Synthesis, Structural Characterization, Cytotoxicity, and Protein/DNA Binding Properties of Pyridoxylidene-Aminoguanidine-Metal (Fe, Co, Zn, Cu) Complexes
Abstract
1. Introduction
2. Results
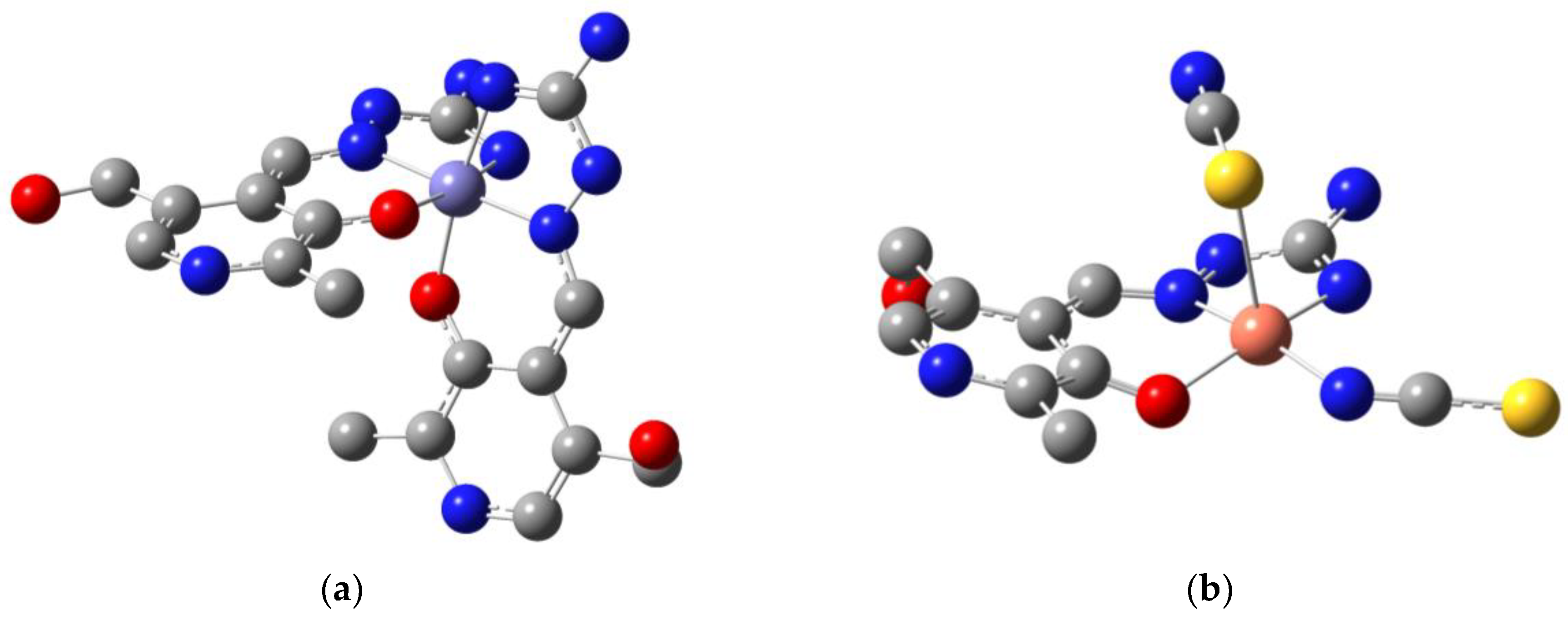
2.1. Crystalographic Structure Analysis
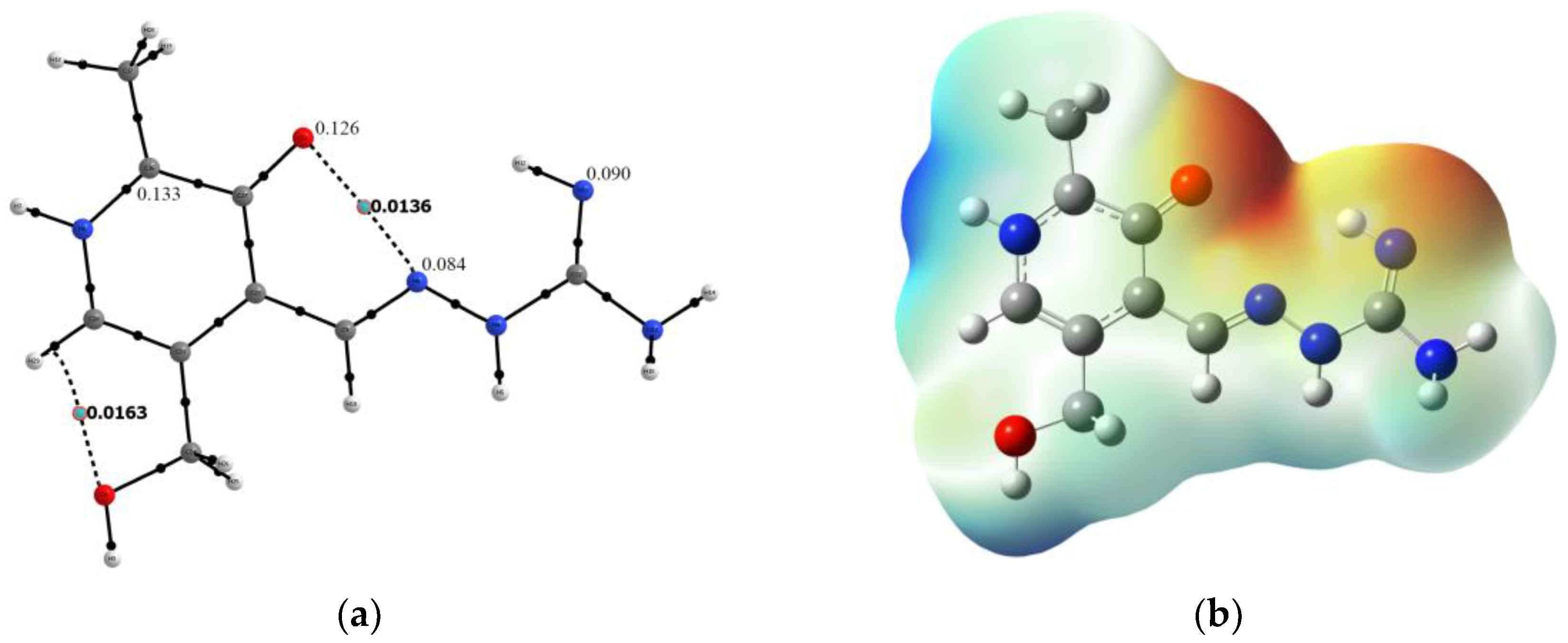
2.2. Hirshfeld Surface Analysis
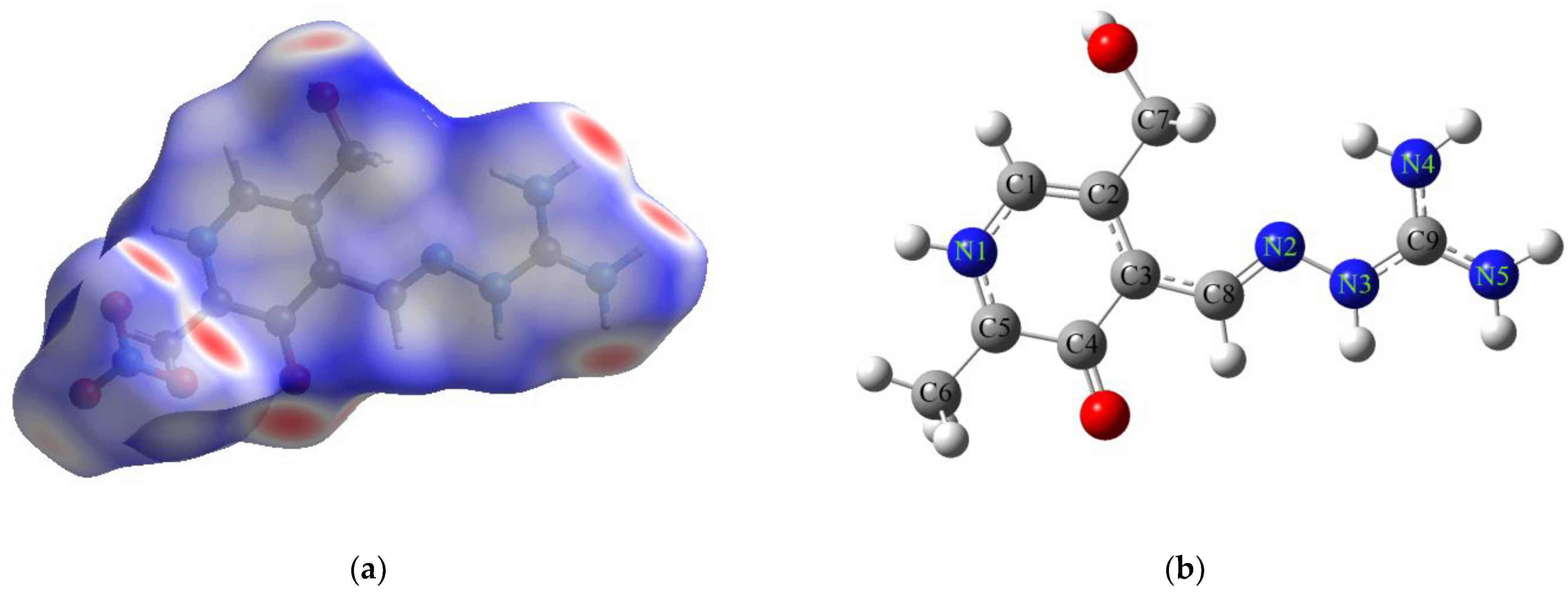
2.3. Theoretical Structural and Reactivity Analysis of PLAG
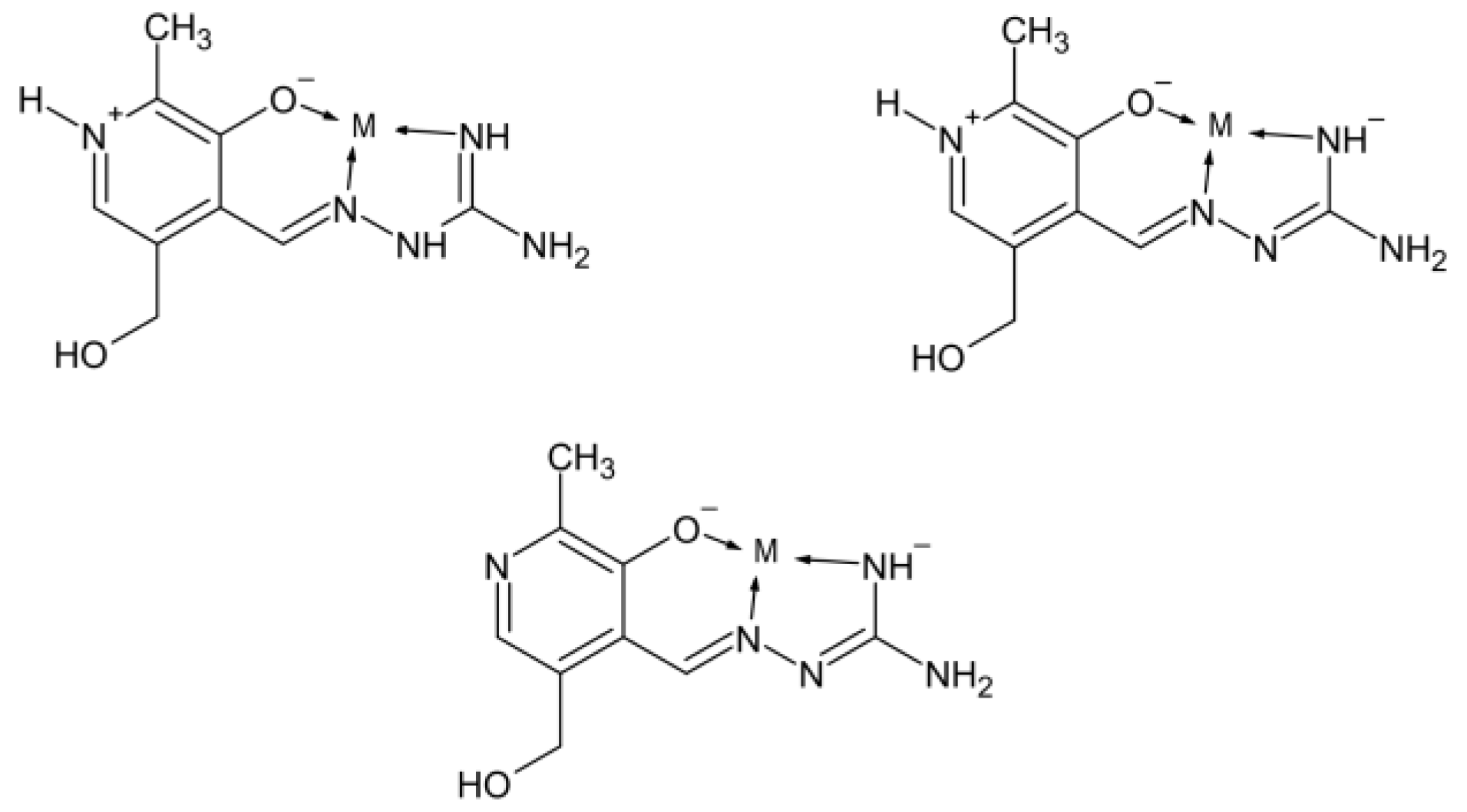
2.4. Theoretical Analysis of Complexes
2.5. Electronic Spectra and Stability of Complexes
2.6. BSA Binding Affinity Analysis
2.7. HSA Binding Affinity Analysis
2.8. DNA Binding Affinity
2.9. Cytotoxicity Analysis
3. Materials and Methods
3.1. Chemicals
3.2. Elemental Analysis
3.3. Synthesis of Pyridoxylidene-Aminoguanidine, PLAG
3.4. Synthesis of [Zn(PLAG)(SO4)(H2O)]H2O Complex
3.5. Synthesis of [Co(PLAG)2]SO4∙2H2O Complex
3.6. Synthesis of [Fe(PLAG)2]SO4∙2H2O Complex
3.7. Synthesis of [Cu(PLAG)(NCS)2]2 Complex
3.8. X-ray Analysis
3.9. Hirshfeld Analysis
3.10. Theoretical Analysis
3.11. Stability Studies
3.12. Spectrofluorimetric Measurements
3.13. Molecular Docking
3.14. Cells
3.15. Viability Assays (MTT and CV)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalt. Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef]
- Yu, C.; Wang, Z.; Sun, Z.; Zhang, L.; Zhang, W.; Xu, Y.; Zhang, J.-J. Platinum-Based Combination Therapy: Molecular Rationale, Current Clinical Uses, and Future Perspectives. J. Med. Chem. 2020, 63, 13397–13412. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Al-Saif, F.A.; Refat, M.S. Ten metal complexes of vitamin B3/niacin: Spectroscopic, thermal, antibacterial, antifungal, cytotoxicity and antitumor studies of Mn(II), Fe(III), Co(II), Ni(II), Cu(II), Zn(II), Pd(II), Cd(II), Pt(IV) and Au(III) complexes. J. Mol. Struct. 2012, 1021, 40–52. [Google Scholar] [CrossRef]
- Mahmoud, N.H.; Elsayed, G.H.; Aboelnaga, A.; Fahim, A.M. Spectroscopic studies, DFT calculations, cytotoxicity activity, and docking stimulation of novel metal complexes of Schiff base ligand of isonicotinohydrazide derivative. Appl. Organomet. Chem. 2022, 36, e6697. [Google Scholar] [CrossRef]
- Telpoukhovskaia, M.A.; Rodríguez-Rodríguez, C.; Scott, L.E.; Page, B.D.G.; Patrick, B.O.; Orvig, C. Synthesis, characterization, and cytotoxicity studies of Cu(II), Zn(II), and Fe(III) complexes of N-derivatized 3-hydroxy-4-pyridiones. J. Inorg. Biochem. 2014, 132, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Franz, K.J.; Metzler-Nolte, N. Introduction: Metals in Medicine. Chem. Rev. 2019, 119, 727–729. [Google Scholar] [CrossRef]
- Eichhorn, T.; Kolbe, F.; Mišić, S.; Dimić, D.; Morgan, I.; Saoud, M.; Milenković, D.; Marković, Z.; Rüffer, T.; Dimitrić Marković, J.; et al. Synthesis, Crystallographic Structure, Theoretical Analysis, Molecular Docking Studies, and Biological Activity Evaluation of Binuclear Ru(II)-1-Naphthylhydrazine Complex. Int. J. Mol. Sci. 2023, 24, 689. [Google Scholar] [CrossRef]
- Jamieson, E.R.; Lippard, S.J. Structure, Recognition, and Processing of Cisplatin−DNA Adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef]
- Todd, R.C.; Lippard, S.J. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009, 1, 280. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, C.M.; Silva, M.M.; Reis, F.S.; Ruiz, A.L.T.G.; de Carvalho, J.E.; Santos, J.C.C.; Figueiredo, I.M.; Alves, R.B.; Modolo, L.V.; de Fátima, Â. Studies on free radical scavenging, cancer cell antiproliferation, and calf thymus DNA interaction of Schiff bases. J. Photochem. Photobiol. B Biol. 2017, 172, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Bharti, S.K.; Nath, G.; Tilak, R.; Singh, S.K. Synthesis, anti-bacterial and anti-fungal activities of some novel Schiff bases containing 2,4-disubstituted thiazole ring. Eur. J. Med. Chem. 2010, 45, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Ronad, P.M.; Noolvi, M.N.; Sapkal, S.; Dharbhamulla, S.; Maddi, V.S. Synthesis and antimicrobial activity of 7-(2-substituted phenylthiazolidinyl)-benzopyran-2-one derivatives. Eur. J. Med. Chem. 2010, 45, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Lin, Z.L.; Huang, J.L. A novel kind of antitumour drugs using sulfonamide as parent compound. Eur. J. Med. Chem. 2001, 36, 863–872. [Google Scholar] [CrossRef]
- Edelstein, D.; Brownlee, M. Mechanistic Studies of Advanced Glycosylation End Product Inhibition by Aminoguanidine. Diabetes 1992, 41, 26–29. [Google Scholar] [CrossRef]
- Giardino, I.; Fard, A.K.; Hatchell, D.L.; Brownlee, M. Aminoguanidine inhibits reactive oxygen species formation, lipid peroxidation, and oxidant-induced apoptosis. Diabetes 1998, 47, 1114–1120. [Google Scholar] [CrossRef]
- Ihm, S.-H.; Yoo, H.J.; Park, S.W.; Ihm, J. Effect of aminoguanidine on lipid peroxidation in streptozotocin-induced diabetic rats. Metabolism 1999, 48, 1141–1145. [Google Scholar] [CrossRef]
- García-Díez, G.; Ramis, R.; Mora-Diez, N. Theoretical Study of the Copper Complexes with Aminoguanidine: Investigating Secondary Antioxidant Activity. ACS Omega 2020, 5, 14502–14512. [Google Scholar] [CrossRef]
- John, R.A. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1995, 1248, 81–96. [Google Scholar] [CrossRef]
- Casas, J.S.; Castiñeiras, A.; Condori, F.; Couce, M.D.; Russo, U.; Sánchez, A.; Seoane, R.; Sordo, J.; Varela, J.M. Diorganotin(IV)-promoted deamination of amino acids by pyridoxal: SnR22+ complexes of pyridoxal 5′-phosphate and of the Schiff base pyridoxal-pyridoxamine (PLPM), and antibacterial activities of PLPM and [SnR2(PLPM-2H)] (R = Me, Et, Bu, Ph). Polyhedron 2003, 22, 53–65. [Google Scholar] [CrossRef]
- Taguchi, T.; Sugiura, M.; Hamada, Y.; Miwa, I. Inhibition of advanced protein glycation by a Schiff base between aminoguanidine and pyridoxal. Eur. J. Pharmacol. 1999, 378, 283–289. [Google Scholar] [CrossRef]
- Taguchi, T.; Sugiura, M.; Hamada, Y.; Miwa, I. In Vivo Formation of a Schiff Base of Aminoguanidine with Pyridoxal Phosphate. Biochem. Pharmacol. 1998, 55, 1667–1671. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Taguchi, T.; Sugiura, M.; Takeuchi, M.; Yanagisawa, K.; Watanabe, Y.; Miwa, I.; Makita, Z.; Koike, T. Aminoguanidine Pyridoxal Adduct is Superior to Aminoguanidine for Preventing Diabetic Nephropathy in Mice. Horm. Metab. Res. 2002, 34, 371–377. [Google Scholar] [CrossRef][Green Version]
- Leovac, V.M.; Joksović, M.D.; Divjaković, V.; Jovanović, L.S.; Šaranović, Ž.; Pevec, A. Synthesis, spectroscopic and X-ray characterization of a copper(II) complex with the Schiff base derived from pyridoxal and aminoguanidine: NMR spectral studies of the ligand. J. Inorg. Biochem. 2007, 101, 1094–1097. [Google Scholar] [CrossRef]
- Leovac, V.M.; Vojinović-Ješić, L.S.; Češljević, V.I.; Novaković, S.B.; Bogdanović, G.A. {4-[(Carbamimidoylhydrazono)methyl-κ2N1, N4]-5-hydroxymethyl-2-methylpyridinium-3-olate-κO}(methanol-κO)copper(II) dinitrate. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2009, 65, m337–m339. [Google Scholar] [CrossRef] [PubMed]
- Radanović, M.M.; Rodić, M.V.; Armaković, S.; Armaković, S.J.; Vojinović-Ješić, L.S.; Leovac, V.M. Pyridoxylidene aminoguanidine and its copper(II) complexes—Syntheses, structure, and DFT calculations. J. Coord. Chem. 2017, 70, 2870–2887. [Google Scholar] [CrossRef]
- Jelić, M.G.; Boukos, N.; Lalović, M.M.; Romčević, N.Ž.; Leovac, V.M.; Hadžić, B.B.; Baloš, S.S.; Jovanović, L.S.; Slankamenac, M.P.; Živanov, M.B.; et al. Synthesis, structure and photoluminescence properties of copper(II) and cobalt(III) complexes with pyridoxalaminoguanidine. Opt. Mater. 2013, 35, 2728–2735. [Google Scholar] [CrossRef]
- Chakraborty, M.; Mohanty, M.; Dinda, R.; Sengupta, S.; Kumar Chattopadhyay, S. Cu(II) complexes of a bio-compatible aminoguanidine Schiff base: Histidine sensing and DNA-binding studies. Polyhedron 2022, 211, 115554. [Google Scholar] [CrossRef]
- Lalović, M.M.; Vojinović-Ješić, L.S.; Jovanović, L.S.; Leovac, V.M.; Češljević, V.I.; Divjaković, V. Synthesis, characterization and crystal structure of square-pyramidal copper(II) complexes with pyridoxylidene aminoguanidine. Inorganica Chim. Acta 2012, 388, 157–162. [Google Scholar] [CrossRef]
- Lalović, M.M.; Jovanović, L.S.; Vojinović-Ješić, L.S.; Leovac, V.M.; Češljević, V.I.; Rodić, M.V.; Divjaković, V. Syntheses, crystal structures, and electrochemical characterizations of two octahedral iron(III) complexes with Schiff base of pyridoxal and aminoguanidine. J. Coord. Chem. 2012, 65, 4217–4229. [Google Scholar] [CrossRef]
- Lalovic, M.; Leovac, V.; Vojinovic-Jesic, L.; Rodic, M.; Jovanovic, L.; Cesljevic, V. Dioxidovanadium(V) complexes with pyridoxalaminoguanidine: Synthesis, spectral and structural characterization. J. Serbian Chem. Soc. 2013, 78, 1161–1170. [Google Scholar] [CrossRef]
- Radanović, M.M.; Jelić, M.G.; Romčević, N.Ž.; Boukos, N.; Vojinović-Ješić, L.S.; Leovac, V.M.; Hadžić, B.B.; Bajac, B.M.; Nađ, L.F.; Chandrinou, C.; et al. Synthesis, structure and photoluminescence of (PLAGH)2[ZnCl4] and comparative analysis of photoluminescence properties with tris(2,2′-bipyridine)ruthenium(II). Mater. Res. Bull. 2015, 70, 951–957. [Google Scholar] [CrossRef]
- Okada, M.; Ayabe, Y. Effects of Aminoguanidine and Pyridoxal Phosphate on Glycation Reaction of Aspartate Aminotransferase and Serum Albumin. J. Nutr. Sci. Vitaminol. 1995, 41, 43–50. [Google Scholar] [CrossRef]
- Ooi, H.; Nasu, R.; Furukawa, A.; Takeuchi, M.; Koriyama, Y. Pyridoxamine and Aminoguanidine Attenuate the Abnormal Aggregation of β-Tubulin and Suppression of Neurite Outgrowth by Glyceraldehyde-Derived Toxic Advanced Glycation End-Products. Front. Pharmacol. 2022, 13, 921611. [Google Scholar] [CrossRef] [PubMed]
- Jevtovic, V.; Alshammari, N.; Latif, S.; Alsukaibi, A.K.D.; Humaidi, J.; Alanazi, T.Y.A.; Abdulaziz, F.; Matalka, S.I.; Pantelić, N.Đ.; Marković, M.; et al. Synthesis, Crystal Structure, Theoretical Calculations, Antibacterial Activity, Electrochemical Behavior, and Molecular Docking of Ni(II) and Cu(II) Complexes with Pyridoxal-Semicarbazone. Molecules 2022, 27, 6322. [Google Scholar] [CrossRef]
- Jevtovic, V.; Alshamari, A.K.; Milenković, D.; Dimitrić Marković, J.; Marković, Z.; Dimić, D. The Effect of Metal Ions (Fe, Co, Ni, and Cu) on the Molecular-Structural, Protein Binding, and Cytotoxic Properties of Metal Pyridoxal-Thiosemicarbazone Complexes. Int. J. Mol. Sci. 2023, 24, 11910. [Google Scholar] [CrossRef]
- Országhová, Z.; Liptáková, A.; Muchová, J.; Uličná, O.; Vančová, O.; Sivoňová, M.; Božek, P.; Čársky, J.; Ďuračková, Z. Influence of pyridoxylidene aminoguanidine on biomarkers of the oxidative stress and selected metabolic parameters of rats with diabetes mellitus. Gen. Physiol. Biophys. 2009, 28, 347–355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vojtaššák, J.; Čársky, J.; Danišovič, L.; Böhmer, D.; Blaško, M.; Braxatorisová, T. Effect of pyridoxylidene aminoguanidine on human diploid cells B-HEF-2: In vitro cytotoxicity test and cytogenetic analysis. Toxicol. In Vitro 2006, 20, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Dimić, D.S.; Kaluđerović, G.N.; Avdović, E.H.; Milenković, D.A.; Živanović, M.N.; Potočňák, I.; Samoľová, E.; Dimitrijević, M.S.; Saso, L.; Marković, Z.S.; et al. Synthesis, Crystallographic, Quantum Chemical, Antitumor, and Molecular Docking/Dynamic Studies of 4-Hydroxycoumarin-Neurotransmitter Derivatives. Int. J. Mol. Sci. 2022, 23, 1001. [Google Scholar] [CrossRef]
- Sakr, M.A.S.; Sherbiny, F.F.; El-Etrawy, A.-A.S. Hydrazone-based Materials; DFT, TD-DFT, NBO Analysis, Fukui Function, MESP Analysis, and Solar Cell Applications. J. Fluoresc. 2022, 32, 1857–1871. [Google Scholar] [CrossRef]
- Mahti, I.; Guillaumont, D.; Berthon, C.; Saint-Louis, G.; Hérès, X.; Berthon, L. Effect of metal complexation on the radiolytic stability of DOTA. Dalt. Trans. 2023, 52, 9952–9963. [Google Scholar] [CrossRef]
- Kostova, I.; Trendafilova, N.; Momekov, G. Theoretical and spectroscopic evidence for coordination ability of 3,3′-benzylidenedi-4-hydroxycoumarin. New neodymium (III) complex and its cytotoxic effect. J. Inorg. Biochem. 2005, 99, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Politzer, P.; Murray, J.S. The fundamental nature and role of the electrostatic potential in atoms and molecules. Theor. Chem. Acc. Theory Comput. Model. (Theor. Chim. Acta) 2002, 108, 134–142. [Google Scholar] [CrossRef]
- Barone, V.; Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Nunes, P.; Correia, I.; Marques, F.; Matos, A.P.; dos Santos, M.M.C.; Azevedo, C.G.; Capelo, J.-L.; Santos, H.M.; Gama, S.; Pinheiro, T.; et al. Copper Complexes with 1,10-Phenanthroline Derivatives: Underlying Factors Affecting Their Cytotoxicity. Inorg. Chem. 2020, 59, 9116–9134. [Google Scholar] [CrossRef] [PubMed]
- Nunes, P.; Correia, I.; Cavaco, I.; Marques, F.; Pinheiro, T.; Avecilla, F.; Pessoa, J.C. Therapeutic potential of vanadium complexes with 1,10-phenanthroline ligands, quo vadis? Fate of complexes in cell media and cancer cells. J. Inorg. Biochem. 2021, 217, 111350. [Google Scholar] [CrossRef] [PubMed]
- Le, M.; Rathje, O.; Levina, A.; Lay, P.A. High cytotoxicity of vanadium(IV) complexes with 1,10-phenanthroline and related ligands is due to decomposition in cell culture medium. JBIC J. Biol. Inorg. Chem. 2017, 22, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Tŏpala, T.; Pascual-Álvarez, A.; Moldes-Tolosa, M.Á.; Bodoki, A.; Castiñeiras, A.; Torres, J.; del Pozo, C.; Borrás, J.; Alzuet-Piña, G. New sulfonamide complexes with essential metal ions [Cu (II), Co (II), Ni (II) and Zn (II)]. Effect of the geometry and the metal ion on DNA binding and nuclease activity. BSA protein interaction. J. Inorg. Biochem. 2020, 202, 110823. [Google Scholar] [CrossRef]
- Rudra, S.; Dasmandal, S.; Patra, C.; Kundu, A.; Mahapatra, A. Binding affinities of Schiff base Fe(II) complex with BSA and calf-thymus DNA: Spectroscopic investigations and molecular docking analysis. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2016, 166, 84–94. [Google Scholar] [CrossRef]
- Duru Kamaci, U.; Kamaci, M.; Peksel, A. Thermally Stable Schiff Base and its Metal Complexes: Molecular Docking and Protein Binding Studies. J. Fluoresc. 2017, 27, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Bera, M.; Das, M.; Dolai, M.; Laha, S.; Islam, M.M.; Samanta, B.C.; Das, A.; Choudhuri, I.; Bhattacharyya, N.; Maity, T. DNA/Protein Binding and Apoptotic-Induced Anticancer Property of a First Time Reported Quercetin–Iron(III) Complex Having a Secondary Anionic Residue: A Combined Experimental and Theoretical Approach. ACS Omega 2023, 8, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Mansouri-Torshizi, H.; Khosravi, F.; Ghahghaei, A.; Shahraki, S.; Zareian-Jahromi, S. Investigation on the Interaction of Newly Designed Potential Antibacterial Zn(II) Complexes with CT-DNA and HSA.; Taylor & Francis: Abingdon, UK, 2018; Volume 36, ISBN 9854334465. [Google Scholar]
- Palchaudhuri, R.; Hergenrother, P.J. DNA as a target for anticancer compounds: Methods to determine the mode of binding and the mechanism of action. Curr. Opin. Biotechnol. 2007, 18, 497–503. [Google Scholar] [CrossRef]
- Li, Y.; Qian, C.; Li, Y.; Yang, Y.; Lin, D.; Liu, X.; Chen, C. Syntheses, crystal structures of two Fe(III) Schiff base complexes with chelating o-vanillin aroylhydrazone and exploration of their bio-relevant activities. J. Inorg. Biochem. 2021, 218, 111405. [Google Scholar] [CrossRef]
- Hassani Moghadam, F.; Taher, M.A.; Karimi-Maleh, H. Doxorubicin Anticancer Drug Monitoring by ds-DNA-Based Electrochemical Biosensor in Clinical Samples. Micromachines 2021, 12, 808. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Asija, S.; Deswal, Y.; Kumar, N. Recent advancements in the anticancer potentials of first row transition metal complexes. J. Indian. Chem. Soc. 2022, 99, 100556. [Google Scholar] [CrossRef]
- Almuzafar, H.M.; AlDuhaisan, N.N.; Ahmed, H.M.; Elsharif, A.M.; Rehman, S.; Akhtar, S.; Alsalem, Z.; Khan, F.A. Synthesis and characterizations of Fe (II) phthalocyanine and Zn phthalocyanine on colon cancer, cervical cancer, and bacterial cells. Arab. J. Basic. Appl. Sci. 2023, 30, 208–220. [Google Scholar] [CrossRef]
- Al-Hakimi, A.N.; Alminderej, F.; Aroua, L.; Alhag, S.K.; Alfaifi, M.Y.; Mahyoub, J.A.; Eldin, I.; Elbehairi, S.; Alnafisah, A.S. Design, synthesis, characterization of zirconium (IV), cadmium (II) and iron (III) complexes derived from Schiff base 2-aminomethylbenzimidazole, 2-hydroxynaphtadehyde and evaluation of their biological activity. Arab. J. Chem. 2020, 13, 7378–7389. [Google Scholar] [CrossRef]
- Lenis-Rojas, O.A.; Cordeiro, S.; Horta-Meireles, M.; Fernández, J.A.A.; Fernández Vila, S.; Rubiolo, J.A.; Cabezas-Sainz, P.; Sanchez, L.; Fernandes, A.R.; Royo, B. N-Heterocyclic Carbene Iron Complexes as Anticancer Agents: In Vitro and In Vivo Biological Studies. Molecules 2021, 26, 5535. [Google Scholar] [CrossRef]
- Kar, K.; Ghosh, D.; Kabi, B.; Chandra, A. A concise review on cobalt Schiff base complexes as anticancer agents. Polyhedron 2022, 222, 115890. [Google Scholar] [CrossRef]
- Pradeepa, S.M.; Bhojya Naik, H.S.; Vinay Kumar, B.; Indira Priyadarsini, K.; Barik, A.; Prabhakara, M.C. DNA binding, photoactivated DNA cleavage and cytotoxic activity of Cu(II) and Co(II) based Schiff-base azo photosensitizers. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 141, 34–42. [Google Scholar] [CrossRef]
- Revathi, N.; Sankarganesh, M.; Rajesh, J.; Raja, J.D. Biologically Active Cu(II), Co(II), Ni(II) and Zn(II) Complexes of Pyrimidine Derivative Schiff Base: DNA Binding, Antioxidant, Antibacterial and In Vitro Anticancer Studies. J. Fluoresc. 2017, 27, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- O’Doherty, C.; Keenan, J.; Horgan, K.; Murphy, R.; O’Sullivan, F.; Clynes, M. Copper-induced non-monotonic dose response in Caco-2 cells. Vitr. Cell. Dev. Biol. Anim. 2019, 55, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, E.; Gandin, V.; Bertani, R.; Sgarbossa, P.; Natarajan, K.; Bhuvanesh, N.S.P.; Venzo, A.; Zoleo, A.; Mozzon, M.; Dolmella, A.; et al. Synthesis, Characterization and Biological Activity of Novel Cu(II) Complexes of 6-Methyl-2-Oxo-1,2-Dihydroquinoline-3-Carbaldehyde-4n-Substituted Thiosemicarbazones. Molecules 2020, 25, 1868. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, P.; Viswanathamurthi, P.; Velmurugan, K.; Nandhakumar, R.; Balakumaran, M.D.; Kalaichelvan, P.T.; Malecki, J.G. Nickel(II) and copper(II) complexes constructed with N2S2 hybrid benzamidine–thiosemicarbazone ligand: Synthesis, X-ray crystal structure, DFT, kinetico-catalytic and in vitro biological applications. RSC Adv. 2015, 5, 103321–103342. [Google Scholar] [CrossRef]
- Predarska, I.; Saoud, M.; Drača, D.; Morgan, I.; Komazec, T.; Eichhorn, T.; Mihajlović, E.; Dunđerović, D.; Mijatović, S.; Maksimović-Ivanić, D.; et al. Mesoporous Silica Nanoparticles Enhance the Anticancer Efficacy of Platinum(IV)-Phenolate Conjugates in Breast Cancer Cell Lines. Nanomaterials 2022, 12, 3767. [Google Scholar] [CrossRef] [PubMed]
- Kordestani, N.; Rudbari, H.A.; Fernandes, A.R.; Raposo, L.R.; Baptista, P.V.; Ferreira, D.; Bruno, G.; Bella, G.; Scopelliti, R.; Braun, J.D.; et al. Antiproliferative Activities of Diimine-Based Mixed Ligand Copper(II) Complexes. ACS Comb. Sci. 2020, 22, 89–99. [Google Scholar] [CrossRef]
- Shahriary, S.; Tafvizi, F.; Khodarahmi, P.; Shaabanzadeh, M. Phyto-mediated synthesis of CuO nanoparticles using aqueous leaf extract of Artemisia deserti and their anticancer effects on A2780-CP cisplatin-resistant ovarian cancer cells. Biomass Convers. Biorefinery 2022. [Google Scholar] [CrossRef]
- CrysAlisPRO; Oxford Diffraction/Agilent Technologies UK Ltd.: Yarnton, UK, 2017.
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17; University of Western Australia: Perth, Australia, 2017. [Google Scholar]
- Spackman, M.A.; Byrom, P.G. A novel definition of a molecule in a crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Jevtović, V.; Hamoud, H.; Al-zahrani, S.; Alenezi, K.; Latif, S.; Alanazi, T.; Abdulaziz, F.; Dimić, D. Synthesis, Crystal Structure, Quantum Chemical Analysis, Electrochemical Behavior, and Antibacterial and Photocatalytic Activity of Co Complex with pyridoxal-(S-methyl)-isothiosemicarbazone ligand. Molecules 2022, 27, 4809. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Dunning, T.H. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J. Chem. Phys. 1989, 90, 1007. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitale. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Reed, A.E.; Weinstock, R.B.; Weinhold, F. Natural population analysis. J. Chem. Phys. 1985, 83, 735–746. [Google Scholar] [CrossRef]
- Behjatmanesh-Ardakani, R. NBO–NEDA, NPA, and QTAIM studies on the interactions between aza-, diaza-, and triaza-12-crown-4 (An-12-crown-4, n = 1, 2, 3) and Li+, Na+, and K+ ions. Comput. Theor. Chem. 2015, 1051, 62–71. [Google Scholar] [CrossRef]
- Bader, R.F.W. A Bond Path: A Universal Indicator of Bonded Interactions. J. Phys. Chem. A 1998, 102, 7314–7323. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules: A Quantum Theory; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Dimic, D.; Petkovic, M. Control of a photoswitching chelator by metal ions: DFT, NBO, and QTAIM analysis. Int. J. Quantum Chem. 2016, 116, 27–34. [Google Scholar] [CrossRef]
- Todd, A. AIMAll; Version 19.10.12; Keith, TK Gristmill Software: Overland Park, KS, USA, 2019. [Google Scholar]
- Yang, W.; Mortier, W.J. The use of global and local molecular parameters for the analysis of the gas-phase basicity of amines. J. Am. Chem. Soc. 1986, 108, 5708–5711. [Google Scholar] [CrossRef]
- De Luca, G.; Sicilia, E.; Russo, N.; Mineva, T. On the Hardness Evaluation in Solvent for Neutral and Charged Systems. J. Am. Chem. Soc. 2002, 124, 1494–1499. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. AMDock: A versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct 2020, 15, 12. [Google Scholar] [CrossRef]
- Canals, A.; Purciolas, M.; Aymamí, J.; Coll, M. The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2005, 61, 1009–1012. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A.; Zielinski, K.; Sekula, B. Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen. Proteins Struct. Funct. Bioinforma. 2014, 82, 2199–2208. [Google Scholar] [CrossRef]
- Fuhrmann, J.; Rurainski, A.; Lenhof, H.-P.; Neumann, D. A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J. Comput. Chem. 2010, 31, 1911–1918. [Google Scholar] [CrossRef]
- Khan, M.F.; Nasr, F.A.; Noman, O.M.; Alyhya, N.A.; Ali, I.; Saoud, M.; Rennert, R.; Dube, M.; Hussain, W.; Green, I.R.; et al. Cichorins D–F: Three New Compounds from Cichorium intybus and Their Biological Effects. Molecules 2020, 25, 4160. [Google Scholar] [CrossRef]
- Morgan, I.; Wessjohann, L.A.; Kaluđerović, G.N. In Vitro Anticancer Screening and Preliminary Mechanistic Study of A-Ring Substituted Anthraquinone Derivatives. Cells 2022, 11, 168. [Google Scholar] [CrossRef] [PubMed]


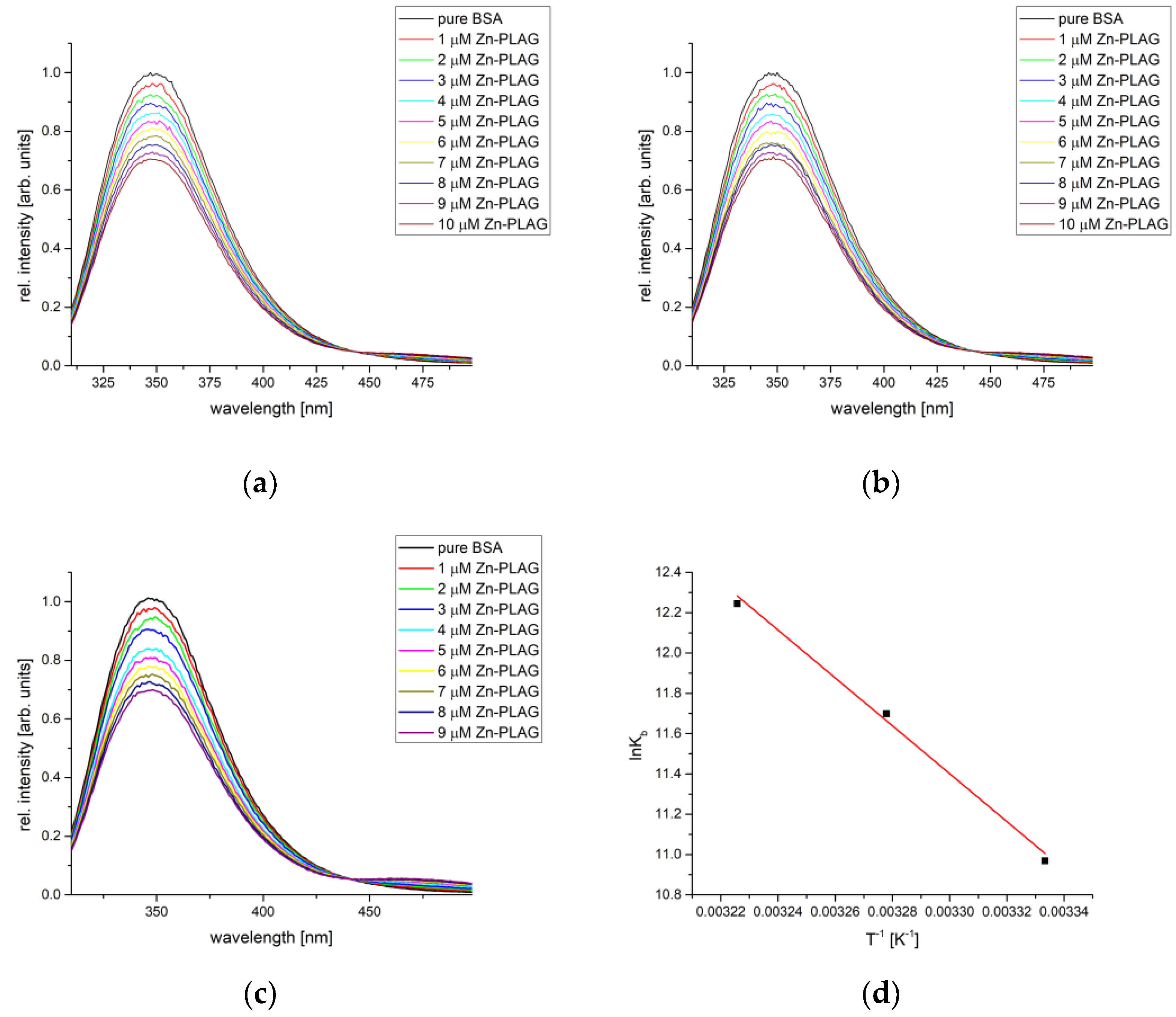

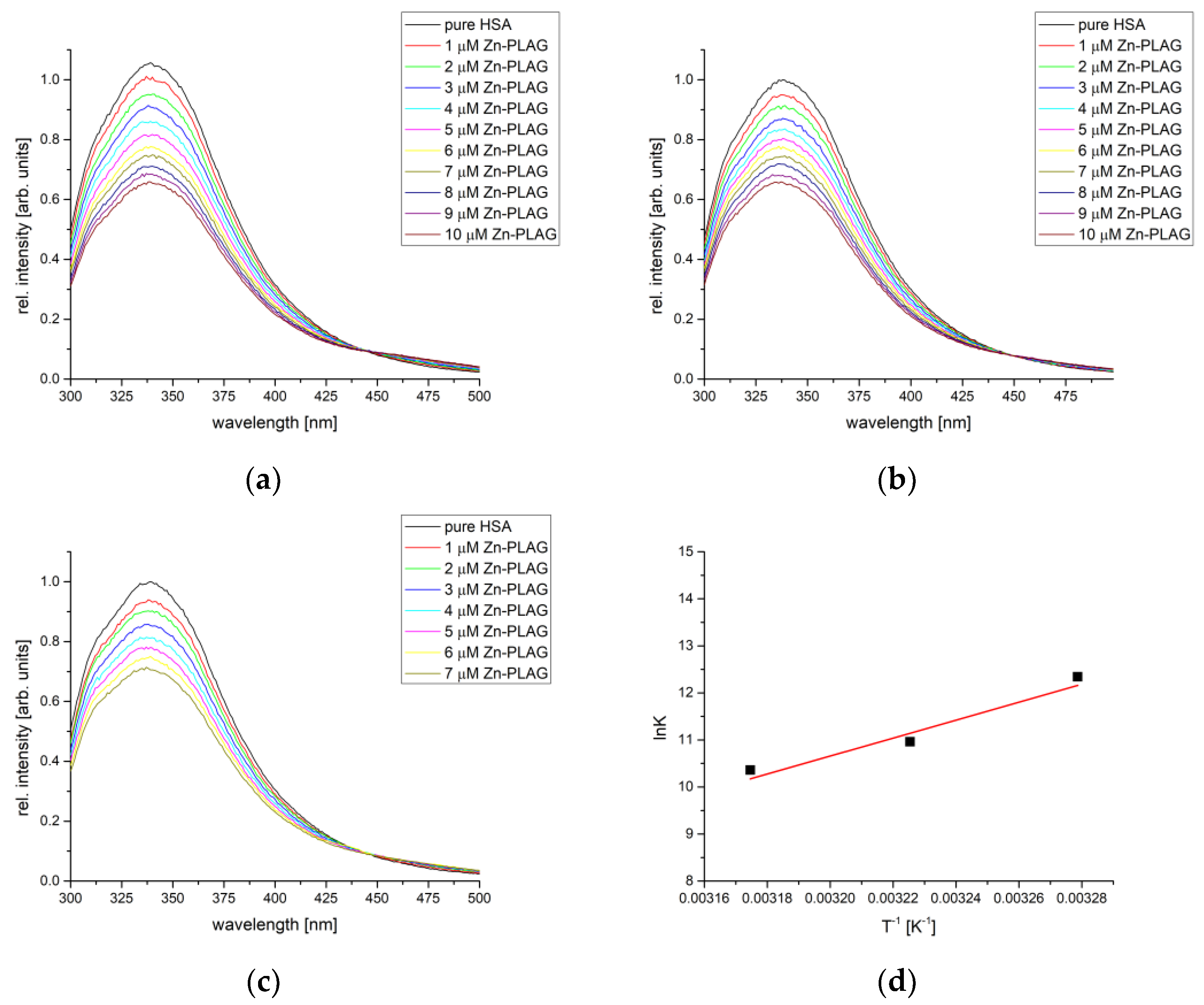

| Compound | T [°C] | Kb [M−1] | n | R2 | ΔHb [kJ mol−1] | ΔSb [J mol−1 K−1] | ΔGb [kJ mol−1] |
|---|---|---|---|---|---|---|---|
| Zn-PLAG | 27 | 5.80 × 104 | 1.13 | 0.999 | 98.78 | 420.77 | −27.45 |
| 32 | 1.24 × 105 | 1.09 | 0.997 | −29.56 | |||
| 35 | 2.08 × 105 | 1.03 | 0.999 | −31.66 | |||
| Co-PLAG | 27 | 1.86 × 104 | 0.97 | 0.998 | 16.01 | 135.08 | −24.51 |
| 32 | 2.04 × 104 | 0.98 | 0.999 | −25.19 | |||
| 35 | 2.29 × 104 | 0.96 | 0.998 | −25.86 | |||
| Fe-PLAG | 27 | 3.18 × 104 | 1.12 | 0.998 | 154.38 | 600.66 | −25.82 |
| 32 | 8.32 × 104 | 1.10 | 0.990 | −28.82 | |||
| 35 | 2.34 × 105 | 1.04 | 0.999 | −31.82 | |||
| Cu-PLAG | 27 | 9.63 × 103 | 1.13 | 0.998 | 404.98 | 1425.02 | −22.52 |
| 32 | 8.97 × 104 | 1.10 | 0.998 | −29.65 | |||
| 35 | 1.82 × 106 | 0.98 | 0.993 | −36.77 |
| Compound | ΔGbind | Ki (µM) | ΔGvdw + hbond + desolv | ΔGelec | ΔGtotal | ΔGtor | ΔGunb |
|---|---|---|---|---|---|---|---|
| Zn-PLAG-BSA | −25.5 | 33.4 | −31.1 | −2.4 | −13.1 | 8.0 | −13.1 |
| Co-PLAG-BSA | −34.4 | 9.4 × 10−1 | −25.6 | −15.7 | 2.6 | 6.9 | 2.6 |
| Fe-PLAG-BSA | −21.0 | 88.4 | −28.3 | −1.7 | 1.4 | 6.9 | 1.4 |
| Cu-PLAG-BSA | −23.1 | 2.1 × 10−2 | −26.8 | −1.0 | −5.1 | 6.9 | −5.1 |
| Compound | T [°C] | Kb [M−1] | n | R2 | ΔHb [kJ mol−1] | ΔSb [J mol−1 K−1] | ΔGb [kJ mol−1] |
|---|---|---|---|---|---|---|---|
| Zn-PLAG | 32 | 3.16 × 104 | 1.11 | 0.998 | −158.66 | −419.11 | −28.65 |
| 37 | 4.78 × 104 | 0.99 | 0.998 | −28.86 | |||
| 42 | 2.29 × 105 | 0.95 | 0.995 | −29.07 | |||
| Co-PLAG | 32 | 9.84 × 103 | 1.05 | 0.999 | 278.64 | 990.11 | −23.34 |
| 37 | 5.94 × 104 | 1.08 | 0.995 | −28.29 | |||
| 42 | 3.22 × 105 | 1.16 | 0.997 | −33.24 | |||
| Fe-PLAG | 32 | 4.37 × 103 | 1.09 | 0.992 | 378.28 | 1309.95 | −21.26 |
| 37 | 4.88 × 104 | 0.90 | 0.990 | −27.81 | |||
| 42 | 4.97 × 105 | 1.12 | 0.992 | −34.36 | |||
| Cu-PLAG | 32 | 7.59 × 103 | 0.91 | 0.990 | 128.56 | 495.17 | −22.47 |
| 37 | 1.38 × 104 | 0.90 | 0.995 | −24.94 | |||
| 42 | 3.80 × 104 | 0.98 | 0.996 | −27.42 |
| Compound | ΔGbind | Ki (µM) | ΔGvdw + hbond + desolv | ΔGelec | ΔGtotal | ΔGtor | ΔGunb |
|---|---|---|---|---|---|---|---|
| Zn-PLAG-DNA | −30.0 | 5.6 | −36.0 | −2.1 | −6.4 | 8.0 | −6.4 |
| Co-PLAG-DNA | −28.2 | 11.5 | −32.3 | −2.8 | 2.3 | 6.9 | 2.3 |
| Fe-PLAG-DNA | −25.7 | 31.6 | −32.3 | −0.3 | 1.5 | 6.9 | 1.5 |
| Cu-PLAG-DNA | −28.0 | 12.5 | −33.5 | −1.4 | −5.4 | 6.9 | −5.0 |
| Cell Line | Assay | Zn-PLAG | Fe-PLAG | Co-PLAG | Cu-PLAG | |
|---|---|---|---|---|---|---|
| HCT116 | Colorectal carcinoma | MTT | >100 | >100 | >100 | 81.5 ± 0.1 |
| CV | >100 | >100 | >100 | 97.4 ± 3.7 | ||
| A375 | melanoma | MTT | >100 | >100 | >100 | 73.6 ± 2.0 |
| CV | >100 | >100 | >100 | 79.8 ± 0.8 | ||
| MCF-7 | Breast cancer | MTT | >100 | >100 | >100 | 48.3 ± 1.5 |
| CV | >100 | >100 | >100 | 65.4 ± 4.8 | ||
| A2780 | Ovarian cancer | MTT | >100 | >100 | >100 | 32.5 ± 2.6 |
| CV | >100 | >100 | >100 | 38.5 ± 2.1 | ||
| Empirical Formula | Zn-PLAG C9H17N5O8SZn | Co-PLAG C18H28CoN10O10.75S | Fe-PLAG C18H28FeN10O9.5S |
|---|---|---|---|
| Molecular mass | 420.70 | 647.49 | 624.41 |
| Crystal system | Triclinic | Triclinic | Monoclinic |
| Space group | P-1 | P-1 | I 1 2/a 1 |
| a (A) | 9.1031 (6) | 11.9844 (3) | 10.9269 (7) |
| b (A) | 9.1317 (5) | 12.5247 (4) | 15.2428 (11) |
| c (A) | 10.0627 (4) | 12.6657 (4) | 29.873 (2) |
| α (°) | 85.167 (4) | 101.946 (3) | 90 |
| β (°) | 80.073 (4) | 113.517 (3) | 91.633 (6) |
| Γ (°) | 65.473 (6) | 110.863 (3) | 90 |
| V (A3) | 749.53 (8) | 1487.60 (9) | 4973.6 (6) |
| Z | 2 | 2 | 2 |
| F(000) | 432.0 | 685.0 | 6.309 |
| Dx, g cm−3 | 1.864 | 1.471 | 1.668 |
| Theta (max) | 87.700 | 80.135 | 80.195 |
| hkl Ranges | 11, 11, 12 | 15, 15, 16 | 13, 19, 38 |
| T (°C) | −100.00 (10) | −150.00 (10) | −150.00 (10) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jevtovic, V.; Alhar, M.S.O.; Milenković, D.; Marković, Z.; Dimitrić Marković, J.; Dimić, D. Synthesis, Structural Characterization, Cytotoxicity, and Protein/DNA Binding Properties of Pyridoxylidene-Aminoguanidine-Metal (Fe, Co, Zn, Cu) Complexes. Int. J. Mol. Sci. 2023, 24, 14745. https://doi.org/10.3390/ijms241914745
Jevtovic V, Alhar MSO, Milenković D, Marković Z, Dimitrić Marković J, Dimić D. Synthesis, Structural Characterization, Cytotoxicity, and Protein/DNA Binding Properties of Pyridoxylidene-Aminoguanidine-Metal (Fe, Co, Zn, Cu) Complexes. International Journal of Molecular Sciences. 2023; 24(19):14745. https://doi.org/10.3390/ijms241914745
Chicago/Turabian StyleJevtovic, Violeta, Munirah Sulaiman Othman Alhar, Dejan Milenković, Zoran Marković, Jasmina Dimitrić Marković, and Dušan Dimić. 2023. "Synthesis, Structural Characterization, Cytotoxicity, and Protein/DNA Binding Properties of Pyridoxylidene-Aminoguanidine-Metal (Fe, Co, Zn, Cu) Complexes" International Journal of Molecular Sciences 24, no. 19: 14745. https://doi.org/10.3390/ijms241914745
APA StyleJevtovic, V., Alhar, M. S. O., Milenković, D., Marković, Z., Dimitrić Marković, J., & Dimić, D. (2023). Synthesis, Structural Characterization, Cytotoxicity, and Protein/DNA Binding Properties of Pyridoxylidene-Aminoguanidine-Metal (Fe, Co, Zn, Cu) Complexes. International Journal of Molecular Sciences, 24(19), 14745. https://doi.org/10.3390/ijms241914745











