian, S. A Novel Small Molecule That Alters Shelterin Integrity and Triggers a DNA-Damage Response at Telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.S.; Gates, A.J.; Waller, Z.A.E. Destabilization of I-Motif DNA at Neutral PH by G-Quadruplex Ligands. Biochemistry 2019, 58, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Pagano, A.; Iaccarino, N.; Abdelhamid, M.A.S.; Brancaccio, D.; Garzarella, E.U.; Di Porzio, A.; Novellino, E.; Waller, Z.A.E.; Pagano, B.; Amato, J.; et al. Common G-Quadruplex Binding Agents Found to Interact With i-Motif-Forming DNA: Unexpected Multi-Target-Directed Compounds. Front. Chem. 2018, 6, 281. [Google Scholar] [CrossRef]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-Quadruplexes: A Promising Target for Cancer Therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef]
- Felsenstein, K.M.; Saunders, L.B.; Simmons, J.K.; Leon, E.; Calabrese, D.R.; Zhang, S.; Michalowski, A.; Gareiss, P.; Mock, B.A.; Schneekloth, J.S., Jr. Small Molecule Microarrays Enable the Identification of a Selective, Quadruplex-Binding Inhibitor of MYC Expression. ACS Chem. Biol. 2016, 11, 139–148. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef]
- Perillo, B.; Ombra, M.N.; Bertoni, A.; Cuozzo, C.; Sacchetti, S.; Sasso, A.; Chiariotti, L.; Malorni, A.; Abbondanza, C.; Avvedimento, E. V DNA Oxidation as Triggered by H3K9me2 Demethylation Drives Estrogen-Induced Gene Expression. Science 2008, 319, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Amente, S.; Bertoni, A.; Morano, A.; Lania, L.; Avvedimento, E.V.; Majello, B. LSD1-Mediated Demethylation of Histone H3 Lysine 4 Triggers Myc-Induced Transcription. Oncogene 2010, 29, 3691–3702. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA Damage Is Epigenetic by Regulating Gene Transcription via Base Excision Repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Amente, S.; Lania, L.; Avvedimento, E.V.; Majello, B. DNA Oxidation Drives Myc Mediated Transcription. Cell Cycle 2010, 9, 3074–3076. [Google Scholar] [CrossRef]
- Amente, S.; Lania, L.; Majello, B. The Histone LSD1 Demethylase in Stemness and Cancer Transcription Programs. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 981–986. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Mo, W.; Zhang, J.; Li, X.; Meng, D.; Gao, Y.; Yang, S.; Wan, X.; Zhou, C.; Guo, F.; Huang, Y.; et al. Identification of Novel AR-Targeted MicroRNAs Mediating Androgen Signalling through Critical Pathways to Regulate Cell Viability in Prostate Cancer. PLoS ONE 2013, 8, e56592. [Google Scholar] [CrossRef]
- Pezone, A.; Taddei, M.L.; Tramontano, A.; Dolcini, J.; Boffo, F.L.; De Rosa, M.; Parri, M.; Stinziani, S.; Comito, G.; Porcellini, A.; et al. Targeted DNA Oxidation by LSD1-SMAD2/3 Primes TGF-Β1/ EMT Genes for Activation or Repression. Nucleic Acids Res. 2020, 48, 8943–8958. [Google Scholar] [CrossRef]
- Sengupta, S.; Wang, H.; Yang, C.; Szczesny, B.; Hegde, M.L.; Mitra, S. Ligand-Induced Gene Activation Is Associated with Oxidative Genome Damage Whose Repair Is Required for Transcription. Proc. Natl. Acad. Sci. USA 2020, 117, 22183–22192. [Google Scholar] [CrossRef]
- Pan, L.; Zhu, B.; Hao, W.; Zeng, X.; Vlahopoulos, S.A.; Hazra, T.K.; Hegde, M.L.; Radak, Z.; Bacsi, A.; Brasier, A.R.; et al. Oxidized Guanine Base Lesions Function in 8-Oxoguanine DNA Glycosylase-1-Mediated Epigenetic Regulation of Nuclear Factor ΚB-Driven Gene Expression. J. Biol. Chem. 2016, 291, 25553–25566. [Google Scholar] [CrossRef]
- Pan, L.; Hao, W.; Zheng, X.; Zeng, X.; Abbasi, A.A.; Boldogh, I.; Ba, X. OGG1-DNA Interactions Facilitate NF-κB Binding to DNA Targets. Sci. Rep. 2017, 7, srep43297. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Zhang, J.; Ellis, L.M.; Semenza, G.L.; Evans, D.B.; Watowich, S.S.; Gallick, G.E. HIF-1α, STAT3, CBP/P300 and Ref-1/APE Are Components of a Transcriptional Complex That Regulates Src-Dependent Hypoxia-Induced Expression of VEGF in Pancreatic and Prostate Carcinomas. Oncogene 2005, 24, 3110–3120. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Yin, M.; Hu, J. G-Quadruplex and 8-Oxo-7,8-Dihydroguanine across the Genome: Methodologies and Crosstalk. Genome Instab. Dis. 2022, 3, 241–254. [Google Scholar] [CrossRef]
- Zhu, J.; Fleming, A.M.; Burrows, C.J. The RAD17 Promoter Sequence Contains a Potential Tail-Dependent G-Quadruplex That Downregulates Gene Expression upon Oxidative Modification. ACS Chem. Biol. 2018, 13, 2577–2584. [Google Scholar] [CrossRef]
- Omaga, C.A.; Fleming, A.M.; Burrows, C.J. The Fifth Domain in the G-Quadruplex-Forming Sequence of the Human NEIL3 Promoter Locks DNA Folding in Response to Oxidative Damage. Biochemistry 2018, 57, 2958–2970. [Google Scholar] [CrossRef]
- Poetsch, A.R.; Boulton, S.J.; Luscombe, N.M. Genomic Landscape of Oxidative DNA Damage and Repair Reveals Regioselective Protection from Mutagenesis 06 Biological Sciences 0604 Genetics. Genome Biol. 2018, 19, 215. [Google Scholar] [CrossRef]
- Wu, J.; McKeague, M.; Sturla, S.J. Nucleotide-Resolution Genome-Wide Mapping of Oxidative DNA Damage by Click-Code-Seq. J. Am. Chem. Soc. 2018, 140, 9783–9787. [Google Scholar] [CrossRef]
- Bielskutė, S.; Plavec, J.; Podbevšek, P. Impact of Oxidative Lesions on the Human Telomeric G-Quadruplex. J. Am. Chem. Soc. 2019, 141, 2594–2603. [Google Scholar] [CrossRef]
- Lee, H.-T.; Sanford, S.; Paul, T.; Choe, J.; Bose, A.; Opresko, P.L.; Myong, S. Position-Dependent Effect of Guanine Base Damage and Mutations on Telomeric G-Quadruplex and Telomerase Extension. Biochemistry 2020, 59, 2627–2639. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These “Spare Tires” Have an Evolved Function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, S.; Pramanik, S.; Harris, H.L.; Tarpley, M.; Sarkar, A.; Spagnol, G.; Sorgen, P.L.; Chowdhury, D.; Band, V.; Klinkebiel, D.; et al. Endogenous Oxidized DNA Bases and APE1 Regulate the Formation of G-Quadruplex Structures in the Genome. Proc. Natl. Acad. Sci. USA 2020, 117, 11409–11420. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, S.; Chen, Y.; Song, H.; Khutsishvili, I.; Marky, L.A.; Ray, S.; Natarajan, A.; Singh, P.K.; Bhakat, K.K. The Human AP-Endonuclease 1 (APE1) Is a DNA G-Quadruplex Structure Binding Protein and Regulates KRAS Expression in Pancreatic Ductal Adenocarcinoma Cells. Nucleic Acids Res. 2022, 50, 3394–3412. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.D.; Marecki, J.C.; Byrd, A.K.; Gao, J.; Raney, K.D. G-Quadruplex Loops Regulate PARP-1 Enzymatic Activation. Nucleic Acids Res. 2021, 49, 416–431. [Google Scholar] [CrossRef]
- Reynolds, P.; Cooper, S.; Lomax, M.; O’Neill, P. Disruption of PARP1 Function Inhibits Base Excision Repair of a Sub-Set of DNA Lesions. Nucleic Acids Res. 2015, 43, 4028–4038. [Google Scholar] [CrossRef]
- Cogoi, S.; Ferino, A.; Miglietta, G.; Pedersen, E.B.; Xodo, L.E. The Regulatory G4 Motif of the Kirsten Ras (KRAS) Gene Is Sensitive to Guanine Oxidation: Implications on Transcription. Nucleic Acids Res. 2018, 46, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Linke, R.; Limmer, M.; Juranek, S.; Heine, A.; Paeschke, K. The Relevance of G-Quadruplexes for DNA Repair. Int. J. Mol. Sci. 2021, 22, 12599. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. G-Quadruplex Folds of the Human Telomere Sequence Alter the Site Reactivity and Reaction Pathway of Guanine Oxidation Compared to Duplex DNA. Chem. Res. Toxicol. 2013, 26, 593–607. [Google Scholar] [CrossRef]
- Scala, G.; Gorini, F.; Ambrosio, S.; Chiariello, A.M.; Nicodemi, M.; Lania, L.; Majello, B.; Amente, S. 8-OxodG Accumulation within Super-Enhancers Marks Fragile CTCF-Mediated Chromatin Loops. Nucleic Acids Res. 2022, 50, 3292–3306. [Google Scholar] [CrossRef]
- Hou, Y.; Li, F.; Zhang, R.; Li, S.; Liu, H.; Qin, Z.S.; Sun, X. Integrative Characterization of G-Quadruplexes in the Three-Dimensional Chromatin Structure. Epigenetics 2019, 14, 894–911. [Google Scholar] [CrossRef]
- Hahm, J.Y.; Park, J.; Jang, E.S.; Chi, S.W. 8-Oxoguanine: From Oxidative Damage to Epigenetic and Epitranscriptional Modification. Exp. Mol. Med. 2022, 54, 1626–1642. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef] [PubMed]
- Hanna, B.M.F.; Helleday, T.; Mortusewicz, O. OGG1 Inhibitor TH5487 Alters OGG1 Chromatin Dynamics and Prevents Incisions. Biomolecules 2020, 10, 1483. [Google Scholar] [CrossRef] [PubMed]
- Baquero, J.M.; Benítez-Buelga, C.; Rajagopal, V.; Zhenjun, Z.; Torres-Ruiz, R.; Müller, S.; Hanna, B.M.F.; Loseva, O.; Wallner, O.; Michel, M.; et al. Small Molecule Inhibitor of OGG1 Blocks Oxidative DNA Damage Repair at Telomeres and Potentiates Methotrexate Anticancer Effects. Sci. Rep. 2021, 11, 3490. [Google Scholar] [CrossRef]
- Tahara, Y.-K.; Auld, D.; Ji, D.; Beharry, A.A.; Kietrys, A.M.; Wilson, D.L.; Jimenez, M.; King, D.; Nguyen, Z.; Kool, E.T. Potent and Selective Inhibitors of 8-Oxoguanine DNA Glycosylase. J. Am. Chem. Soc. 2018, 140, 2105–2114. [Google Scholar] [CrossRef]
- Caimi, P.F.; Cooper, B.W.; William, B.M.; Dowlati, A.; Barr, P.M.; Fu, P.; Pink, J.; Xu, Y.; Lazarus, H.M.; de Lima, M.; et al. Phase I Clinical Trial of the Base Excision Repair Inhibitor Methoxyamine in Combination with Fludarabine for Patients with Advanced Hematologic Malignancies. Oncotarget 2017, 8, 79864–79875. [Google Scholar] [CrossRef]
- Gordon, M.S.; Rosen, L.S.; Mendelson, D.; Ramanathan, R.K.; Goldman, J.; Liu, L.; Xu, Y.; Gerson, S.L.; Anthony, S.P.; Figg, W.D.; et al. A Phase 1 Study of TRC102, an Inhibitor of Base Excision Repair, and Pemetrexed in Patients with Advanced Solid Tumors. Investig. New Drugs 2013, 31, 714–723. [Google Scholar] [CrossRef]
- Grundy, G.J.; Parsons, J.L. Base excision repair and its implications to cancer therapy. Essays Biochem. 2020, 64, 831–843. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorini, F.; Ambrosio, S.; Lania, L.; Majello, B.; Amente, S. The Intertwined Role of 8-oxodG and G4 in Transcription Regulation. Int. J. Mol. Sci. 2023, 24, 2031. https://doi.org/10.3390/ijms24032031
Gorini F, Ambrosio S, Lania L, Majello B, Amente S. The Intertwined Role of 8-oxodG and G4 in Transcription Regulation. International Journal of Molecular Sciences. 2023; 24(3):2031. https://doi.org/10.3390/ijms24032031
Chicago/Turabian StyleGorini, Francesca, Susanna Ambrosio, Luigi Lania, Barbara Majello, and Stefano Amente. 2023. "The Intertwined Role of 8-oxodG and G4 in Transcription Regulation" International Journal of Molecular Sciences 24, no. 3: 2031. https://doi.org/10.3390/ijms24032031
APA StyleGorini, F., Ambrosio, S., Lania, L., Majello, B., & Amente, S. (2023). The Intertwined Role of 8-oxodG and G4 in Transcription Regulation. International Journal of Molecular Sciences, 24(3), 2031. https://doi.org/10.3390/ijms24032031






