miR-889-3p Facilitates the Browning Process of White Adipocyte Precursors by Targeting the SON Gene
Abstract
:1. Introduction
2. Results
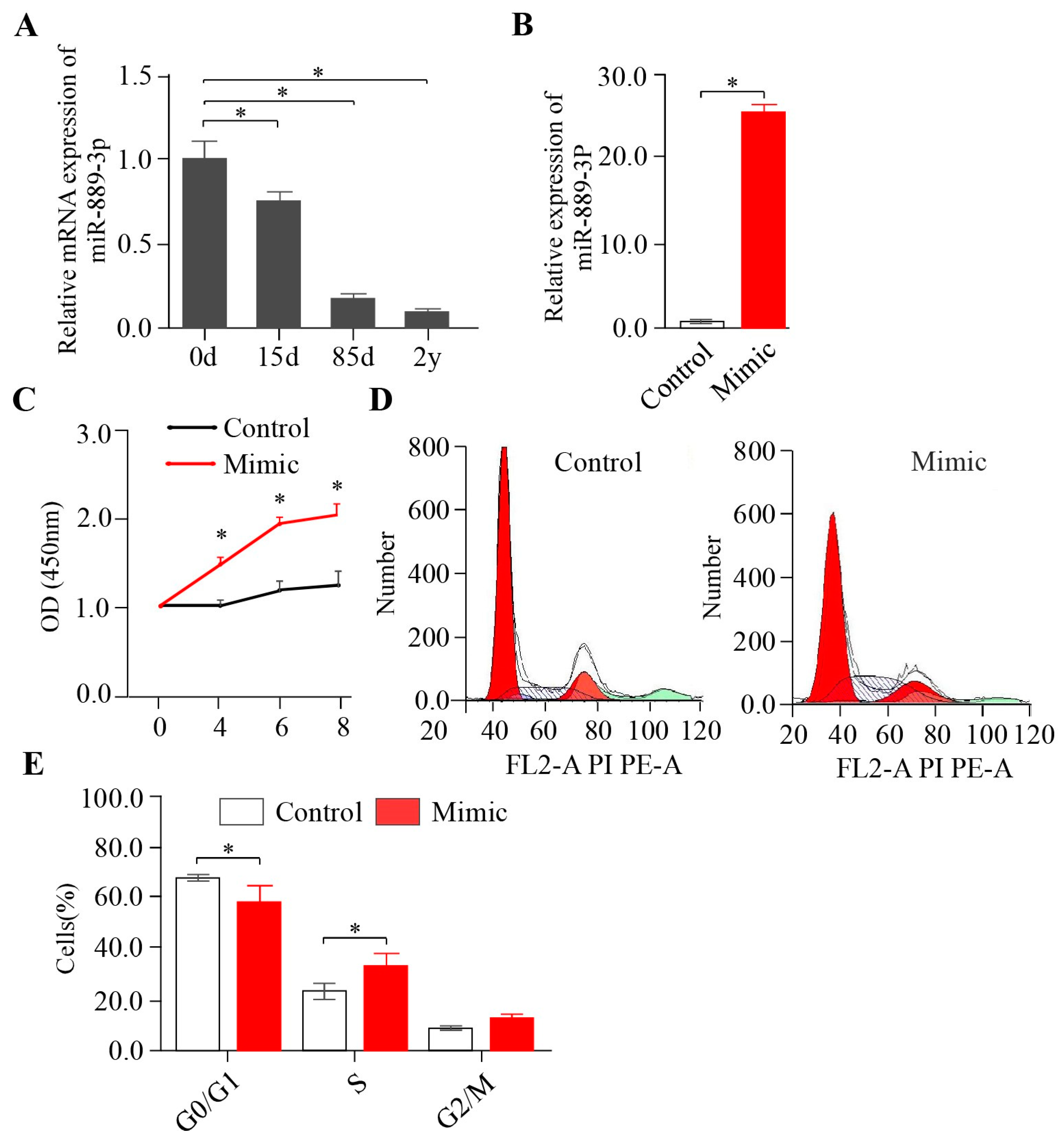
2.1. miR-889-3p Promotes Rabbit Preadipocyte Proliferation
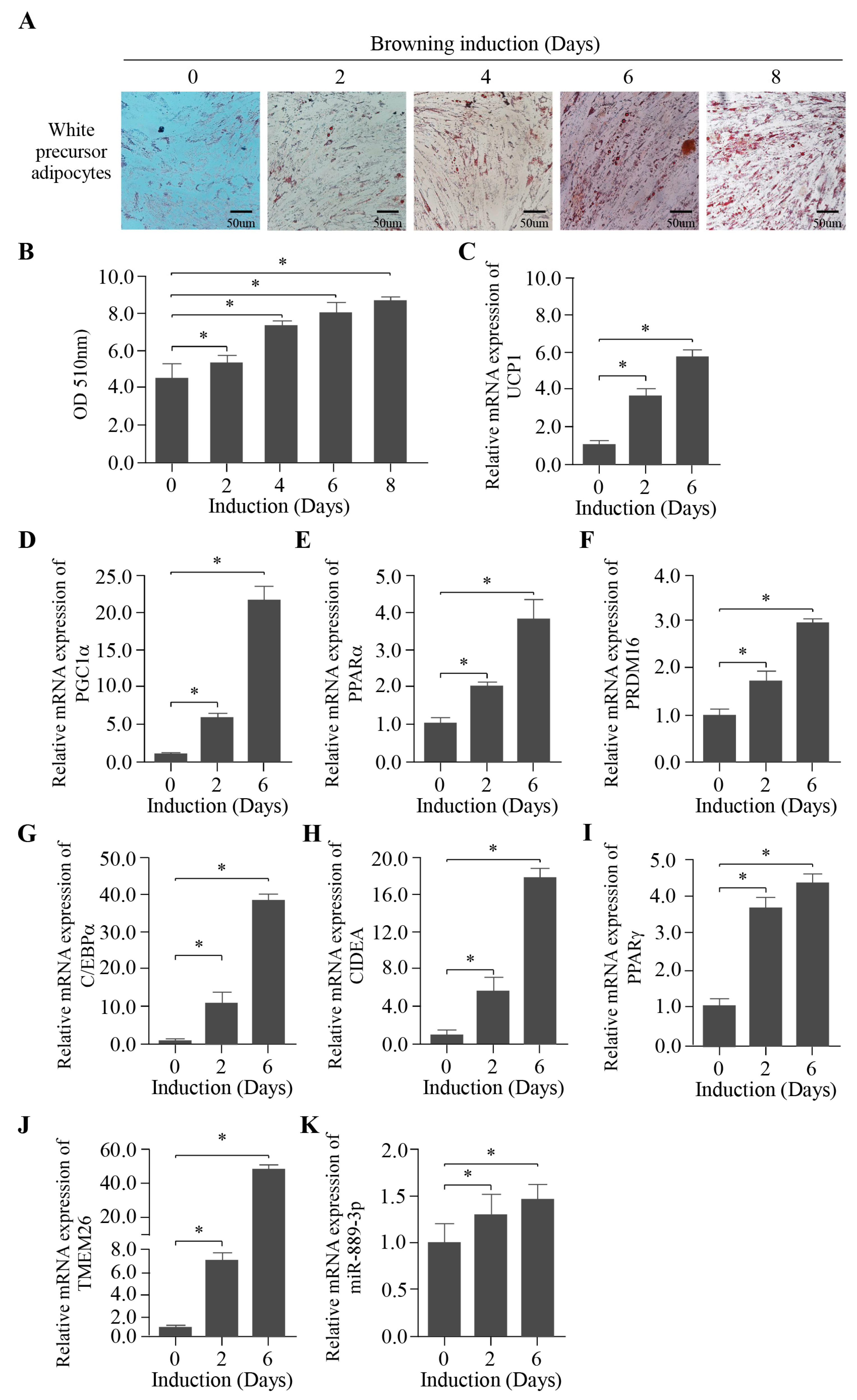
2.2. Establishment of the Browning Induction of Rabbit White Preadipocytes In Vitro
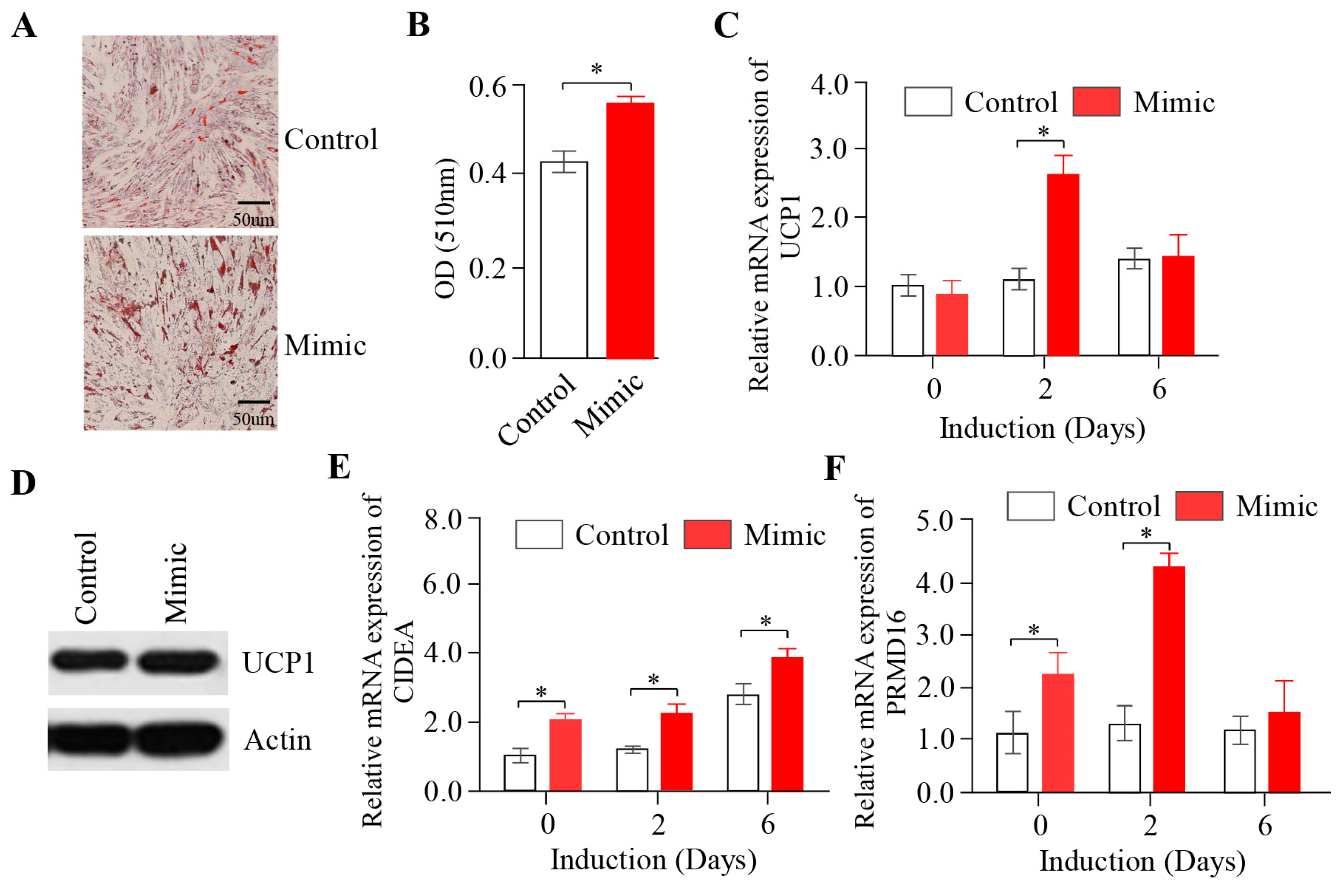
2.3. The Overexpression of miR-889-3p Promotes the Browning of Rabbit Precursor White Adipocytes
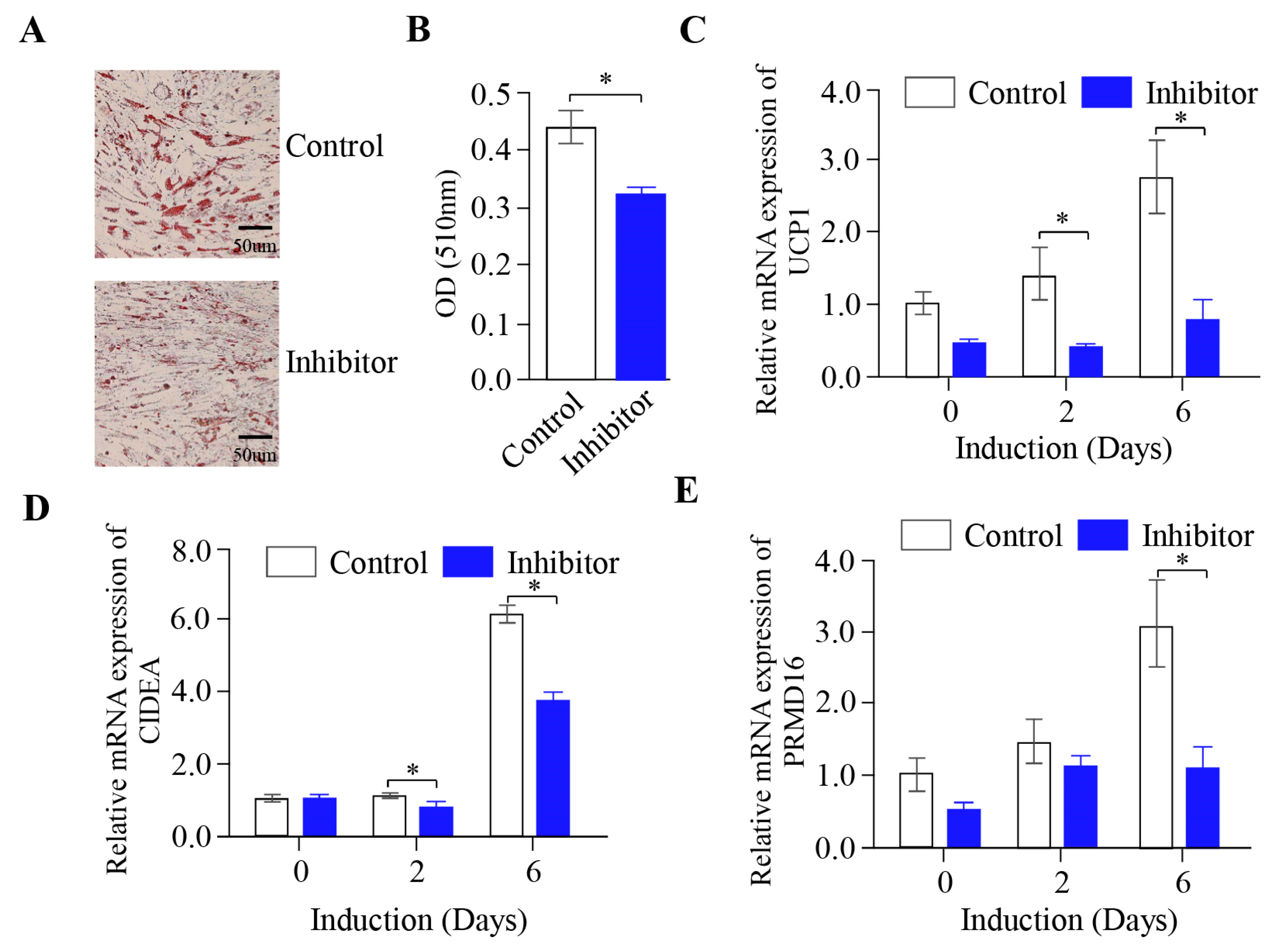
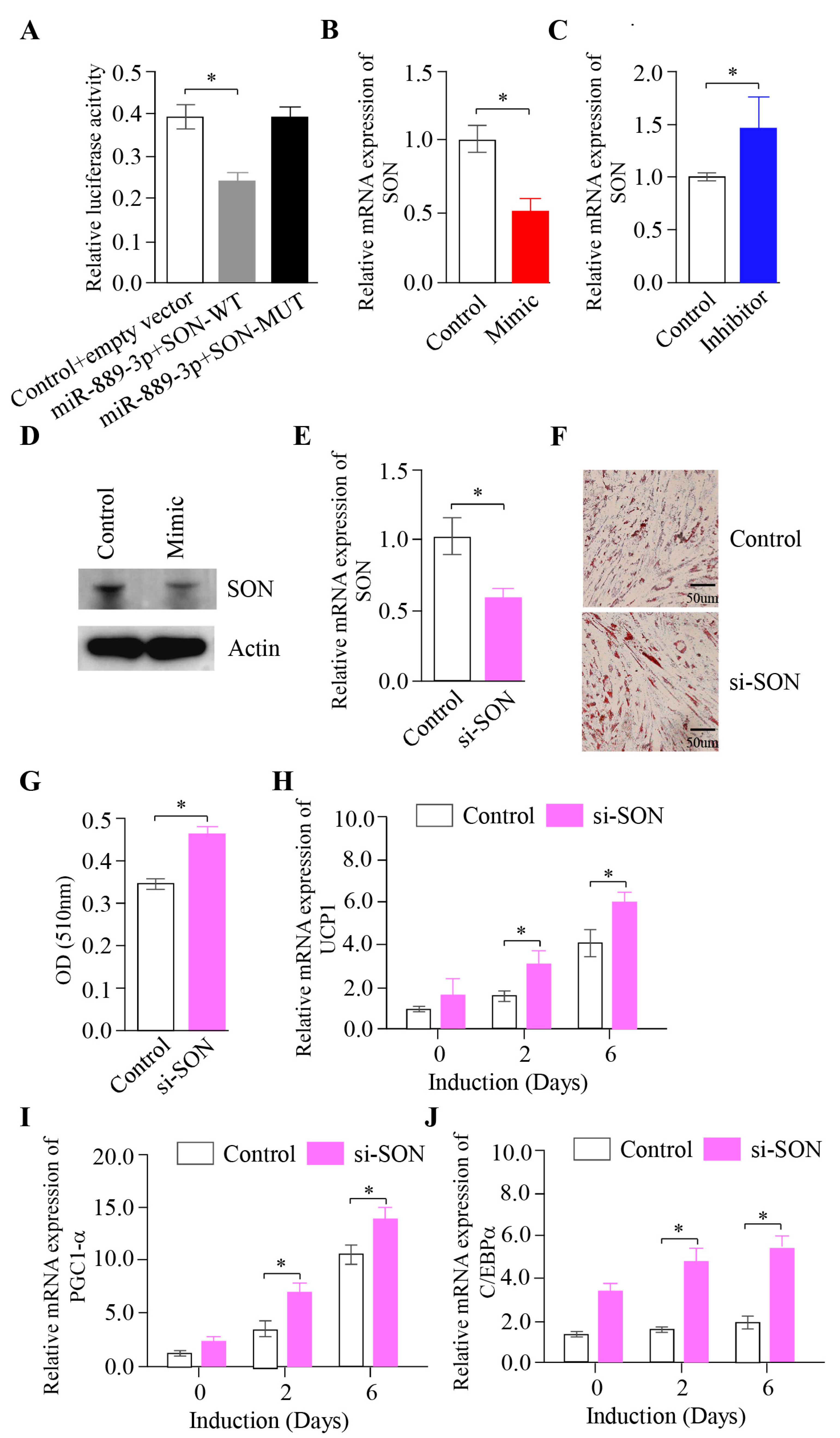
2.4. The Inhibition of miR-889-3p Expression Inhibits the Browning of Rabbit Precursor White Adipocytes
2.5. The Effect of Inhibiting the SON Gene on the Browning of Rabbit White Precursor Adipocytes In Vitro
3. Discussion
4. Materials and Methods
4.1. Animal Tissues and Sequencing
4.2. Cell Culture and Browning Induction
4.3. RNA Extraction and RT-qPCR
4.4. Oil Red O Staining
4.5. Cell Transfection
4.6. Western Blotting
4.7. Cell Cycle Analysis
4.8. Cell Counting Kit-8 Assay
4.9. Validation of the Target Gene by Double Luciferase Report
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef]
- Cancello, R.; Zingaretti, M.C.; Sarzani, R.; Ricquier, D.; Cinti, S. Leptin and UCP1 genes are reciprocally regulated in brown adipose tissue. Endocrinology 1998, 139, 4747–4750. [Google Scholar] [CrossRef]
- Cousin, B.; Cinti, S.; Morroni, M.; Raimbault, S.; Ricquier, D.; Penicaud, L.; Casteilla, L. Occurrence of brown adipocytes in rat white adipose tissue: Molecular and morphological characterization. J. Cell Sci. 1992, 103 Pt 4, 931–942. [Google Scholar] [CrossRef]
- Ikeda, K.; Maretich, P.; Kajimura, S. The Common and Distinct Features of Brown and Beige Adipocytes. Trends Endocrinol. Metab. 2018, 29, 191–200. [Google Scholar] [CrossRef]
- Wu, J.; Cohen, P.; Spiegelman, B.M. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013, 27, 234–250. [Google Scholar] [CrossRef]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef]
- Gharanei, S.; Shabir, K.; Brown, J.E.; Weickert, M.O.; Barber, T.M.; Kyrou, I.; Randeva, H.S. Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells 2020, 9, 2489. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, Y.; Li, L.; Guo, M.; Zou, C.; Xu, Y.; Yang, Z. MicroRNA-889-3p restrains the proliferation and epithelial-mesenchymal transformation of lung cancer cells via down-regulation of Homeodomain-interacting protein kinase 1. Bioengineered 2021, 12, 10945–10958. [Google Scholar] [CrossRef]
- Zhao, X.; Dong, W.; Luo, G.; Xie, J.; Liu, J.; Yu, F. Silencing of hsa_circ_0009035 Suppresses Cervical Cancer Progression and Enhances Radiosensitivity through MicroRNA 889-3p-Dependent Regulation of HOXB7. Mol. Cell. Biol. 2021, 41, e0063120. [Google Scholar] [CrossRef]
- Hickey, C.J.; Kim, J.H.; Ahn, E.Y. New discoveries of old SON: A link between RNA splicing and cancer. J. Cell. Biochem. 2014, 115, 224–231. [Google Scholar] [CrossRef]
- Sharma, A.; Takata, H.; Shibahara, K.; Bubulya, A.; Bubulya, P.A. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 2010, 21, 650–663. [Google Scholar] [CrossRef]
- Furukawa, T.; Tanji, E.; Kuboki, Y.; Hatori, T.; Yamamoto, M.; Shimizu, K.; Shibata, N.; Shiratori, K. Targeting of MAPK-associated molecules identifies SON as a prime target to attenuate the proliferation and tumorigenicity of pancreatic cancer cells. Mol. Cancer 2012, 11, 88. [Google Scholar] [CrossRef]
- Du, K.; Bai, X.; Yang, L.; Shi, Y.; Chen, L.; Wang, H.; Cai, M.; Wang, J.; Chen, S.; Jia, X.; et al. De Novo Reconstruction of Transcriptome Identified Long Non-Coding RNA Regulator of Aging-Related Brown Adipose Tissue Whitening in Rabbits. Biology 2021, 10, 1176. [Google Scholar] [CrossRef]
- Asano, H.; Kanamori, Y.; Higurashi, S.; Nara, T.; Kato, K.; Matsui, T.; Funaba, M. Induction of beige-like adipocytes in 3T3-L1 cells. J. Vet. Med. Sci. 2014, 76, 57–64. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Cannon, B.; Ching, T.M.; Lindberg, O. Oxidative metabolism in cells isolated from brown adipose tissue. 2. Catecholamine regulated respiratory control. Eur. J. Biochem. 1968, 7, 51–57. [Google Scholar] [CrossRef]
- Klingenberg, M. Uncoupling protein—A useful energy dissipator. J. Bioenerg. Biomembr. 1999, 31, 419–430. [Google Scholar] [CrossRef]
- Nedergaard, J.; Cannon, B.; Lindberg, O. Microcalorimetry of isolated mammalian cells. Nature 1977, 267, 518–520. [Google Scholar] [CrossRef]
- Lin, J.; Wu, P.H.; Tarr, P.T.; Lindenberg, K.S.; St-Pierre, J.; Zhang, C.Y.; Mootha, V.K.; Jager, S.; Vianna, C.R.; Reznick, R.M.; et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 2004, 119, 121–135. [Google Scholar] [CrossRef]
- Uldry, M.; Yang, W.; St-Pierre, J.; Lin, J.; Seale, P.; Spiegelman, B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006, 3, 333–341. [Google Scholar] [CrossRef]
- Liu, C.; Lin, J.D. PGC-1 coactivators in the control of energy metabolism. Acta Biochim. Biophys. Sin. 2011, 43, 248–257. [Google Scholar] [CrossRef]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Tiraby, C.; Tavernier, G.; Lefort, C.; Larrouy, D.; Bouillaud, F.; Ricquier, D.; Langin, D. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003, 278, 33370–33376. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.; Choi, C.; Song, C.; Im, H.; Cho, Y.K.; Son, J.S.; Joo, S.; Joh, Y.; Lee, Y.J.; Seong, J.K.; et al. Development of CIDEA reporter mouse model and its application for screening thermogenic drugs. Sci. Rep. 2021, 11, 18429. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.A.; Lin, H.; Yu, J.Y.; Zhang, H.L.; Zhang, J.F.; Wang, C.Q.; Gu, J. MiR-21-3p Inhibits Adipose Browning by Targeting FGFR1 and Aggravates Atrial Fibrosis in Diabetes. Oxid. Med. Cell. Longev. 2021, 2021, 9987219. [Google Scholar] [CrossRef] [PubMed]
- Vonhogen, I.G.C.; El Azzouzi, H.; Olieslagers, S.; Vasilevich, A.; de Boer, J.; Tinahones, F.J.; da Costa Martins, P.A.; de Windt, L.J.; Murri, M. MiR-337-3p Promotes Adipocyte Browning by Inhibiting TWIST1. Cells 2020, 9, 1056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, L.; Bai, M.; Liu, Y.; Zhan, Y.; Deng, T.; Yang, H.; Sun, W.; Wang, X.; Zhu, K.; et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer 2019, 144, 2501–2515. [Google Scholar] [CrossRef]
- Saha, P.K.; Hamilton, M.P.; Rajapakshe, K.; Putluri, V.; Felix, J.B.; Masschelin, P.; Cox, A.R.; Bajaj, M.; Putluri, N.; Coarfa, C.; et al. miR-30a targets gene networks that promote browning of human and mouse adipocytes. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E667–E677. [Google Scholar] [CrossRef]
- Ye, C.; Duan, J.; Zhang, X.; Yao, L.; Song, Y.; Wang, G.; Li, Q.; Wang, B.; Ai, D.; Wang, C.; et al. Cold-induced Yes-associated-protein expression through miR-429 mediates the browning of white adipose tissue. Sci. China Life Sci. 2021, 64, 404–418. [Google Scholar] [CrossRef]
- Tan, X.; Zhu, T.; Zhang, L.; Fu, L.; Hu, Y.; Li, H.; Li, C.; Zhang, J.; Liang, B.; Liu, J. miR-669a-5p promotes adipogenic differentiation and induces browning in preadipocytes. Adipocyte 2022, 11, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Trajkovski, M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism 2014, 63, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Ng, H.H.; Bubulya, P.A. The role of SON in splicing, development, and disease. Wiley Interdiscip. Rev. RNA 2014, 5, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Loth, S.; Kuhlen, M.; Ginzel, S.; Schaper, J.; Rosenbaum, T.; Pietsch, T.; Borkhardt, A.; Hoell, J.I. Mutated SON putatively causes a cancer syndrome comprising high-risk medulloblastoma combined with cafe-au-lait spots. Fam. Cancer 2019, 18, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Baddoo, M.C.; Park, E.Y.; Stone, J.K.; Park, H.; Butler, T.W.; Huang, G.; Yan, X.; Pauli-Behn, F.; Myers, R.M.; et al. SON and Its Alternatively Spliced Isoforms Control MLL Complex-Mediated H3K4me3 and Transcription of Leukemia-Associated Genes. Mol. Cell 2016, 61, 859–873. [Google Scholar] [CrossRef] [PubMed]
- Tokita, M.J.; Braxton, A.A.; Shao, Y.; Lewis, A.M.; Vincent, M.; Kury, S.; Besnard, T.; Isidor, B.; Latypova, X.; Bezieau, S.; et al. De Novo Truncating Variants in SON Cause Intellectual Disability, Congenital Malformations, and Failure to Thrive. Am. J. Hum. Genet. 2016, 99, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Shinde, D.N.; Reijnders, M.R.F.; Hauser, N.S.; Belmonte, R.L.; Wilson, G.R.; Bosch, D.G.M.; Bubulya, P.A.; Shashi, V.; Petrovski, S.; et al. De Novo Mutations in SON Disrupt RNA Splicing of Genes Essential for Brain Development and Metabolism, Causing an Intellectual-Disability Syndrome. Am. J. Hum. Genet. 2016, 99, 711–719. [Google Scholar] [CrossRef]
- Sun, C.T.; Lo, W.Y.; Wang, I.H.; Lo, Y.H.; Shiou, S.R.; Lai, C.K.; Ting, L.P. Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J. Biol. Chem. 2001, 276, 24059–24067. [Google Scholar] [CrossRef]
- Pepino, M.Y.; Kuda, O.; Samovski, D.; Abumrad, N.A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014, 34, 281–303. [Google Scholar] [CrossRef]
- Febbraio, M.; Abumrad, N.A.; Hajjar, D.P.; Sharma, K.; Cheng, W.; Pearce, S.F.; Silverstein, R.L. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 1999, 274, 19055–19062. [Google Scholar] [CrossRef]
- Luo, G.; Hu, S.; Lai, T.; Wang, J.; Wang, L.; Lai, S. MiR-9-5p promotes rabbit preadipocyte differentiation by suppressing leptin gene expression. Lipids Health Dis. 2020, 19, 126. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Primer Sequence (5′-3′) |
|---|---|
| ACTB | GTGCTTCTAGGCGGACTGTT |
| CGGCCACATTGCAGAACTTT | |
| UCP1 | CCGGGACAATATGCGAGTGT |
| CACAGTCCACAGTCTGCCTT | |
| PRDM16 | CCCCGGAAGAACCACGTCTA |
| TCTCCACCCTGCTGTAGATGG | |
| C/EBPα | CAAGAACAGCAACGAGTACCG |
| GTCACTGGTCAACTCCAGCAC | |
| CIDEA | TAGGGGACAACACGCACTTC |
| CTCTGGCGATTCCCGATCTC | |
| PPARα | ACAGGATTGATGCTGCCGAA |
| AAACGGATGTGCTGCCCTAA | |
| PGC1α | AAAAGCTTGACTGGCGTCAC |
| ACTGCACCACTTGAGTCCAC | |
| TMEM26 | TCTATGCCATTCTAATTTTATGGACTTGG |
| GATGAAGACGCTGATTCCAATGTTCC | |
| PPARg | GAGGACATCCAGGACAACC |
| GTCCGTCTCCGTCTTCTTT | |
| SON | ACAGCAGTAGTGCCGAAGTC |
| TGGCTCTAGGGCTTTTGCTC | |
| miR-889-3p | TTAATATCGGACAACCATTGT |
| GGAACGATACAGAGAAGATTAGC | |
| U6 | GGAACGATACAGAGAAGATTAGC |
| TGGAACGCTTCACGAATTTGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Zhang, X.; Bai, X.; Du, K.; Chen, L.; Wang, H.; Jia, X.; Lai, S. miR-889-3p Facilitates the Browning Process of White Adipocyte Precursors by Targeting the SON Gene. Int. J. Mol. Sci. 2023, 24, 17580. https://doi.org/10.3390/ijms242417580
Sun W, Zhang X, Bai X, Du K, Chen L, Wang H, Jia X, Lai S. miR-889-3p Facilitates the Browning Process of White Adipocyte Precursors by Targeting the SON Gene. International Journal of Molecular Sciences. 2023; 24(24):17580. https://doi.org/10.3390/ijms242417580
Chicago/Turabian StyleSun, Wenqiang, Xiaoxiao Zhang, Xue Bai, Kun Du, Li Chen, Haoding Wang, Xianbo Jia, and Songjia Lai. 2023. "miR-889-3p Facilitates the Browning Process of White Adipocyte Precursors by Targeting the SON Gene" International Journal of Molecular Sciences 24, no. 24: 17580. https://doi.org/10.3390/ijms242417580
APA StyleSun, W., Zhang, X., Bai, X., Du, K., Chen, L., Wang, H., Jia, X., & Lai, S. (2023). miR-889-3p Facilitates the Browning Process of White Adipocyte Precursors by Targeting the SON Gene. International Journal of Molecular Sciences, 24(24), 17580. https://doi.org/10.3390/ijms242417580





