Bacterial Virus Forcing of Bacterial O-Antigen Shields: Lessons from Coliphages
Abstract
:1. OPS Can Serve as Physical Barriers to Phage Adsorption
1.1. Exopolysaccharides and Surface Polysaccharides of Enterobacteria
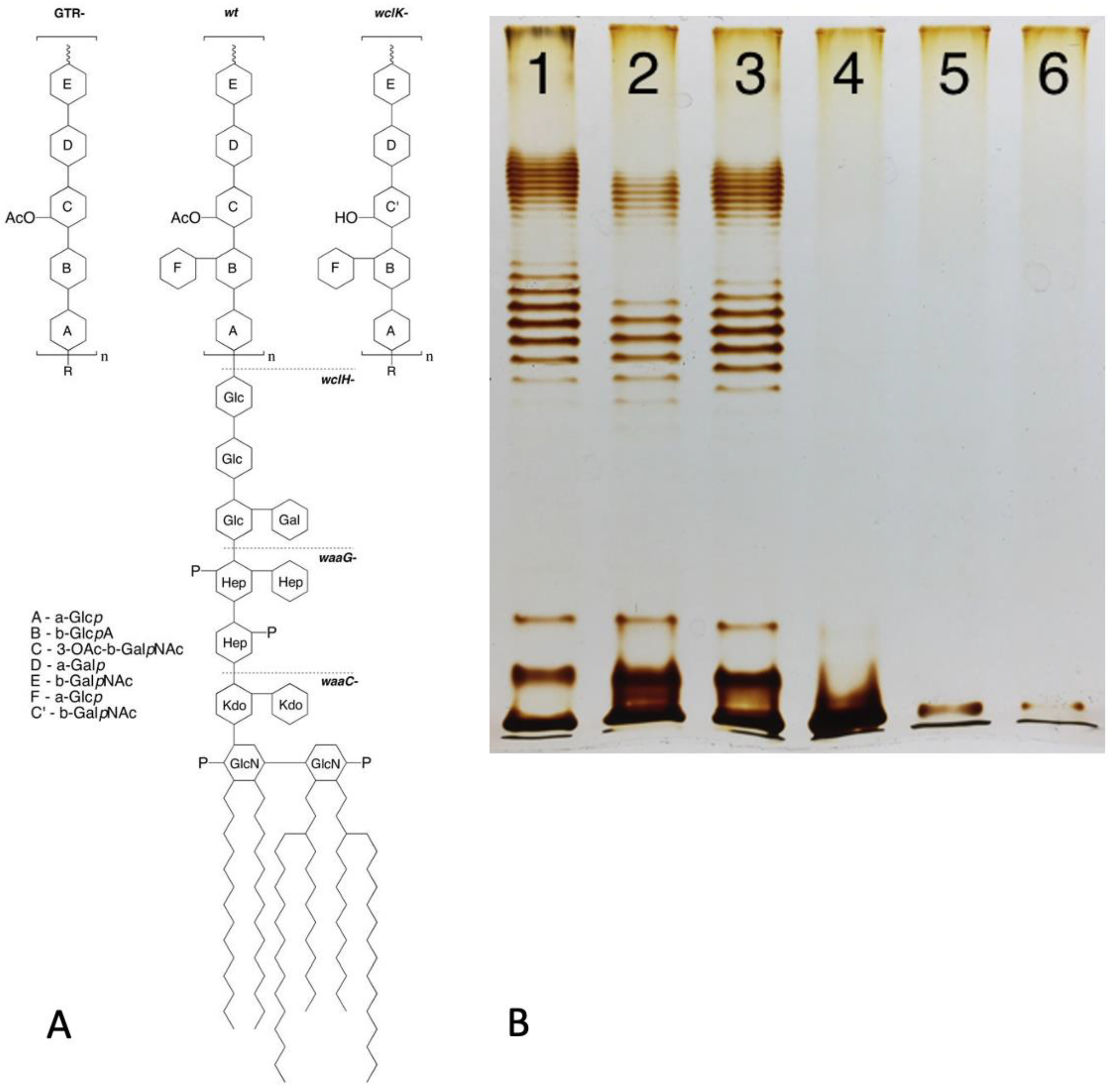
1.2. OPS Synthesis
1.3. Other Surface Polysaccharides of E. coli
2. OPS-Mediated OM Protection
OPS Shields the Outer Membrane Surface
3. Strategies of Host Cell Recognition by Bacteriophages and Properties of Phage-Resistant Mutants
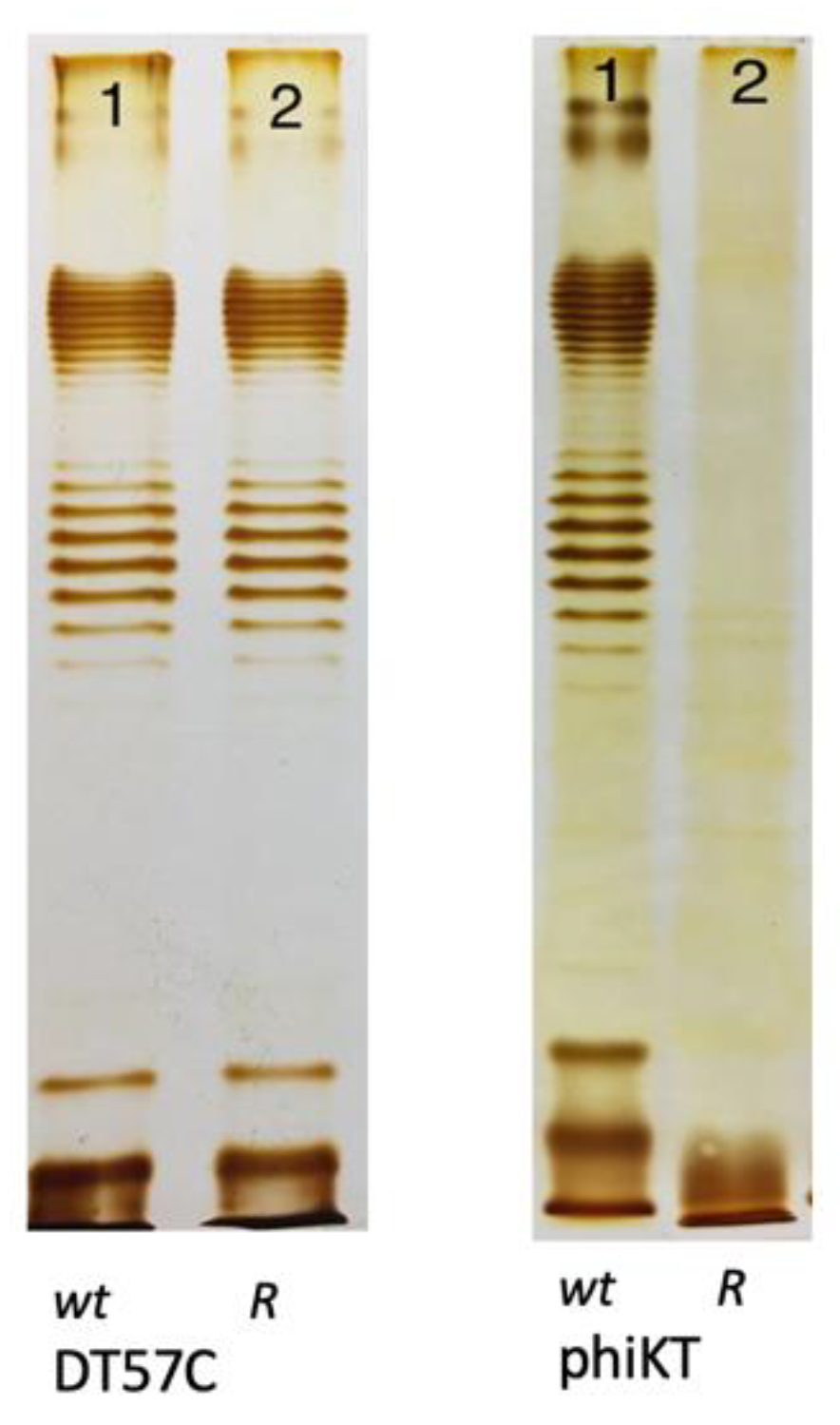
4. How Strong Is the O-Antigen Barrier in Enterobacteria?
4.1. Blocking of Lytic Phage Infection by OPS
4.2. OPS Restricts Prophage Acquisition
4.3. OPS-Mediated Protection in Other Species
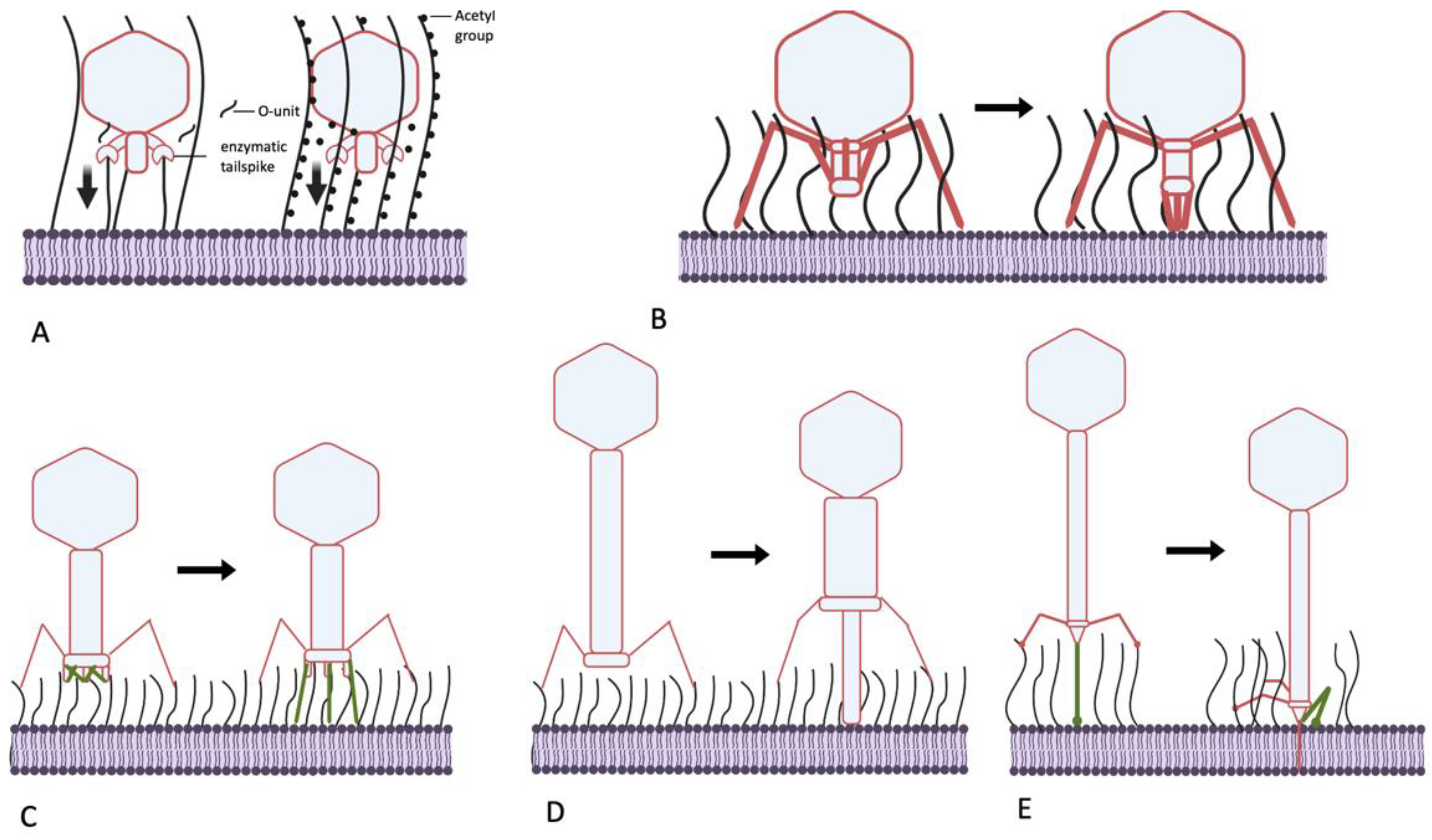
5. Mechanisms Used by Bacteriophages to Penetrate the O-Antigen Barrier
5.1. Podoviruses: Cut or Pull?
5.2. Can Podoviruses Push?
5.3. Myoviruses—Clutch and Push
5.4. Siphoviruses—Grab and Drag
6. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Kaur, N.; Dey, P. Bacterial exopolysaccharides as emerging bioactive macromolecules: From fundamentals to applications. Res. Microbiol. 2023, 174, 104024. [Google Scholar] [CrossRef]
- Rajagopal, M.; Walker, S. Envelope Structures of Gram-Positive Bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Aftalion, M.; Tidhar, A.; Vagima, Y.; Gur, D.; Zauberman, A.; Holtzman, T.; Makovitzki, A.; Chitlaru, T.; Mamroud, E.; Levy, Y. Rapid Induction of Protective Immunity against Pneumonic Plague by Yersinia pestis Polymeric F1 and LcrV Antigens. Vaccines 2023, 11, 581. [Google Scholar] [CrossRef]
- Secor, P.R.; Burgener, E.B.; Kinnersley, M.; Jennings, L.K.; Roman-Cruz, V.; Popescu, M.; Van Belleghem, J.D.; Haddock, N.; Copeland, C.; Michaels, L.A.; et al. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front. Immunol. 2020, 11, 244. [Google Scholar] [CrossRef]
- Ray, R.R. Role of Virus on Oral Biofilm: Inducer or Eradicator? Appl. Biochem. Biotechnol. 2023; Online ahead of print. [Google Scholar] [CrossRef]
- Meneses, L.; Brandao, A.C.; Coenye, T.; Braga, A.C.; Pires, D.P.; Azeredo, J. A systematic review of the use of bacteriophages for in vitro biofilm control. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage Adsorption: Likelihood of Virion Encounter with Bacteria and Other Factors Affecting Rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef] [PubMed]
- Caffalette, C.A.; Kuklewicz, J.; Spellmon, N.; Zimmer, J. Biosynthesis and Export of Bacterial Glycolipids. Annu. Rev. Biochem. 2020, 89, 741–768. [Google Scholar] [CrossRef]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef]
- Kulikov, E.E.; Golomidova, A.K.; Prokhorov, N.S.; Ivanov, P.A.; Letarov, A.V. High-throughput LPS profiling as a tool for revealing of bacteriophage infection strategies. Sci. Rep. 2019, 9, 2958. [Google Scholar] [CrossRef] [PubMed]
- Lundstedt, E.; Kahne, D.; Ruiz, N. Assembly and Maintenance of Lipids at the Bacterial Outer Membrane. Chem. Rev. 2021, 121, 5098–5123. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, A.; Kahne, D.E.; Silhavy, T.J. Outer Membrane Biogenesis. Annu. Rev. Microbiol. 2017, 71, 539–556. [Google Scholar] [CrossRef] [PubMed]
- Cian, M.B.; Giordano, N.P.; Masilamani, R.; Minor, K.E.; Dalebroux, Z.D. Salmonella enterica Serovar Typhimurium Uses PbgA/YejM to Regulate Lipopolysaccharide Assembly during Bacteremia. Infect. Immun. 2019, 88, e00758.19. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Mitchell, A.M. Enterobacterial Common Antigen: Synthesis and Function of an Enigmatic Molecule. mBio 2020, 11, e01914.20. [Google Scholar] [CrossRef]
- Whitfield, C.; Williams, D.M.; Kelly, S.D. Lipopolysaccharide O-antigens-bacterial glycans made to measure. J. Biol. Chem. 2020, 295, 10593–10609. [Google Scholar] [CrossRef]
- Weckener, M.; Woodward, L.S.; Clarke, B.R.; Liu, H.; Ward, P.N.; Le Bas, A.; Bhella, D.; Whitfield, C.; Naismith, J.H. The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase, Wzz, from enterobacterial common antigen biosynthesis form a complex. Open Biol. 2023, 13, 220373. [Google Scholar] [CrossRef]
- Sellner, B.; Prakapaite, R.; van Berkum, M.; Heinemann, M.; Harms, A.; Jenal, U. A New Sugar for an Old Phage: A c-di-GMP-Dependent Polysaccharide Pathway Sensitizes Escherichia coli for Bacteriophage Infection. mBio 2021, 12, e0324621. [Google Scholar] [CrossRef]
- Junkermeier, E.H.; Hengge, R. A Novel Locally c-di-GMP-Controlled Exopolysaccharide Synthase Required for Bacteriophage N4 Infection of Escherichia coli. mBio 2021, 12, e0324921. [Google Scholar] [CrossRef]
- Kiino, D.R.; Rothman-Denes, L.B. Genetic analysis of bacteriophage N4 adsorption. J. Bacteriol. 1989, 171, 4595–4602. [Google Scholar] [CrossRef]
- Thongsomboon, W.; Serra, D.O.; Possling, A.; Hadjineophytou, C.; Hengge, R.; Cegelski, L. Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 2018, 359, 334–338. [Google Scholar] [CrossRef]
- Ross, P.; Mayer, R.; Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 1991, 55, 35–58. [Google Scholar] [CrossRef]
- Boehm, A.; Steiner, S.; Zaehringer, F.; Casanova, A.; Hamburger, F.; Ritz, D.; Keck, W.; Ackermann, M.; Schirmer, T.; Jenal, U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 2009, 72, 1500–1516. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Q.; Reeves, P.R. The variation of O antigens in gram-negative bacteria. Subcell. Biochem. 2010, 53, 123–152. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Reperant-Ferter, M.; Gitton, C.; Germon, P. Shielding Effect of Escherichia coli O-Antigen Polysaccharide on J5-Induced Cross-Reactive Antibodies. mSphere 2021, 6, e01227.20. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Medina, C.C.; Perez-Toledo, M.; Schager, A.E.; Marshall, J.L.; Cook, C.N.; Bobat, S.; Hwang, H.; Chun, B.J.; Logan, E.; Bryant, J.A.; et al. Outer membrane protein size and LPS O-antigen define protective antibody targeting to the Salmonella surface. Nat. Commun. 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Bentley, A.T.; Klebba, P.E. Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. J. Bacteriol. 1988, 170, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Pluschke, G.; Mayden, J.; Achtman, M.; Levine, R.P. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. Infect. Immun. 1983, 42, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Golomidova, A.K.; Efimov, A.D.; Kulikov, E.E.; Kuznetsov, A.S.; Belalov, I.S.; Letarov, A.V. O antigen restricts lysogenization of non-O157 Escherichia coli strains by Stx-converting bacteriophage phi24B. Sci. Rep. 2021, 11, 3035. [Google Scholar] [CrossRef]
- Krzyzewska-Dudek, E.; Kotimaa, J.; Kapczynska, K.; Rybka, J.; Meri, S. Lipopolysaccharides and outer membrane proteins as main structures involved in complement evasion strategies of non-typhoidal Salmonella strains. Mol. Immunol. 2022, 150, 67–77. [Google Scholar] [CrossRef]
- Heiman, C.M.; Maurhofer, M.; Calderon, S.; Dupasquier, M.; Marquis, J.; Keel, C.; Vacheron, J. Pivotal role of O-antigenic polysaccharide display in the sensitivity against phage tail-like particles in environmental Pseudomonas kin competition. ISME J. 2022, 16, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, A.; Vitiello, G.; Wanke, A.; Cavasso, D.; Clifton, L.A.; Mahdi, L.; Campanero-Rhodes, M.A.; Solis, D.; Wuhrer, M.; Nicolardi, S.; et al. Lipopolysaccharide O-antigen molecular and supramolecular modifications of plant root microbiota are pivotal for host recognition. Carbohydr. Polym. 2022, 277, 118839. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhang, H.; Huang, X.; Ma, J.; Logue, C.M.; Nolan, L.K.; Li, G. O-specific polysaccharide confers lysozyme resistance to extraintestinal pathogenic Escherichia coli. Virulence 2018, 9, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, E.E.; Majewska, J.; Prokhorov, N.S.; Golomidova, A.K.; Tatarskiy, E.V.; Letarov, A.V. Effect of O-acetylation of O antigen of Escherichia coli lipopolysaccharide on the nonspecific barrier function of the outer membrane. Microbiology 2017, 86, 310–316. [Google Scholar] [CrossRef]
- Vila, J.; Saez-Lopez, E.; Johnson, J.R.; Romling, U.; Dobrindt, U.; Canton, R.; Giske, C.G.; Naas, T.; Carattoli, A.; Martinez-Medina, M.; et al. Escherichia coli: An old friend with new tidings. FEMS Microbiol. Rev. 2016, 40, 437–463. [Google Scholar] [CrossRef]
- Luthje, P.; Brauner, A. Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Microb. Physiol. 2014, 65, 337–372. [Google Scholar] [CrossRef]
- Wildschutte, H.; Lawrence, J.G. Differential Salmonella survival against communities of intestinal amoebae. Microbiology 2007, 153, 1781–1789. [Google Scholar] [CrossRef]
- van der Ley, P.; de Graaff, P.; Tommassen, J. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J. Bacteriol. 1986, 168, 449–451. [Google Scholar] [CrossRef]
- Letarov, A.; Kulikov, E. Adsorption of bacteriophages on bacterial cells. Biochemistry 2017, 82, 1632–1658. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Ayariga, J.A.; Venkatesan, K.; Ward, R.; Wu, H.; Jackson, D.; Villafane, R. Initiation of P22 infection at the phage centennial. Front. Sci. Technol. Eng. Math. 2018, 2, 64–81. [Google Scholar]
- Golomidova, A.K.; Kulikov, E.E.; Prokhorov, N.S.; Guerrero-Ferreira Rcapital Es, C.; Knirel, Y.A.; Kostryukova, E.S.; Tarasyan, K.K.; Letarov, A.V. Branched Lateral Tail Fiber Organization in T5-Like Bacteriophages DT57C and DT571/2 is Revealed by Genetic and Functional Analysis. Viruses 2016, 8, 26. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Ivanov, P.A.; Senchenkova, S.N.; Naumenko, O.I.; Ovchinnikova, O.O.; Shashkov, A.S.; Golomidova, A.K.; Babenko, V.V.; Kulikov, E.E.; Letarov, A.V. Structure and gene cluster of the O antigen of Escherichia coli F17, a candidate for a new O-serogroup. Int. J. Biol. Macromol. 2019, 124, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.A.; Prokhorov, N.S.; Shashkov, A.S.; Ovchinnikova, O.G.; Zdorovenko, E.L.; Liu, B.; Kostryukova, E.S.; Larin, A.K.; Golomidova, A.K.; Letarov, A.V. Variations in O-antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4s. J. Bacteriol. 2015, 197, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 2017, 105, 385–398. [Google Scholar] [CrossRef]
- Letarov, A.V.; Letarova, M.A.; Kulikov, E.E. Revised Sequence and Annotation of Burkholderia pseudomallei/thailandensis Bacteriophage Bp-AMP1—A Potential Agent of Natural Biocontrol of the Populations of the Melioidosis Causative Agent. Microbiology 2019, 88, 756–758. [Google Scholar] [CrossRef]
- Lukianova, A.A.; Evseev, P.V.; Shneider, M.M.; Dvoryakova, E.A.; Tokmakova, A.D.; Shpirt, A.M.; Kabilov, M.R.; Obraztsova, E.A.; Shashkov, A.S.; Ignatov, A.N.; et al. Pectobacterium versatile Bacteriophage Possum: A Complex Polysaccharide-Deacetylating Tail Fiber as a Tool for Host Recognition in Pectobacterial Schitoviridae. Int. J. Mol. Sci. 2022, 23, 11043. [Google Scholar] [CrossRef]
- Lukianova, A.A.; Shneider, M.M.; Evseev, P.V.; Shpirt, A.M.; Bugaeva, E.N.; Kabanova, A.P.; Obraztsova, E.A.; Miroshnikov, K.K.; Senchenkova, S.N.; Shashkov, A.S.; et al. Morphologically Different Pectobacterium brasiliense Bacteriophages PP99 and PP101: Deacetylation of O-Polysaccharide by the Tail Spike Protein of Phage PP99 Accompanies the Infection. Front. Microbiol. 2019, 10, 3147. [Google Scholar] [CrossRef]
- Efimov, A.D.; Golomidova, A.K.; Kulikov, E.E.; Belalov, I.S.; Ivanov, P.A.; Letarov, A.V. RB49-like Bacteriophages Recognize O Antigens as One of the Alternative Primary Receptors. Int. J. Mol. Sci. 2022, 23, 11329. [Google Scholar] [CrossRef]
- Olivenza, D.R.; Casadesus, J.; Ansaldi, M. Epigenetic biosensors for bacteriophage detection and phage receptor discrimination. Environ. Microbiol. 2020, 22, 3126–3142. [Google Scholar] [CrossRef]
- Cota, I.; Sanchez-Romero, M.A.; Hernandez, S.B.; Pucciarelli, M.G.; Garcia-Del Portillo, F.; Casadesus, J. Epigenetic Control of Salmonella enterica O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance. PLoS Genet. 2015, 11, e1005667. [Google Scholar] [CrossRef]
- Heller, K.; Braun, V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F., resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 1979, 139, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Heller, K.; Braun, V. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J. Virol. 1982, 41, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Heller, K.J.; Bryniok, D. O antigen-dependent mutant of bacteriophage T5. J. Virol. 1984, 49, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Golomidova, A.K.; Naumenko, O.I.; Senchenkova, S.N.; Knirel, Y.A.; Letarov, A.V. The O-polysaccharide of Escherichia coli F5, which is structurally related to that of E. coli O28ab, provides cells only weak protection against bacteriophage attack. Arch. Virol. 2019, 164, 2783–2787. [Google Scholar] [CrossRef]
- Golomidova, A.K.; Kulikov, E.E.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Ksenzenko, V.N.; Tarasyan, K.K.; Letarov, A.V. Complete genome sequences of T5-related Escherichia coli bacteriophages DT57C and DT571/2 isolated from horse feces. Arch. Virol. 2015, 160, 3133–3137. [Google Scholar] [CrossRef]
- Golomidova, A.K.; Kulikov, E.E.; Babenko, V.V.; Ivanov, P.A.; Prokhorov, N.S.; Letarov, A.V. Escherichia coli bacteriophage Gostya9, representing a new species within the genus T5virus. Arch. Virol. 2019, 164, 879–884. [Google Scholar] [CrossRef]
- Kulikov, E.E.; Golomidova, A.K.; Efimov, A.D.; Belalov, I.S.; Letarova, M.A.; Zdorovenko, E.L.; Knirel, Y.A.; Dmitrenok, A.S.; Letarov, A.V. Equine Intestinal O-Seroconverting Temperate Coliphage Hf4s: Genomic and Biological Characterization. Appl. Environ. Microbiol. 2021, 87, e0112421. [Google Scholar] [CrossRef]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic exploration of Escherichia coli phage-host interactions with the BASEL phage collection. PLoS Biol. 2021, 19, e3001424. [Google Scholar] [CrossRef]
- James, C.E.; Stanley, K.N.; Allison, H.E.; Flint, H.J.; Stewart, C.S.; Sharp, R.J.; Saunders, J.R.; McCarthy, A.J. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 2001, 67, 4335–4337. [Google Scholar] [CrossRef]
- Yerigeri, K.; Kadatane, S.; Mongan, K.; Boyer, O.; Burke, L.L.G.; Sethi, S.K.; Licht, C.; Raina, R. Atypical Hemolytic-Uremic Syndrome: Genetic Basis, Clinical Manifestations, and a Multidisciplinary Approach to Management. J. Multidiscip. Healthc. 2023, 16, 2233–2249. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rubio, L.; Haarmann, N.; Schwidder, M.; Muniesa, M.; Schmidt, H. Bacteriophages of Shiga Toxin-Producing Escherichia coli and Their Contribution to Pathogenicity. Pathogens 2021, 10, 404. [Google Scholar] [CrossRef] [PubMed]
- Berryhill, B.A.; Garcia, R.; McCall, I.C.; Manuel, J.A.; Chaudhry, W.; Petit, M.A.; Levin, B.R. The book of Lambda does not tell us that naturally occurring lysogens of Escherichia coli are likely to be resistant as well as immune. Proc. Natl. Acad. Sci. USA 2023, 120, e2212121120. [Google Scholar] [CrossRef]
- Latka, A.; Leiman, P.G.; Drulis-Kawa, Z.; Briers, Y. Modeling the Architecture of Depolymerase-Containing Receptor Binding Proteins in Klebsiella Phages. Front. Microbiol. 2019, 10, 2649. [Google Scholar] [CrossRef] [PubMed]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Falenczyk, B.; Wegrzyn, G.; Wegrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics 2021, 10, 175. [Google Scholar] [CrossRef]
- Reuter, M.; Kruger, D.H. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Genes 2020, 56, 136–149. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2019, 10, 2949. [Google Scholar] [CrossRef]
- Fernandes, S.; Sao-Jose, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Weintraub, A.; Seckler, R. Self-Competitive Inhibition of the Bacteriophage P22 Tailspike Endorhamnosidase by O-Antigen Oligosaccharides. Biochemistry 2020, 59, 4845–4855. [Google Scholar] [CrossRef]
- Ouyang, R.; Costa, A.R.; Cassidy, C.K.; Otwinowska, A.; Williams, V.C.J.; Latka, A.; Stansfeld, P.J.; Drulis-Kawa, Z.; Briers, Y.; Pelt, D.M.; et al. High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers. Nat. Commun. 2022, 13, 7241. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E.; Raya, R.; Carlson, C. Molecular mechanisms of phage infection. In Bacteriophages: Biology and Applications; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, Fl, USA; London, UK; New York, NY, USA; Washington, DC, USA, 2004; pp. 165–222. [Google Scholar]
- Volozhantsev, N.V.; Borzilov, A.I.; Shpirt, A.M.; Krasilnikova, V.M.; Verevkin, V.V.; Denisenko, E.A.; Kombarova, T.I.; Shashkov, A.S.; Knirel, Y.A.; Dyatlov, I.A. Comparison of the therapeutic potential of bacteriophage KpV74 and phage-derived depolymerase (beta-glucosidase) against Klebsiella pneumoniae capsular type K2. Virus Res. 2022, 322, 198951. [Google Scholar] [CrossRef]
- Drobiazko, A.Y.; Kasimova, A.A.; Evseev, P.V.; Shneider, M.M.; Klimuk, E.I.; Shashkov, A.S.; Dmitrenok, A.S.; Chizhov, A.O.; Slukin, P.V.; Skryabin, Y.P.; et al. Capsule-Targeting Depolymerases Derived from Acinetobacter baumannii Prophage Regions. Int. J. Mol. Sci. 2022, 23, 4971. [Google Scholar] [CrossRef]
- Parent, K.N.; Erb, M.L.; Cardone, G.; Nguyen, K.; Gilcrease, E.B.; Porcek, N.B.; Pogliano, J.; Baker, T.S.; Casjens, S.R. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol. Microbiol. 2014, 92, 47–60. [Google Scholar] [CrossRef]
- Subramanian, S.; Dover, J.A.; Parent, K.N.; Doore, S.M. Host Range Expansion of Shigella Phage Sf6 Evolves through Point Mutations in the Tailspike. J. Virol. 2022, 96, e0092922. [Google Scholar] [CrossRef]
- Andres, D.; Hanke, C.; Baxa, U.; Seul, A.; Barbirz, S.; Seckler, R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J. Biol. Chem. 2010, 285, 36768–36775. [Google Scholar] [CrossRef]
- Wang, C.; Tu, J.; Liu, J.; Molineux, I.J. Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol. 2019, 4, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Babenko, V.V.; Golomidova, A.K.; Ivanov, P.A.; Letarova, M.A.; Kulikov, E.E.; Manolov, A.I.; Prokhorov, N.S.; Kostrukova, E.S.; Matyushkina, D.M.; Prilipov, A.G. Phages associated with horses provide new insights into the dominance of lateral gene transfer in virulent bacteriophages evolution in natural systems. bioRxiv 2019, bioRxiv:542787. [Google Scholar]
- Kiino, D.R.; Singer, M.S.; Rothman-Denes, L.B. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. J. Bacteriol. 1993, 175, 7081–7085. [Google Scholar] [CrossRef]
- McPartland, J.; Rothman-Denes, L.B. The tail sheath of bacteriophage N4 interacts with the Escherichia coli receptor. J. Bacteriol. 2009, 191, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Dolgalev, G.V.; Safonov, T.A.; Arzumanian, V.A.; Kiseleva, O.I.; Poverennaya, E.V. Estimating Total Quantitative Protein Content in Escherichia coli, Saccharomyces cerevisiae, and HeLa Cells. Int. J. Mol. Sci. 2023, 24, 32081. [Google Scholar] [CrossRef] [PubMed]
- Golomidova, A.K.; Kulikov, E.E.; Kudryavtseva, A.V.; Letarov, A.V. Complete genome sequence of Escherichia coli bacteriophage PGT2. Genome Announc. 2018, 6, e01370.17. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 2013, 339, 576–579. [Google Scholar] [CrossRef]
- Swanson, N.A.; Lokareddy, R.K.; Li, F.; Hou, C.D.; Leptihn, S.; Pavlenok, M.; Niederweis, M.; Pumroy, R.A.; Moiseenkova-Bell, V.Y.; Cingolani, G. Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 A resolution. Mol. Cell 2021, 81, 3145–3159.e7. [Google Scholar] [CrossRef] [PubMed]
- Swanson, N.A.; Hou, C.D.; Cingolani, G. Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts. Microorganisms 2022, 10, 504. [Google Scholar] [CrossRef]
- Siborova, M.; Fuzik, T.; Prochazkova, M.; Novacek, J.; Benesik, M.; Nilsson, A.S.; Plevka, P. Tail proteins of phage SU10 reorganize into the nozzle for genome delivery. Nat. Commun. 2022, 13, 5622. [Google Scholar] [CrossRef]
- Taylor, N.M.I.; van Raaij, M.; Leiman, P.G. Contractile injection system of bacteriophages and related systems. Mol. Microbiol. 2018, 108, 6–15. [Google Scholar] [CrossRef]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key “Blueprint” for Reprogramming Phage Host Range. Int. J. Mol. Sci. 2022, 23, 12146. [Google Scholar] [CrossRef]
- Trojet, S.N.; Caumont-Sarcos, A.; Perrody, E.; Comeau, A.M.; Krisch, H.M. The gp38 adhesins of the T4 superfamily: A complex modular determinant of the phage’s host specificity. Genome Biol. Evol. 2011, 3, 674–686. [Google Scholar] [CrossRef]
- Dunne, M.; Denyes, J.M.; Arndt, H.; Loessner, M.J.; Leiman, P.G.; Klumpp, J. Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition. Structure 2018, 26, 1573–1582.e4. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc. Natl. Acad. Sci. USA 2015, 112, E4919–E4928. [Google Scholar] [CrossRef] [PubMed]
- Efimov, A.; Kulikov, E.; Golomidova, A.; Belalov, I.; Babenko, V.; Letarov, A. Isolation and sequencing of three RB49-like bacteriophages infecting O antigen-producing E. coli strains. F1000Research 2021, 10, 1113. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, L.; Abdelgader, S.A.; Yu, L.; Xu, J.; Yao, H.; Lu, C.; Zhang, W. Alterations in gp37 Expand the Host Range of a T4-Like Phage. Appl. Environ. Microbiol. 2017, 83, e01576.17. [Google Scholar] [CrossRef]
- Li, M.; Shi, D.; Li, Y.; Xiao, Y.; Chen, M.; Chen, L.; Du, H.; Zhang, W. Recombination of T4-like Phages and Its Activity against Pathogenic Escherichia coli in Planktonic and Biofilm Forms. Virol. Sin. 2020, 35, 651–661. [Google Scholar] [CrossRef]
- Hogins, J.; Sudarshan, S.; Zimmern, P.; Reitzer, L. Facile transduction with P1 phage in Escherichia coli associated with urinary tract infections. J. Microbiol. Methods 2023, 208, 106722. [Google Scholar] [CrossRef]
- Huan, Y.W.; Fa-Arun, J.; Wang, B. The Role of O-antigen in P1 Transduction of Shigella flexneri and Escherichia coli with its Alternative S’ Tail Fibre. J. Mol. Biol. 2022, 434, 167829. [Google Scholar] [CrossRef]
- Ho, T.D.; Waldor, M.K. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infect. Immun. 2007, 75, 1661–1666. [Google Scholar] [CrossRef]
- Yang, F.; Wang, L.; Zhou, J.; Xiao, H.; Liu, H. In Situ Structures of the Ultra-Long Extended and Contracted Tail of Myoviridae Phage P1. Viruses 2023, 15, 1267. [Google Scholar] [CrossRef]
- Liu, J.; Chen, C.Y.; Shiomi, D.; Niki, H.; Margolin, W. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology 2011, 417, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Linares, R.; Arnaud, C.A.; Degroux, S.; Schoehn, G.; Breyton, C. Structure, function and assembly of the long, flexible tail of siphophages. Curr. Opin. Virol. 2020, 45, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.R.; Cardarelli, L.; Pell, L.G.; Radford, D.R.; Maxwell, K.L. Long noncontractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012, 726, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Rabsch, W.; Ma, L.; Wiley, G.; Najar, F.Z.; Kaserer, W.; Schuerch, D.W.; Klebba, J.E.; Roe, B.A.; Laverde Gomez, J.A.; Schallmey, M.; et al. FepA- and TonB-dependent bacteriophage H8: Receptor binding and genomic sequence. J. Bacteriol. 2007, 189, 5658–5674. [Google Scholar] [CrossRef]
- Linares, R.; Arnaud, C.A.; Effantin, G.; Darnault, C.; Epalle, N.H.; Boeri Erba, E.; Schoehn, G.; Breyton, C. Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation. Sci. Adv. 2023, 9, eade9674. [Google Scholar] [CrossRef]
- van den Berg, B.; Silale, A.; Basle, A.; Brandner, A.F.; Mader, S.L.; Khalid, S. Structural basis for host recognition and superinfection exclusion by bacteriophage T5. Proc. Natl. Acad. Sci. USA 2022, 119, e2211672119. [Google Scholar] [CrossRef]
- Ayala, R.; Moiseenko, A.V.; Chen, T.-H.; Kulikov, E.E.; Golomidova, A.K.; Orekhov, P.S.; Street, M.A.; Sokolova, O.S.; Letarov, A.V.; Wolf, M. Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers. Nat. Commun. 2023; in press. [Google Scholar]
- Wang, C.; Duan, J.; Gu, Z.; Ge, X.; Zeng, J.; Wang, J. Architecture of the bacteriophage lambda tail. Structure 2023. [Google Scholar] [CrossRef]
- Letarov, A.; Kulikov, E. The bacteriophages in human- and animal body-associated microbial communities. J. Appl. Microbiol. 2009, 107, 1–13. [Google Scholar] [CrossRef]
- Golomidova, A.; Kulikov, E.; Isaeva, A.; Manykin, A.; Letarov, A. The diversity of coliphages and coliforms in horse feces reveals a complex pattern of ecological interactions. Appl. Environ. Microbiol. 2007, 73, 5975–5981. [Google Scholar] [CrossRef]
- Romling, U. The power of unbiased phenotypic screens—Cellulose as a first receptor for the Schitoviridae phage S6 of Erwinia amylovora. Environ. Microbiol. 2022, 24, 3316–3321. [Google Scholar] [CrossRef]
- Knecht, L.E.; Heinrich, N.; Born, Y.; Felder, K.; Pelludat, C.; Loessner, M.J.; Fieseler, L. Bacteriophage S6 requires bacterial cellulose for Erwinia amylovora infection. Environ. Microbiol. 2022, 24, 3436–3450. [Google Scholar] [CrossRef] [PubMed]
- Letarov, A.V.; Letarova, M.A. The Burden of Survivors: How Can Phage Infection Impact Non-Infected Bacteria? Int. J. Mol. Sci. 2023, 24, 2733. [Google Scholar] [CrossRef] [PubMed]
- Luthe, T.; Kever, L.; Thormann, K.; Frunzke, J. Bacterial multicellular behavior in antiviral defense. Curr. Opin. Microbiol. 2023, 74, 102314. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Letarov, A.V. Bacterial Virus Forcing of Bacterial O-Antigen Shields: Lessons from Coliphages. Int. J. Mol. Sci. 2023, 24, 17390. https://doi.org/10.3390/ijms242417390
Letarov AV. Bacterial Virus Forcing of Bacterial O-Antigen Shields: Lessons from Coliphages. International Journal of Molecular Sciences. 2023; 24(24):17390. https://doi.org/10.3390/ijms242417390
Chicago/Turabian StyleLetarov, Andrey V. 2023. "Bacterial Virus Forcing of Bacterial O-Antigen Shields: Lessons from Coliphages" International Journal of Molecular Sciences 24, no. 24: 17390. https://doi.org/10.3390/ijms242417390
APA StyleLetarov, A. V. (2023). Bacterial Virus Forcing of Bacterial O-Antigen Shields: Lessons from Coliphages. International Journal of Molecular Sciences, 24(24), 17390. https://doi.org/10.3390/ijms242417390




