Impact of Conventional and Potential New Metal-Based Drugs on Lipid Metabolism in Osteosarcoma MG-63 Cells
Abstract
:1. Introduction
2. Results and Discussion
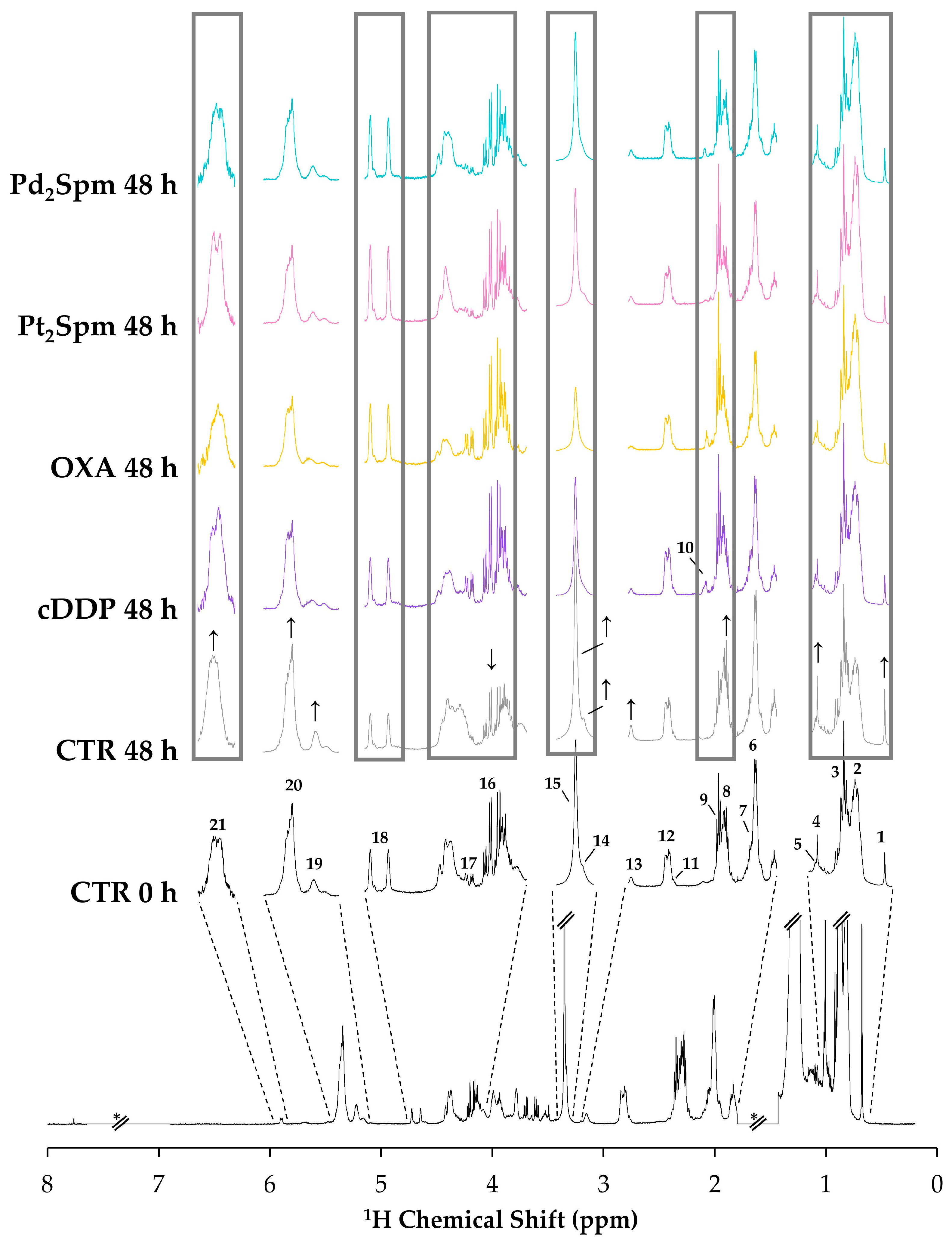
2.1. NMR Spectra of Lipidic Extracts of OS Cells Exposed to the Different Pt(II)/Pd(II) Drugs
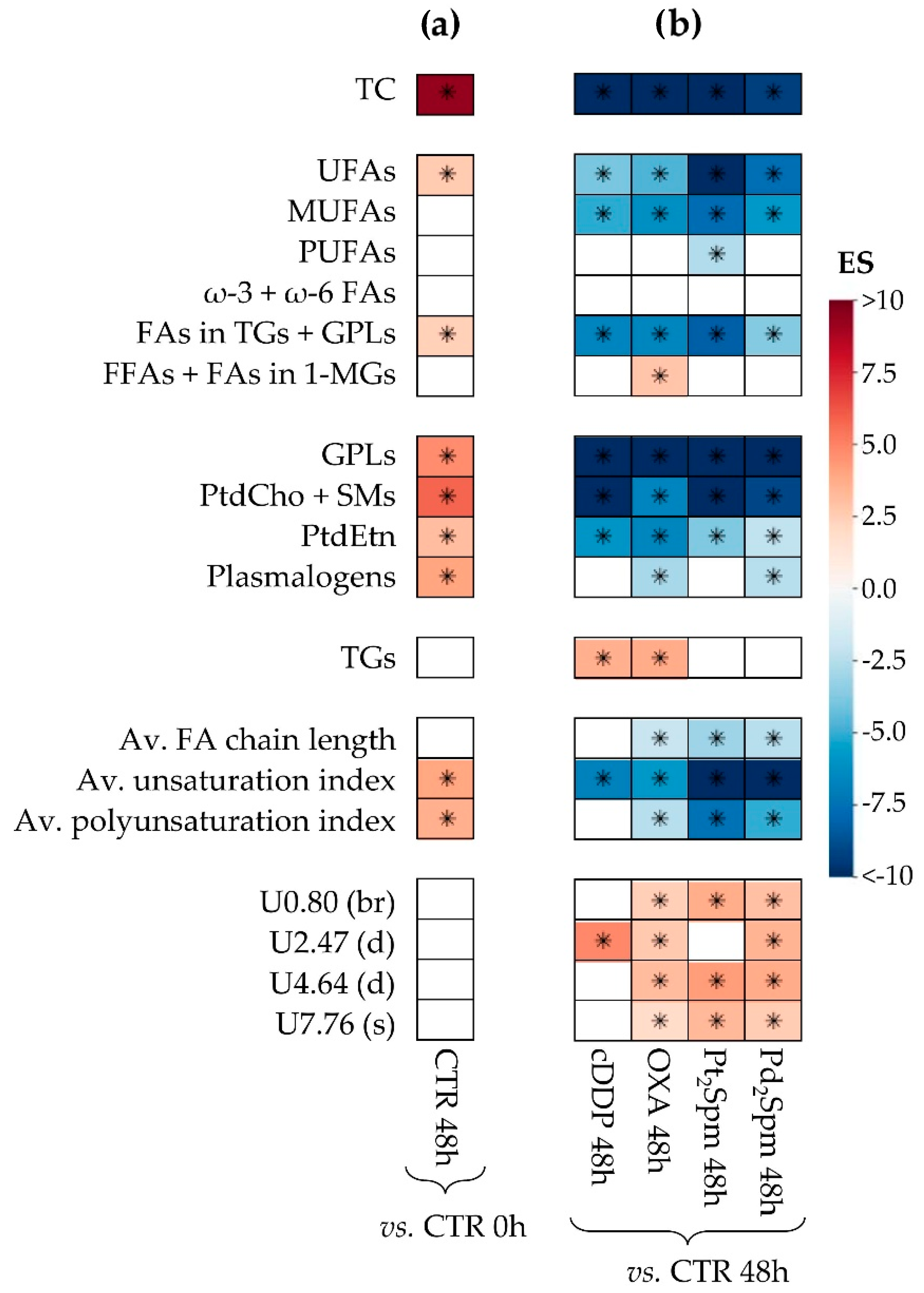
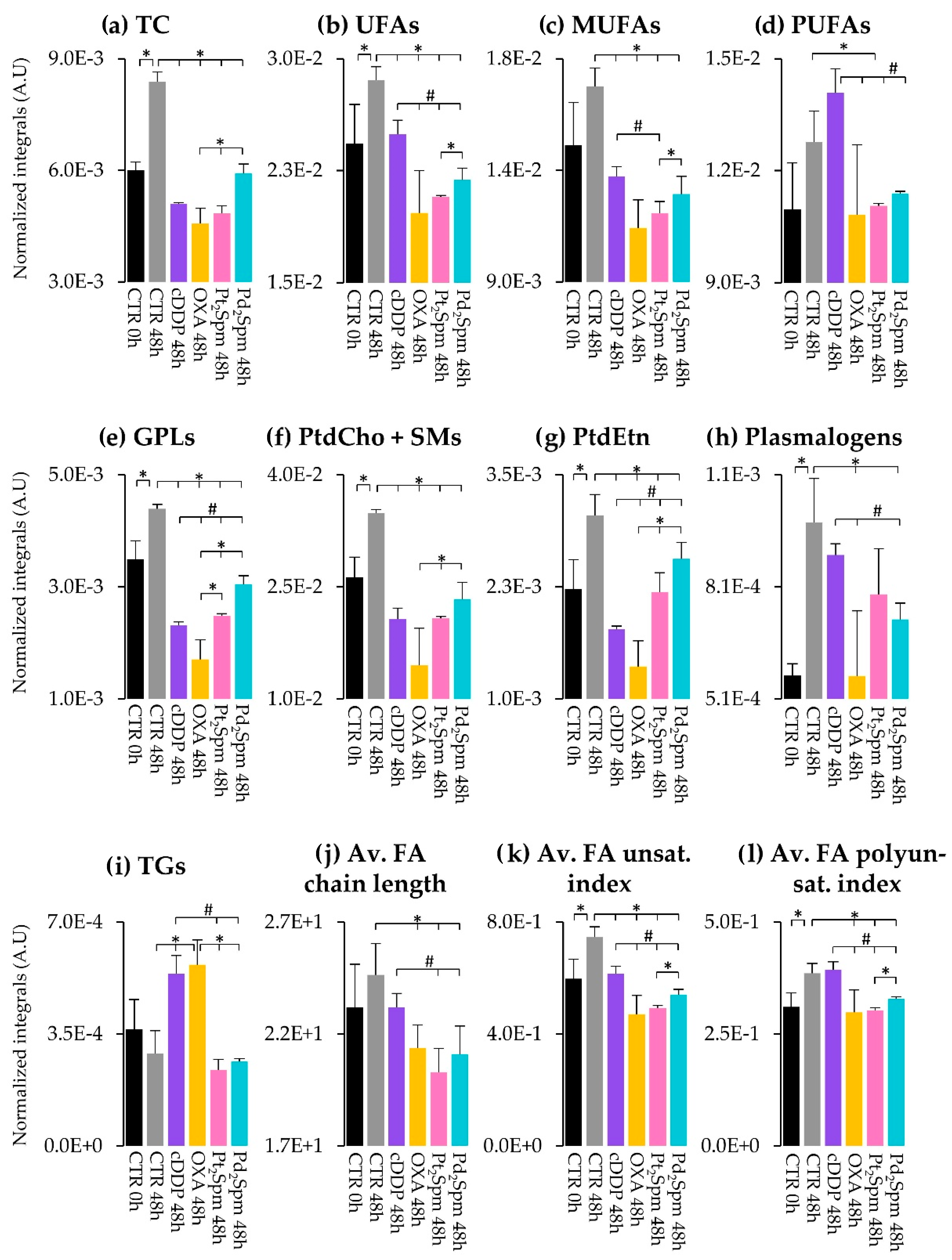
2.2. Multivariate and Univariate Statistical Analysis of NMR Data
2.2.1. Multivariate Models of Lipidic Extracts of MG-63 Cells after 48 h of Exposure Time to Pt(II)/Pd(II) Drugs
2.2.2. Lipidic Changes in Untreated Proliferating MG-63 Cells (0–48 h Period)
2.2.3. Lipidic Changes in MG-63 Cells Treated with Pt(II) Complexes cDDP, OXA and Pt2Spm (Same Metal, Distinct Ligands)
2.2.4. Lipidic Changes in MG-63 Cells Treated with Pt2Spm and Pd2Spm (Same Ligand, Distinct Metal Centers)
3. Materials and Methods
3.1. Chemicals and Solutions
3.2. Cell Culture
3.3. Sample Preparation
3.4. Cell Extraction
3.5. NMR Spectroscopy
3.6. Data Processing and Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taran, S.J.; Taran, R.; Malipatil, N.B. Pediatric Osteosarcoma: An Updated Review. Indian J. Med. Paediatr. Oncol. 2017, 38, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Isakoff, M.S.; Bielack, S.S.; Meltzer, P.; Gorlick, R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J. Clin. Oncol. 2015, 33, 3029–3035. [Google Scholar] [CrossRef] [PubMed]
- Tsang, R.Y.; Al-Fayea, T.; Au, H.-J. Cisplatin Overdose: Toxicities and Management. Drug Saf. 2009, 32, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Janeway, K.A.; Grier, H.E. Sequelae of Osteosarcoma Medical Therapy: A Review of Rare Acute Toxicities and Late Effects. Lancet Oncol. 2010, 11, 670–678. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Zhao, N.; Wang, C.; Kamar, S.; Zhou, Y.; He, Z.; Yang, J.; Sun, B.; Shi, X.; et al. Progress in the Chemotherapeutic Treatment of Osteosarcoma. Oncol. Lett. 2018, 16, 6228–6237. [Google Scholar] [CrossRef]
- Rosenberg, B.; Van Camp, L.; Krigas, T. Inhibition of Cell Division in Escherichia Coli by Electrolysis Products from a Platinum Electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Wiltshaw, E. Cisplatin in the Treatment of Cancer: The First Metal Antitumor Drug. Platin. Met. Rev. 1979, 23, 90–98. [Google Scholar]
- Kelland, L. The Resurgence of Platinum-Based Cancer Chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in Cancer Therapy: Molecular Mechanisms of Action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular Mechanisms of Cisplatin Resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The Side Effects of Platinum-Based Chemotherapy Drugs: A Review for Chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Lippard, S.J. Direct Cellular Responses to Platinum-Induced DNA Damage. Chem. Rev. 2007, 107, 1387–1407. [Google Scholar] [CrossRef] [PubMed]
- Mauri, G.; Gori, V.; Bonazzina, E.; Amatu, A.; Tosi, F.; Bencardino, K.; Ruggieri, L.; Patelli, G.; Arena, S.; Bardelli, A.; et al. Oxaliplatin Retreatment in Metastatic Colorectal Cancer: Systematic Review and Future Research Opportunities. Cancer Treat. Rev. 2020, 91, 102112. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.P.M. Platinum and Palladium Polyamine Complexes as Anticancer Agents: The Structural Factor. Int. Sch. Res. Not. 2013, 2013, e287353. [Google Scholar] [CrossRef]
- Vojtek, M.; Marques, M.P.M.; Ferreira, I.M.P.L.V.O.; Mota-Filipe, H.; Diniz, C. Anticancer Activity of Palladium-Based Complexes against Triple-Negative Breast Cancer. Drug Discov. Today 2019, 24, 1044–1058. [Google Scholar] [CrossRef] [PubMed]
- Lamego, I.; Marques, M.P.M.; Duarte, I.F.; Martins, A.S.; Oliveira, H.; Gil, A.M. Impact of the Pd2Spermine Chelate on Osteosarcoma Metabolism: An NMR Metabolomics Study. J. Proteome Res. 2017, 16, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.S.; Batista de Carvalho, A.L.M.; Lamego, I.; Marques, M.; Paula, M.; Gil, A.M. Cytotoxicity of Platinum and Palladium Chelates against Osteosarcoma. ChemistrySelect 2020, 5, 5993–6000. [Google Scholar] [CrossRef]
- Martins, A.S.; Batista de Carvalho, A.L.M.; Marques, M.P.M.; Gil, A.M. Response of Osteosarcoma Cell Metabolism to Platinum and Palladium Chelates as Potential New Drugs. Molecules 2021, 26, 4805. [Google Scholar] [CrossRef]
- Huang, Z.; Tong, Y.; Wang, J.; Huang, Y. NMR Studies of the Relationship between the Changes of Membrane Lipids and the Cisplatin-Resistance of A549/DDP Cells. Cancer Cell Int. 2003, 3, 5. [Google Scholar] [CrossRef]
- Lee, S.; Kang, S.; Park, S. Comparison of Metabolic Profiles of Normal and Cancer Cells in Response to Cytotoxic Agents. J. Korean Magn. Reson. Soc. 2017, 21, 31–43. [Google Scholar] [CrossRef]
- Vermathen, M.; von Tengg-Kobligk, H.; Hungerbühler, M.N.; Vermathen, P.; Ruprecht, N. 1H HR-MAS NMR Based Metabolic Profiling of Lung Cancer Cells with Induced and De-Induced Cisplatin Resistance to Reveal Metabolic Resistance Adaptations. Mol. Basel Switz. 2021, 26, 6766. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, M.; Spenger, C.; Jarvet, J.; Gräslund, A.; Kogner, P. Predicting Resistance or Response to Chemotherapy by Proton Magnetic Resonance Spectroscopy in Neuroblastoma. J. Natl. Cancer Inst. 2004, 96, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wilson, M.; Mirbahai, L.; McConville, C.; Arvanitis, T.N.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. In Vitro Metabonomic Study Detects Increases in UDP-GlcNAc and UDP-GalNAc, as Early Phase Markers of Cisplatin Treatment Response in Brain Tumor Cells. J. Proteome Res. 2011, 10, 3493–3500. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wilson, M.; McConville, C.; Arvanitis, T.N.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. Increased Unsaturation of Lipids in Cytoplasmic Lipid Droplets in DAOY Cancer Cells in Response to Cisplatin Treatment. Metabolomics 2013, 9, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colnago, L.A. Impact of Chemotherapy on Metabolic Reprogramming: Characterization of the Metabolic Profile of Breast Cancer MDA-MB-231 Cells Using 1H HR-MAS NMR Spectroscopy. J. Pharm. Biomed. Anal. 2017, 146, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colnago, L.A. Effects of Doxorubicin, Cisplatin, and Tamoxifen on the Metabolic Profile of Human Breast Cancer MCF-7 Cells As Determined by 1H High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance. Biochemistry 2017, 56, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Del Coco, L.; Majellaro, M.; Boccarelli, A.; Cellamare, S.; Altomare, C.D.; Fanizzi, F.P. Novel Antiproliferative Biphenyl Nicotinamide: NMR Metabolomic Study of Its Effect on the MCF-7 Cell in Comparison with Cisplatin and Vinblastine. Molecules 2020, 25, 3502. [Google Scholar] [CrossRef]
- Resendiz-Acevedo, K.; García-Aguilera, M.E.; Esturau-Escofet, N.; Ruiz-Azuara, L. 1H -NMR Metabolomics Study of the Effect of Cisplatin and Casiopeina IIgly on MDA-MB-231 Breast Tumor Cells. Front. Mol. Biosci. 2021, 8, 742859. [Google Scholar] [CrossRef]
- De Castro, F.; Benedetti, M.; Antonaci, G.; Del Coco, L.; De Pascali, S.A.; Muscella, A.; Marsigliante, S.; Fanizzi, F.P. Response of Cisplatin Resistant Skov-3 Cells to [Pt(O,O′-Acac)(γ-Acac)(DMS)] Treatment Revealed by a Metabolomic 1H-NMR Study. Molecules 2018, 23, 2301. [Google Scholar] [CrossRef]
- Duarte, I.F.; Lamego, I.; Marques, J.; Marques, M.P.M.; Blaise, B.J.; Gil, A.M. Nuclear Magnetic Resonance (NMR) Study of the Effect of Cisplatin on the Metabolic Profile of MG-63 Osteosarcoma Cells. J. Proteome Res. 2010, 9, 5877–5886. [Google Scholar] [CrossRef]
- Lamego, I.; Duarte, I.F.; Marques, M.P.M.; Gil, A.M. Metabolic Markers of MG-63 Osteosarcoma Cell Line Response to Doxorubicin and Methotrexate Treatment: Comparison to Cisplatin. J. Proteome Res. 2014, 13, 6033–6045. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Dong, J.; Wei, Z.; Cheng, K.-K.; Li, J.; You, S.; Liu, Y.; Wang, X.; Chen, Z. 1H NMR-Based Metabolic Profiles Delineate the Anticancer Effect of Vitamin C and Oxaliplatin on Hepatocellular Carcinoma Cells. J. Proteome Res. 2020, 19, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by Numbers: Acquiring and Understanding Global Metabolite Data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Wallingford CT Gaussian 16, Revision C.01 2016; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Santini, M.T.; Romano, R.; Rainaldi, G.; Indovina, P.; Ferrante, A.; Motta, A.; Indovina, P.L. Temporal Dynamics of 1H-NMR-Visible Metabolites during Radiation-Induced Apoptosis in MG-63 Human Osteosarcoma Spheroids. Radiat. Res. 2006, 166, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Santini, M.T.; Romano, R.; Rainaldi, G.; Ferrante, A.; Indovina, P.; Motta, A.; Indovina, P.L. 1H-NMR Evidence for a Different Response to the Same Dose (2 Gy) of Ionizing Radiation of MG-63 Human Osteosarcoma Cells and Three-Dimensional Spheroids. Anticancer Res. 2006, 26, 267–281. [Google Scholar] [PubMed]
- Santini, M.T.; Romano, R.; Rainaldi, G.; Ferrante, A.; Motta, A.; Indovina, P.L. Increases in 1H-NMR Mobile Lipids Are Not Always Associated with Overt Apoptosis: Evidence from MG-63 Human Osteosarcoma Three-Dimensional Spheroids Exposed to a Low Dose (2 Gy) of Ionizing Radiation. Radiat. Res. 2006, 165, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Hac-Wydro, K.; Wydro, P. The Influence of Fatty Acids on Model Cholesterol/Phospholipid Membranes. Chem. Phys. Lipids 2007, 150, 66–81. [Google Scholar] [CrossRef]
- Szlasa, W.; Zendran-Zahorska, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid Composition of the Cancer Cell Membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef]
- Mayengbam, S.S.; Singh, A.; Pillai, A.D.; Bhat, M.K. Influence of Cholesterol on Cancer Progression and Therapy. Transl. Oncol. 2021, 14, 101043. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.M.; Engberg, S. Effect Size Estimation: Methods and Examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef]
- Mosconi, E.; Fontanella, M.; Sima, D.M.; Van Huffel, S.; Fiorini, S.; Sbarbati, A.; Marzola, P. Investigation of Adipose Tissues in Zucker Rats Using in Vivo and Ex Vivo Magnetic Resonance Spectroscopy. J. Lipid Res. 2011, 52, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Borel, M.; Degoul, F.; Communal, Y.; Mounetou, E.; Bouchon, B.; C-Gaudreault, R.; Madelmont, J.C.; Miot-Noirault, E. N-(4-Iodophenyl)-N′-(2-Chloroethyl)Urea as a Microtubule Disrupter: In Vitro and in Vivo Profiling of Antitumoral Activity on CT-26 Murine Colon Carcinoma Cell Line Cultured and Grafted to Mice. Br. J. Cancer 2007, 96, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Codina, G.; Caubet, A.; López, C.; Moreno, V.; Molins, E. Palladium(II) and Platinum(II) Polyamine Complexes: X-Ray Crystal Structures of (SP-4-2)-ChloroN-[(3-Amino-κN)Propyl]Propane-1,3-Diamine-κN,κN′palladium(1+) Tetrachloropalladate (2–) (2 : 1) and (R,S)-Tetrachloro[μ-(Spermine)]Dipalladium(II) (=μ-N,N′-Bis[(3-Amino-κN)Propyl]Butane-1,4-Diamine-κN:κN′tetrachlorodipalladium). Helv. Chim. Acta 1999, 82, 1025–1037. [Google Scholar] [CrossRef]
- Fiuza, S.M.; Amado, A.M.; Parker, S.F.; Marques, M.P.M.; Batista de Carvalho, L.A.E. Conformational Insights and Vibrational Study of a Promising Anticancer Agent: The Role of the Ligand in Pd(II)–Amine Complexes. New J. Chem. 2015, 39, 6274–6283. [Google Scholar] [CrossRef]
- Dettmer, K.; Nürnberger, N.; Kaspar, H.; Gruber, M.A.; Almstetter, M.F.; Oefner, P.J. Metabolite Extraction from Adherently Growing Mammalian Cells for Metabolomics Studies: Optimization of Harvesting and Extraction Protocols. Anal. Bioanal. Chem. 2011, 399, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR Analysis of Intra- and Extracellular Metabolism of Mammalian Cells: A Tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Oostendorp, M.; Engelke, U.F.; Willemsen, M.A.; Wevers, R.A. Diagnosing Inborn Errors of Lipid Metabolism with Proton Nuclear Magnetic Resonance Spectroscopy. Clin. Chem. 2006, 52, 1395–1405. [Google Scholar] [CrossRef]
- Srivastava, N.K. Proton Nuclear Magnetic Resonance (1H NMR) Spectroscopy-Based Analysis of Lipid Components in Serum/Plasma of Patients with Duchenne Muscular Dystrophy (DMD). In Duchenne Muscular Dystrophy: Methods and Protocols; Bernardini, C., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 195–204. ISBN 978-1-4939-7374-3. [Google Scholar]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High Resolution NMR Spectroscopy as a Structural and Analytical Tool for Unsaturated Lipids in Solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef]
- Subramanian, A.; Shankar Joshi, B.; Roy, A.D.; Roy, R.; Gupta, V.; Dang, R.S. NMR Spectroscopic Identification of Cholesterol Esters, Plasmalogen and Phenolic Glycolipids as Fingerprint Markers of Human Intracranial Tuberculomas. NMR Biomed. 2008, 21, 272–288. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Veselkov, K.A.; Lindon, J.C.; Ebbels, T.M.D.; Crockford, D.; Volynkin, V.V.; Holmes, E.; Davies, D.B.; Nicholson, J.K. Recursive Segment-Wise Peak Alignment of Biological 1H NMR Spectra for Improved Metabolic Biomarker Recovery. Anal. Chem. 2009, 81, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in Metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Bridge, P.D.; Sawilowsky, S.S. Increasing Physicians’ Awareness of the Impact of Statistics on Research Outcomes: Comparative Power of the t-Test and Wilcoxon Rank-Sum Test in Small Samples Applied Research. J. Clin. Epidemiol. 1999, 52, 229–235. [Google Scholar] [CrossRef] [PubMed]


| Compound | cDDP 48 h | OXA 48 h | Pt2Spm 48 h | Pd2Spm 48 h | ||||
|---|---|---|---|---|---|---|---|---|
| ES (Error (%)) | p-Value | ES (Error (%)) | p-Value | ES (Error (%)) | p-Value | ES (Error (%)) | p-Value | |
| TC | −13.3 (50.4) | <0.05 | −11.5 (45.2) | <0.05 | −14.4 (44.7) | <0.05 | −9.3 (48.5) | <0.05 |
| UFAs | −4.0 (63.5) | <0.05 | −4.7 (51.4) | <0.05 | −10.8 (45.4) | <0.05 | −7.6 (49.6) | <0.05 |
| MUFAs | −5.1 (58.2) | <0.05 | −6.2 (48.3) | <0.05 | −7.7 (46.8) | <0.05 | −5.8 (52.0) | <0.05 |
| PUFAs | - | - | - | - | −2.6 (65.7) | <0.05 | - | - |
| FAs in TGs + GPLs | −6.7 (54.5) | <0.05 | −6.6 (47.9) | <0.05 | −8.2 (46.5) | <0.05 | −3.7 (59.4) | <0.05 |
| FFAs + FAs in 1-MGs | - | - | 2.9 (62.3) | <0.05 | - | - | - | - |
| GPLs | −27.3 (49.4) | <0.05 | −12 (45.1) | <0.05 | −29.1 (44.0) | <0.05 | −12.9 (47.4) | <0.05 |
| PtdCho + SMs | −20.4 (49.6) | <0.05 | −6.6 (47.8) | <0.05 | −37.5 (44.0) | <0.05 | −9 (48.7) | <0.05 |
| PtdEtn | −5.9 (55.9) | <0.05 | −6.6 (47.8) | <0.05 | −3.8 (55.2) | <0.05 | −2.2 (78.0) | <0.05 |
| Pls | - | - | −2.9 (62.1) | <0.05 | - | - | −2.5 (72.2) | <0.05 |
| TGs | 3.6 (66.1) | <0.05 | 3.8 (55.2) | <0.05 | - | - | - | - |
| Av. FA chain length | - | - | −2.1 (74.7) | <0.05 | −3.1 (60.3) | <0.05 | −2.5 (72.5) | <0.05 |
| Av. FA unsat. index | −6.8 (54.3) | <0.05 | −5.8 (48.9) | <0.05 | −20.7 (44.2) | <0.05 | −13.9 (47.3) | <0.05 |
| Av. FA polyunsat. index | - | - | −2.5 (67.5) | <0.05 | −7.4 (47.0) | <0.05 | −5 (53.8) | <0.05 |
| Uδ0.80 (br) | - | - | 2.4 (68.0) | <0.05 | 3.7 (55.5) | <0.05 | 3 (65.0) | <0.05 |
| Uδ2.47 (d) | 4.9 (58.9) | <0.05 | 2.7 (64.1) | <0.05 | - | - | 3.4 (61.3) | <0.05 |
| Uδ4.64 (d) | - | - | 3.2 (59.2) | <0.05 | 4.3 (52.9) | <0.05 | 3.8 (58.9) | <0.05 |
| Uδ7.76 (s) | - | - | 1.9 (79.9) | <0.05 | 3.3 (58.3) | <0.05 | 2.6 (71.1) | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bispo, D.S.C.; Correia, M.; Carneiro, T.J.; Martins, A.S.; Reis, A.A.N.; Carvalho, A.L.M.B.d.; Marques, M.P.M.; Gil, A.M. Impact of Conventional and Potential New Metal-Based Drugs on Lipid Metabolism in Osteosarcoma MG-63 Cells. Int. J. Mol. Sci. 2023, 24, 17556. https://doi.org/10.3390/ijms242417556
Bispo DSC, Correia M, Carneiro TJ, Martins AS, Reis AAN, Carvalho ALMBd, Marques MPM, Gil AM. Impact of Conventional and Potential New Metal-Based Drugs on Lipid Metabolism in Osteosarcoma MG-63 Cells. International Journal of Molecular Sciences. 2023; 24(24):17556. https://doi.org/10.3390/ijms242417556
Chicago/Turabian StyleBispo, Daniela S. C., Marlene Correia, Tatiana J. Carneiro, Ana S. Martins, Aliana A. N. Reis, Ana L. M. Batista de Carvalho, Maria P. M. Marques, and Ana M. Gil. 2023. "Impact of Conventional and Potential New Metal-Based Drugs on Lipid Metabolism in Osteosarcoma MG-63 Cells" International Journal of Molecular Sciences 24, no. 24: 17556. https://doi.org/10.3390/ijms242417556
APA StyleBispo, D. S. C., Correia, M., Carneiro, T. J., Martins, A. S., Reis, A. A. N., Carvalho, A. L. M. B. d., Marques, M. P. M., & Gil, A. M. (2023). Impact of Conventional and Potential New Metal-Based Drugs on Lipid Metabolism in Osteosarcoma MG-63 Cells. International Journal of Molecular Sciences, 24(24), 17556. https://doi.org/10.3390/ijms242417556










