The Link of mRNA and rRNA Transcription by PUF60/FIR through TFIIH/P62 as a Novel Therapeutic Target for Cancer
Abstract
:1. Introduction
2. Results
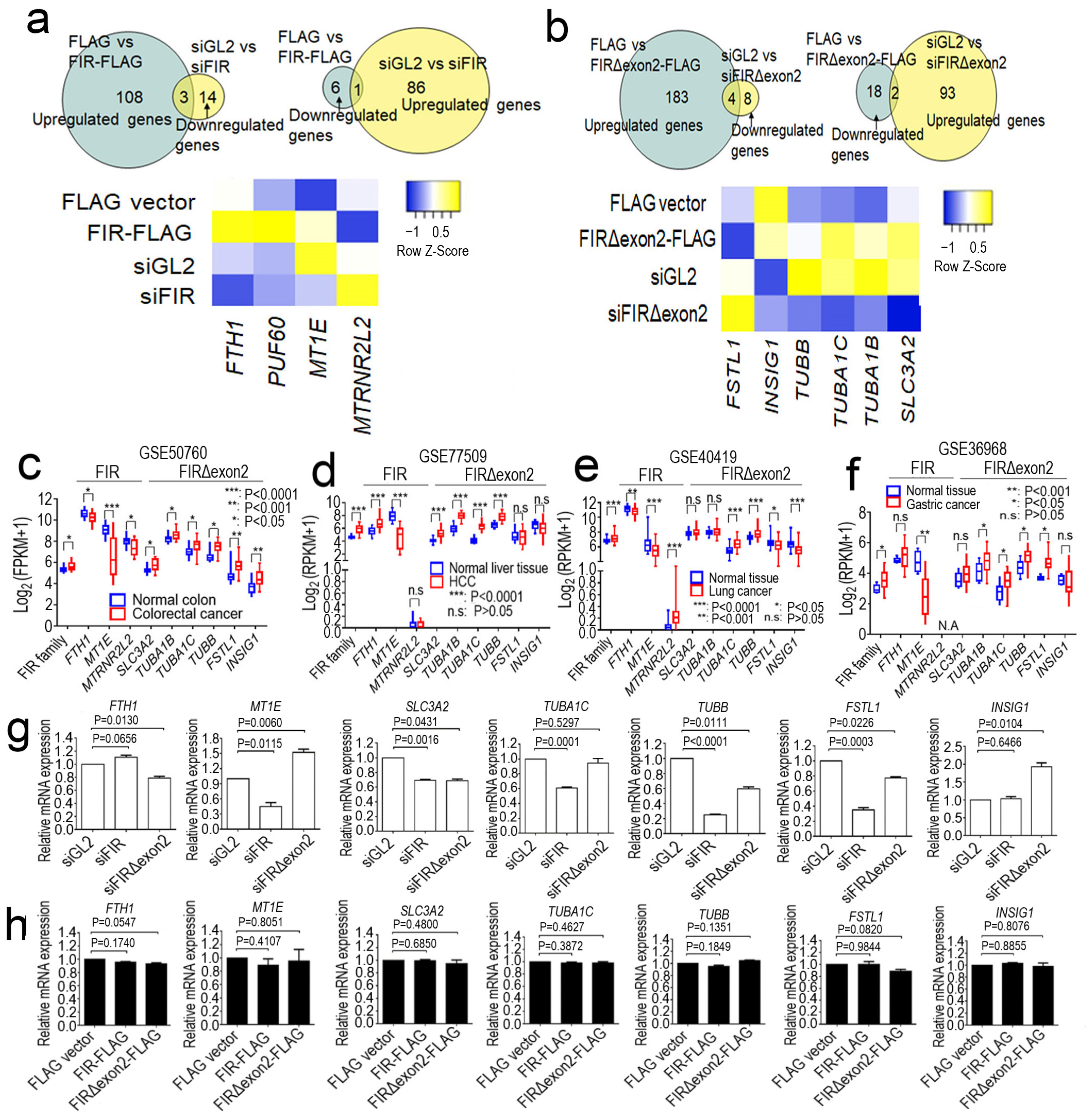
2.1. FIR and FIRΔexon2 Engaged in the Transcription of Various Cancer-Related mRNAs
2.2. FIR and FIRΔexon2 Affected Transcription of Various rRNAs
2.3. The Majority of rRNA Expressions by FIR and FIRΔexon2 Were Independent of c-Myc Activation
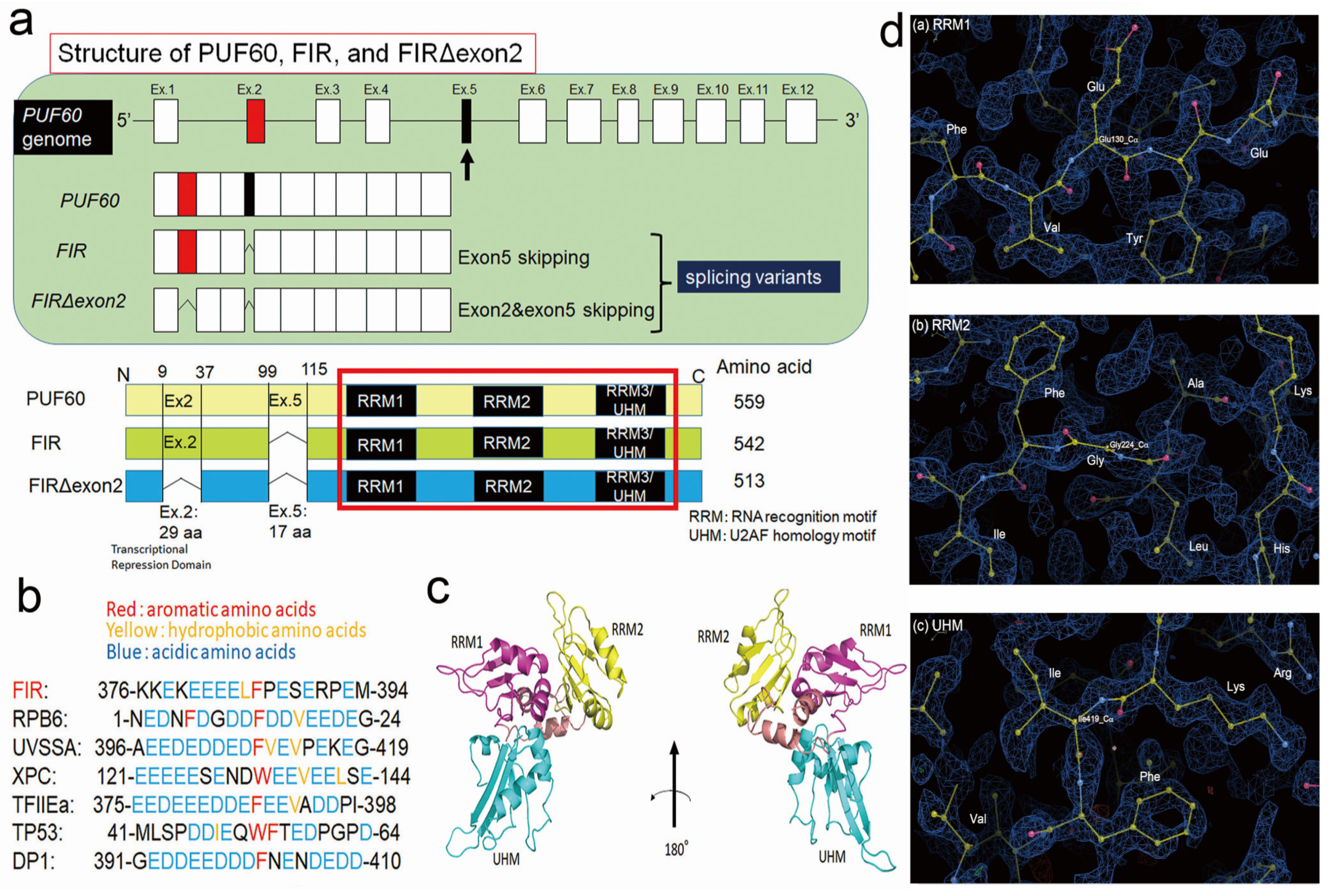
2.4. FIR and FIRΔexon2 Have Highly Homologous Sequence with RPB6
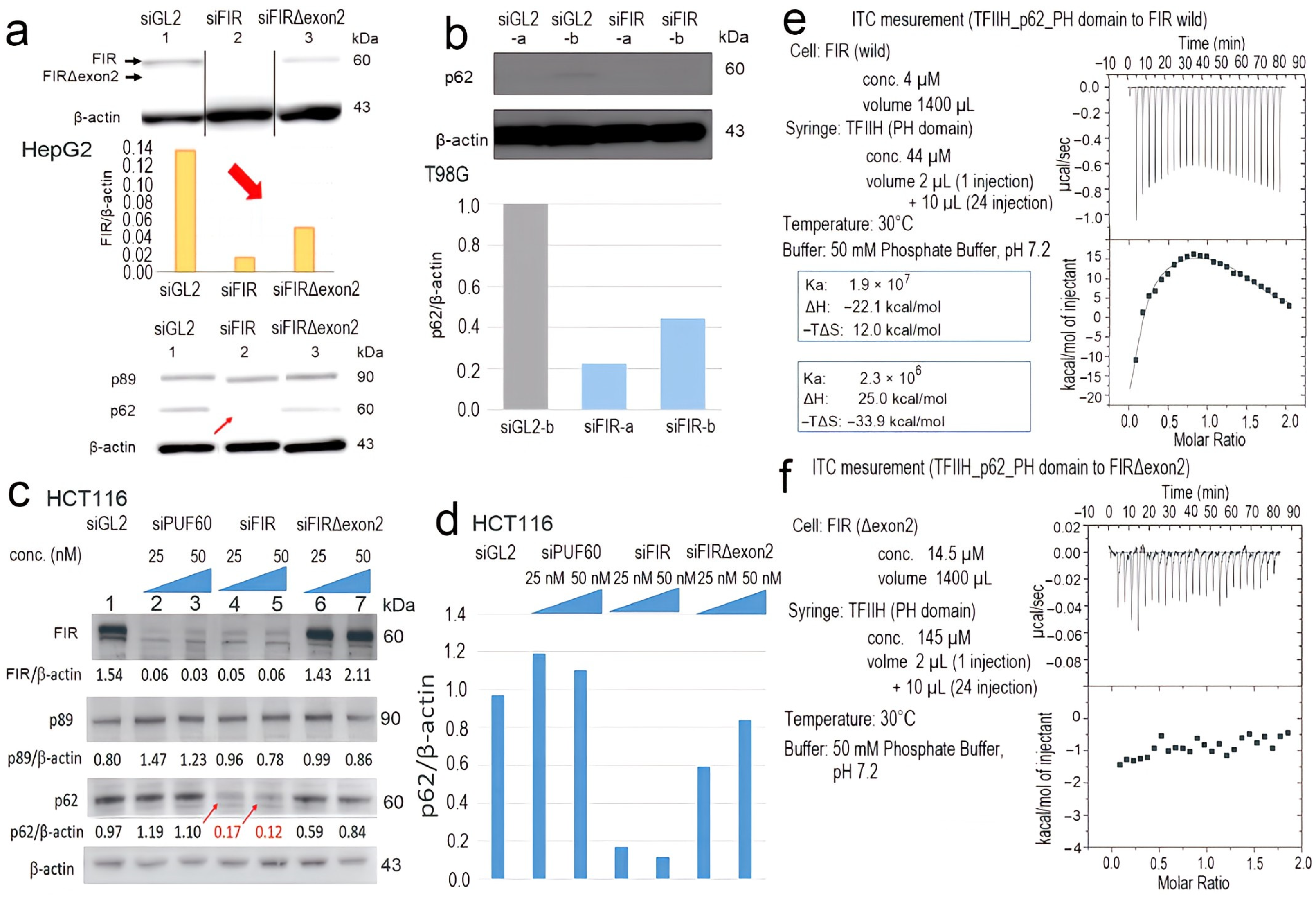
2.5. FIR, but Not FIRΔexon2, Directly Interacted with P62 of TFIIH
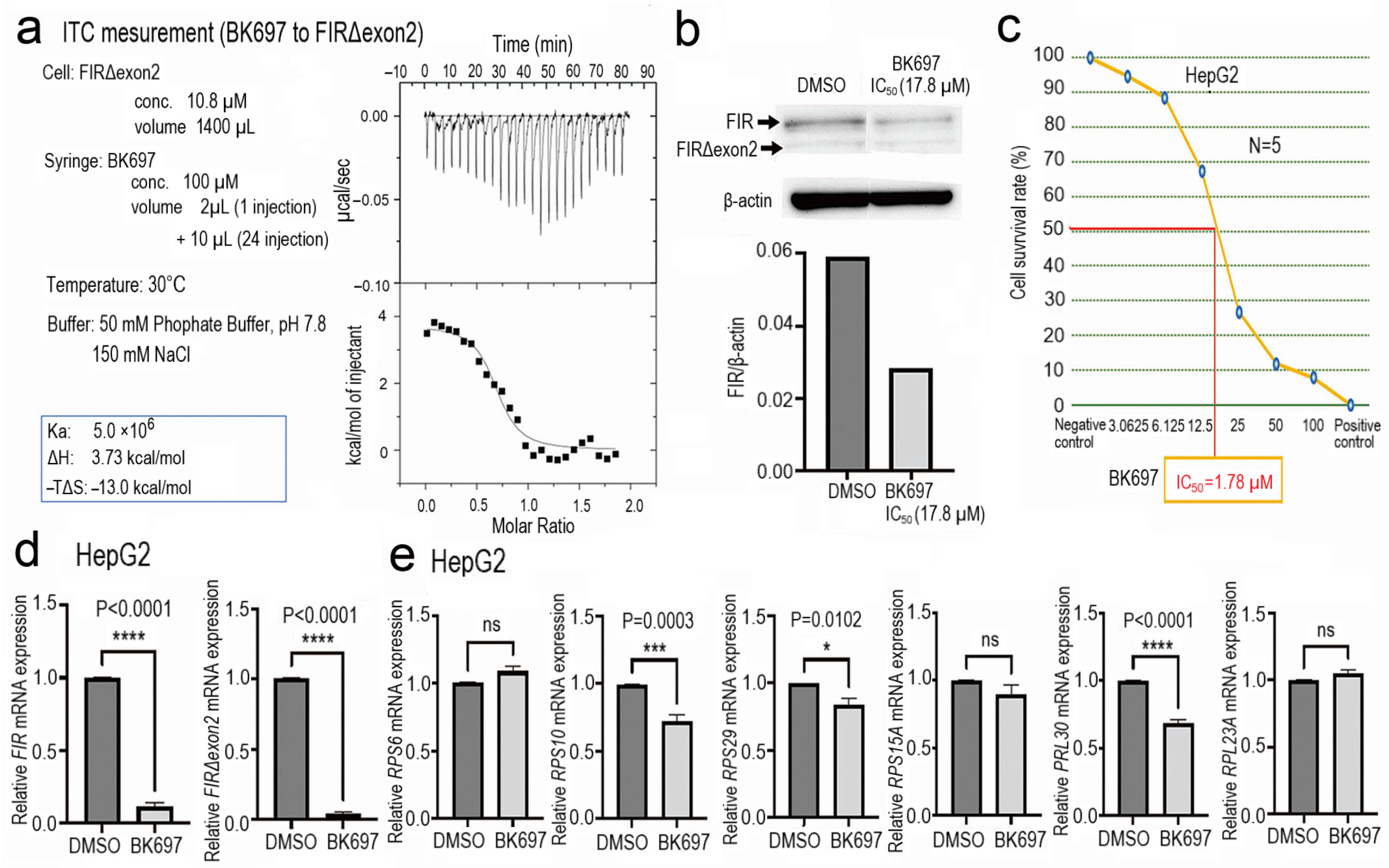
2.6. Cell Growth Inhibition with Altered Expression of rRNA by a Low-Molecular-Weight Chemical BK697 against FIRΔexon2
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Plasmids, and Plasmids Transfection
4.2. RNA Extraction and Quantitative Reverse Transcription (qRT)-PCR
4.3. RNA-Seq and Data Analysis
4.4. Western Blotting
4.5. siRNAs for Specific Proteins Expression
4.6. X-ray Crystal Structure Analysis
4.7. Protein Expression and Purification of FIR, FIR∆exon2, and p62 PH Domain
4.8. ITC Measurement
4.9. Procedure of In Silico Screening of Small Molecular Chemical Compounds BK697 against FIRΔexon2
4.10. MTS Assay (Cell Proliferation Assay)
4.11. Yeast Two-Hybrid Analysis
4.12. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piazzi, M.; Bavelloni, A.; Gallo, A.; Faenza, I.; Blalock, W.L. Signal Transduction in Ribosome Biogenesis: A Recipe to Avoid Disaster. Int. J. Mol. Sci. 2019, 20, 2718. [Google Scholar] [CrossRef]
- Assfalg, R.; Lebedev, A.; Gonzalez, O.G.; Schelling, A.; Koch, S. Iben STFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012, 40, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Suwa, T.; Suzuki, H.; Yamaguchi, Y.; Nishimura, Y. Three human RNA polymerases interact with TFIIH via a common RPB6 subunit. Nucleic Acids Res. 2022, 50, 1–16. [Google Scholar] [CrossRef]
- Abraham, K.J.; Khosraviani, N.; Chan, J.N.Y.; Gorthi, A.; Samman, A.; Zhao, D.Y.; Wang, M.; Bokros, M.; Vidya, E.; Ostrowski, L.A.; et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 2020, 585, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Doudna Cate, J.H. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef] [PubMed]
- Wandrey, F.; Montellese, C.; Koos, K.; Badertscher, L.; Bammert, L.; Cook, A.G.; Zemp, I.; Horvath, P.; Kutay, U. The NF45/NF90 heterodimer contributes to the biogenesis of 60S ribosomal subunits and influences nucleolar morphology. Mol. Cell Biol. 2015, 35, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Poot, M.; Hochstenbach, R. Prevalence and Phenotypic Impact of Robertsonian Translocations. Mol. Syndr. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Worton, R.G.; Sutherland, J.; Sylvester, J.E.; Willard, H.F.; Bodrug, S.; Dubé, I.; Duff, C.; Kean, V.; Ray, P.N.; Schmickel, R.D. Human ribosomal RNA genes: Orientation of the tandem array and conservation of the 5′ end. Science 1988, 239, 64–68. [Google Scholar] [CrossRef]
- Liu, J.; He, L.; Collins, I.; Ge, H.; Libutti, D.; Li, J.; Egly, J.-M.; Levens, D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell 2000, 5, 331–334. [Google Scholar] [CrossRef]
- Liu, J.; Akoulitchev, S.; Weber, A.; Ge, H.; Chuikov, S.; Libutti, D.; Wang, X.W.; Conaway, J.W.; Harris, C.C.; Conaway, R.C.; et al. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 2001, 104, 353–363. [Google Scholar] [CrossRef]
- Page-McCaw, P.S.; Amonlirdviman, K.; Sharp, P.A. PUF60: A novel U2AF65-related splicing activity. RNA 1999, 5, 1548–1560. [Google Scholar] [CrossRef]
- Cukier, C.D.; Hollingworth, D.; Martin, S.R.; Kelly, G.; Díaz-Moreno, I.; Ramos, A. Molecular basis of FIR-mediated c-myc transcriptional control. Nat. Struct. Mol. Biol. 2010, 17, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Tomonaga, T.; Shimada, H.; Shioya, A.; Higashi, M.; Matsubara, H.; Harigaya, K.; Nomura, F.; Libutti, D.; Levens, D.; et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006, 66, 1409–1417. [Google Scholar] [CrossRef]
- Kajiwara, T.; Matsushita, K.; Itoga, S.; Tamura, M.; Tanaka, N.; Tomonaga, T.; Matsubara, H.; Shimada, H.; Habara, Y.; Matsuo, M.; et al. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. 2013, 104, 149–156. [Google Scholar] [CrossRef]
- Matsushita, K.; Kajiwara, T.; Tamura, M.; Satoh, M.; Tanaka, N.; Tomonaga, T.; Matsubara, H.; Shimada, H.; Yoshimoto, R.; Ito, A.; et al. SAP155-mediated splicing of FUSE-binding protein-interacting repressor (FIR) serves as a molecular switch for c-myc gene expression. Mol. Cancer Res. 2012, 10, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Corsini, L.; Hothorn, M.; Stier, G.; Rybin, V.; Scheffzek, K.; Gibson, T.J.; Sattler, M. Dimerization and Protein Binding Specificity of the U2AF Homology Motif of the Splicing Factor Puf60. J. Biol. Chem. 2009, 284, 630–639. [Google Scholar] [CrossRef]
- Han, H.; Best, A.J.; Braunschweig, U.; Mikolajewicz, N.; Li, J.D.; Roth, J.; Chowdhury, F.; Mantica, F.; Nabeel-Shah, S.; Parada, G.; et al. Systematic exploration of dynamic splicing networks reveals conserved multistage regulators of neurogenesis. Mol. Cell 2022, 82, 2982–2999. [Google Scholar] [CrossRef] [PubMed]
- Ailiken, G.; Kitamura, K.; Hoshino, T.; Satoh, M.; Tanaka, N.; Minamoto, T.; Rahmutulla, B.; Kobayashi, S.; Kano, M.; Tanaka, T.; et al. Posttranscriptional regulation of BRG1 by FIRΔexon2 in gastric cancer. Oncogenesis 2020, 9, 26. [Google Scholar] [CrossRef]
- Kanoh, N.; Kumashiro, S.; Simizu, S.; Kondoh, Y.; Hatakeyama, S.; Tashiro, H.; Osada, H. Immobilization of natural products on glass slides by using a photoaffinity reaction and the detection of protein-small-molecule interactions. Angew. Chem. Int. Ed. Engl. 2003, 42, 5584–5587. [Google Scholar] [CrossRef]
- Osada, H. Introduction of new tools for chemical biology research on microbial metabolites. Biosci. Biotechnol. Biochem. 2010, 74, 1135–1140. [Google Scholar] [CrossRef]
- Kawatani, M.; Fukushima, Y.; Kondoh, Y.; Honda, K.; Sekine, T.; Yamaguchi, Y.; Taniguchi, N.; Osada, H. Identification of matrix metalloproteinase inhibitors by chemical arrays. Biosci. Biotechnol. Biochem. 2015, 79, 1597–1602. [Google Scholar] [CrossRef] [PubMed]
- Kampen, K.R.; Sulima, S.O.; Vereecke, S.; Keersmaecker, K.D. Hallmarks of ribosomopathies. Nucleic Acids Res. 2020, 48, 1013–1028. [Google Scholar] [CrossRef]
- Kobayashi, S.; Hoshino, T.; Hiwasa, T.; Satoh, M.; Rahmutulla, B.; Tsuchida, S.; Komukai, Y.; Tanaka, T.; Matsubara, H.; Shimada, H.; et al. Anti-FIRs (PUF60) auto-antibodies are detected in the sera of early-stage colon cancer patients. Oncotarget 2016, 7, 82493–82503. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, D.F.; Presby, M.; Baer, A.N.; Petri, M.; Rieger, K.E.; Soloski, M.; Rosen, A.; Mammen, A.L.; Christopher-Stine, L.; Casciola-Rosen, L. Puf60: A prominent new target of the autoimmune response in dermatomyositis and sjogren’s syndrome. Ann. Rheum. Dis. 2015, 75, 1145–1151. [Google Scholar] [CrossRef]
- Sun, Y.; Wei, G.; Luo, H.; Wu, W.; Skogerbø, G.; Luo, J.; Chen, R. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene 2017, 36, 6774–6783. [Google Scholar] [CrossRef]
- Pagon, R.A.; Graham, J.M.; Jr Zonana, J.; Yong, S.L. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J. Pediatr. 1981, 99, 223–227. [Google Scholar] [CrossRef]
- El Chehadeh, S.; Kerstjens-Frederikse, W.S.; Thevenon, J.; Kuentz, P.; Bruel, A.-L.; Thauvin-Robinet, C.; Bensignor, C.; Dollfus, H.; Laugel, V.; Rivière, J.-B.; et al. Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur. J. Hum. Genet. 2016, 25, 43–51. [Google Scholar] [CrossRef]
- Verheij, J.; de Munnik, S.; Dijkhuizen, T.; de Leeuw, N.; Weghuis, D.O.; Hoek, G.v.D.; Rijlaarsdam, R.; Thomasse, Y.; Dikkers, F.; Marcelis, C.; et al. An 8.35 Mb overlapping interstitial deletion of 8q24 in two patients with coloboma, congenital heart defect, limb abnormalities, psychomotor retardation, and convulsions. Eur. J. Med. Genet. 2009, 52, 353–357. [Google Scholar] [CrossRef]
- Dauber, A.; Golzio, C.; Guenot, C.; Jodelka, F.M.; Kibaek, M.; Kjaergaard, S.; Leheup, B.; Martinet, D.; Nowaczyk, M.J.; Rosenfeld, J.A.; et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 2018, 93, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. c-Myc binds to human ribosomal DNA stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef]
- Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Kitamura, K.; Ailiken, G.; Satoh, M.; Minamoto, T.; Tanaka, N.; Nomura, F.; Matsushita, K. FIR haplodeficiency promotes splicing to pyruvate kinase M2 in mice thymic lymphoma tissues revealed by six-plex tandem mass tag quantitative proteomic analysis. Oncotarget 2017, 8, 67955–67965. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xiong, X.; Sun, Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci. China Life Sci. 2016, 59, 656–672. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, J.M.; Dash, A.P.; Prochownik, E.V. Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 2017, 18, 275. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Nag, S.; Zhang, X.; Wang, M.; Wang, H.; Zhou, J.; Zhang, R. Ribosomal proteins and human diseases: Pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 2015, 35, 225–285. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Z.; Liu, N.; Chen, Y. Ribosomal protein L10 in mitochondria serves as a regulator for ROS level in pancreatic cancer cells. Redox Biol. 2018, 19, 158–165. [Google Scholar] [CrossRef]
- Russo, A.; Pagliara, V.; Albano, F.; Esposito, D.; Sagar, V.; Loreni, F.; Irace, C.; Santamaria, R.; Russo, G. Regulatory role of rpL3 in cell response to nucleolar stress induced by Act D in tumor cells lacking functional p53. Cell Cycle 2016, 15, 41–51. [Google Scholar] [CrossRef]
- Chen, J.; Crutchley, J.; Zhang, D.; Owzar, K.; Kastan, M.B. Identification of a DNA Damage-Induced Alternative Splicing Pathway That Regulates p53 and Cellular Senescence Markers. Cancer Discov. 2017, 7, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Xu, L.; Pelosof, L.; Wang, R.; McFarland, H.I.; Wu, W.W.; Phue, J.-N.; Lee, C.-T.; Shen, R.-F.; Juhl, H.; Wu, L.-H.; et al. NGS Evaluation of Colorectal Cancer Reveals Interferon Gamma Dependent Expression of Immune Checkpoint Genes and Identification of Novel IFNγ Induced Genes. Front. Immunol. 2020, 11, 224. [Google Scholar] [CrossRef]
- Crichlow, G.V.; Zhou, H.; Hsiao, H.-H.; Frederick, K.B.; Debrosse, M.; Yang, Y.; Folta-Stogniew, E.J.; Chung, H.-J.; Fan, C.; De La Cruz, E.M.; et al. Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. EMBO J. 2008, 27, 277–289. [Google Scholar] [CrossRef]
- Rahmutulla, B.; Matsushita, K.; Satoh, M.; Seimiya, M.; Tsuchida, S.; Kubo, S.; Shimada, H.; Ohtsuka, M.; Miyazaki, M.; Nomura, F. Alternative splicing of FBP-interacting repressor coordinates c-Myc, P27Kip1/cyclinE and Ku86/XRCC5 expression as a molecular sensor for bleomycin-induced DNA damage pathway. Oncotarget 2014, 5, 2404–2417. [Google Scholar] [CrossRef]
- Levens, D.; Baranello, L.; Kouzine, F. Controlling gene expression by DNA mechanics: Emerging insights and challenges. Biophys. Rev. 2016, 8, 259–268. [Google Scholar] [CrossRef]
- Zentner, G.E.; Hurd, E.A.; Schnetz, M.P.; Handoko, L.; Wang, C.; Wang, Z.; Wei, C.; Tesar, P.J.; Hatzoglou, M.; Martin, D.M.; et al. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum. Mol. Genet. 2010, 19, 3491–3501. [Google Scholar] [CrossRef]
- Marino, M.M.; Rega, C.; Russo, R.; Valletta, M.; Gentile, M.T.; Esposito, S.; Baglivo, I.; De Feis, I.; Angelini, C.; Xiao, T.; et al. Interactome mapping defines BRG1 a component of the SWI/SNF chromatin remodeling complex as a new partner of the transcriptional regulator CTCF. J. Biol. Chem. 2018, 294, 861–873. [Google Scholar] [CrossRef]
- Zhai, N.; Zhao, Z.-L.; Cheng, M.-B.; Di, Y.-W.; Yan, H.-X.; Cao, C.-Y.; Dai, H.; Zhang, Y.; Shen, Y.-F. Human PIH1 associates with histone H4 to mediate the glucose-dependent enhancement of pre-rRNA synthesis. J. Mol. Cell Biol. 2012, 4, 231–241. [Google Scholar] [CrossRef]
- Yang, P.; Oldfield, A.; Kim, T.; Yang, A.; Yang, J.Y.H.; Ho, J.W.K. Integrative analysis identifies co-dependent gene expression regulation of BRG1 and CHD7 at distal regulatory sites in embryonic stem cells. Bioinformatics 2017, 33, 1916–1920. [Google Scholar] [CrossRef] [PubMed]
- Chau, K.F.; Shannon, M.L.; Fame, R.M.; Fonseca, E.; Mullan, H.; Johnson, M.B.; Sendamarai, A.K.; Springel, M.W.; Laurent, B.; Lehtinen, M.K. Downregulation of ribosome biogenesis during early forebrain development. eLife 2018, 7, e36998. [Google Scholar] [CrossRef]
- Ogura, Y.; Hoshino, T.; Tanaka, N.; Ailiken, G.; Kobayashi, S.; Kitamura, K.; Rahmutulla, B.; Kano, M.; Murakami, K.; Akutsu, Y.; et al. Disturbed alternative splicing of FIR (PUF60) directed cyclin E overexpression in esophageal cancers. Oncotarget 2018, 9, 22929–22944. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitamura, K.; Hoshino, T.; Okabe, A.; Fukuyo, M.; Rahmutulla, B.; Tanaka, N.; Kobayashi, S.; Tanaka, T.; Shida, T.; Ueda, M.; et al. The Link of mRNA and rRNA Transcription by PUF60/FIR through TFIIH/P62 as a Novel Therapeutic Target for Cancer. Int. J. Mol. Sci. 2023, 24, 17341. https://doi.org/10.3390/ijms242417341
Kitamura K, Hoshino T, Okabe A, Fukuyo M, Rahmutulla B, Tanaka N, Kobayashi S, Tanaka T, Shida T, Ueda M, et al. The Link of mRNA and rRNA Transcription by PUF60/FIR through TFIIH/P62 as a Novel Therapeutic Target for Cancer. International Journal of Molecular Sciences. 2023; 24(24):17341. https://doi.org/10.3390/ijms242417341
Chicago/Turabian StyleKitamura, Kouichi, Tyuji Hoshino, Atsushi Okabe, Masaki Fukuyo, Bahityar Rahmutulla, Nobuko Tanaka, Sohei Kobayashi, Tomoaki Tanaka, Takashi Shida, Mashiro Ueda, and et al. 2023. "The Link of mRNA and rRNA Transcription by PUF60/FIR through TFIIH/P62 as a Novel Therapeutic Target for Cancer" International Journal of Molecular Sciences 24, no. 24: 17341. https://doi.org/10.3390/ijms242417341
APA StyleKitamura, K., Hoshino, T., Okabe, A., Fukuyo, M., Rahmutulla, B., Tanaka, N., Kobayashi, S., Tanaka, T., Shida, T., Ueda, M., Minamoto, T., Matsubara, H., Kaneda, A., Ishii, H., & Matsushita, K. (2023). The Link of mRNA and rRNA Transcription by PUF60/FIR through TFIIH/P62 as a Novel Therapeutic Target for Cancer. International Journal of Molecular Sciences, 24(24), 17341. https://doi.org/10.3390/ijms242417341






