Epigenomic Profiling Advises Therapeutic Potential of Leukotriene Receptor Inhibitors for a Subset of Triple-Negative Breast Tumors
Abstract
1. Introduction
2. Results
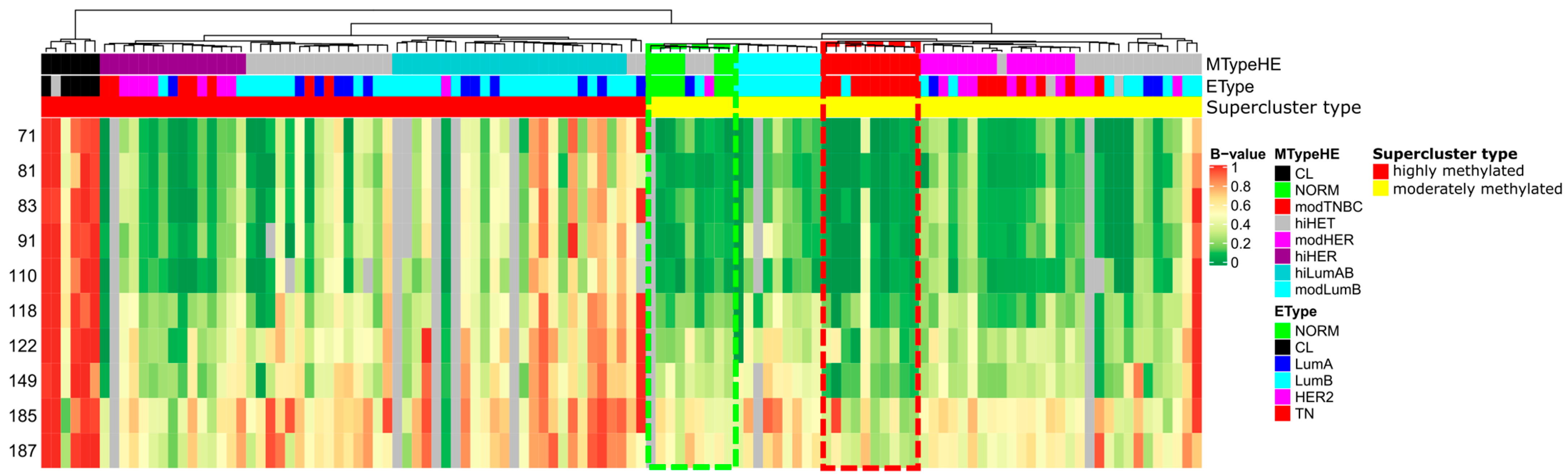
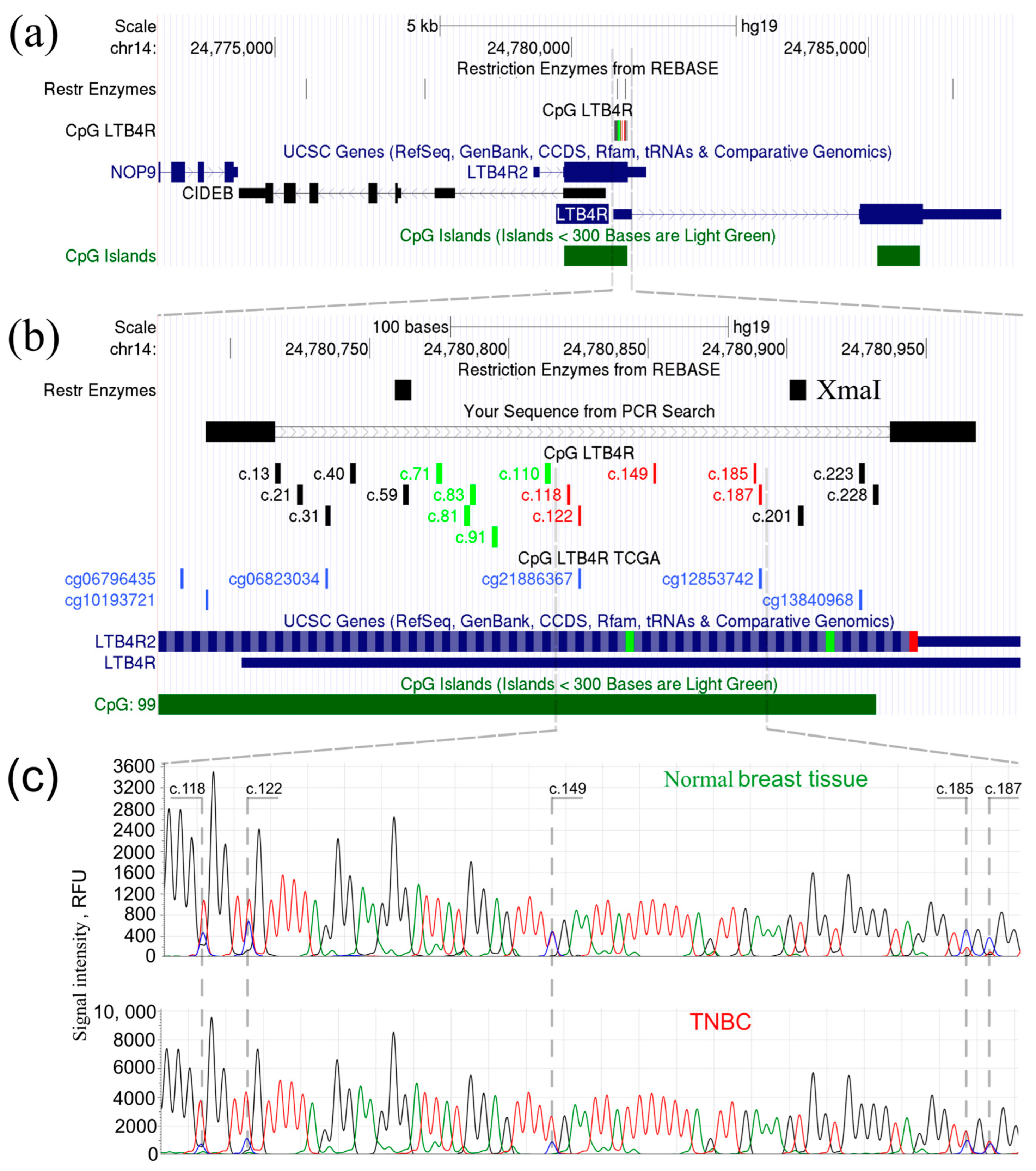
2.1. LTB4R/LTB4R2 Genes Are Abnormally Hypomethylated in a Subset of TNBC Samples
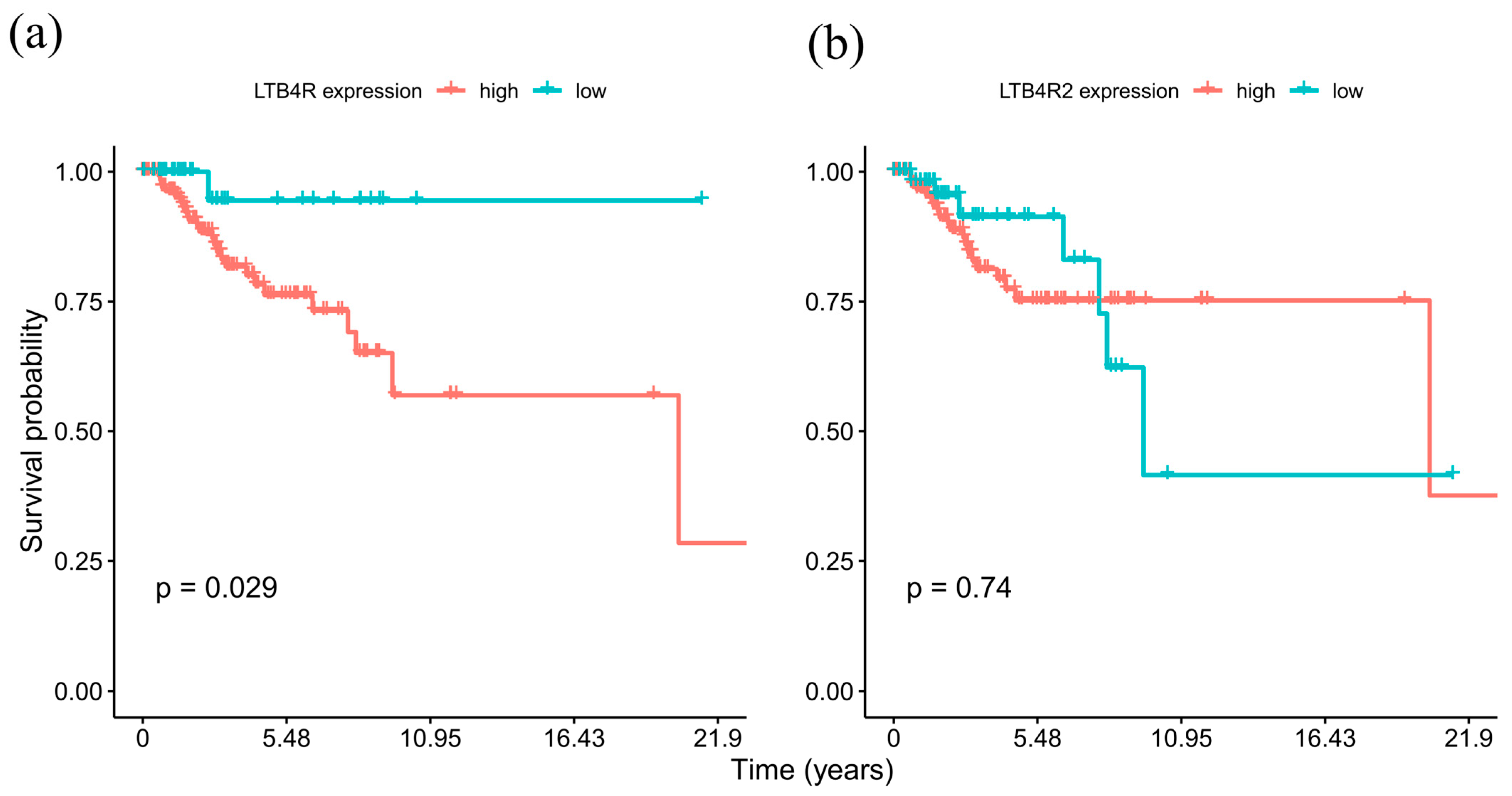
2.2. High LTB4R Expression Is Associated with Poor Prognosis in TNBC
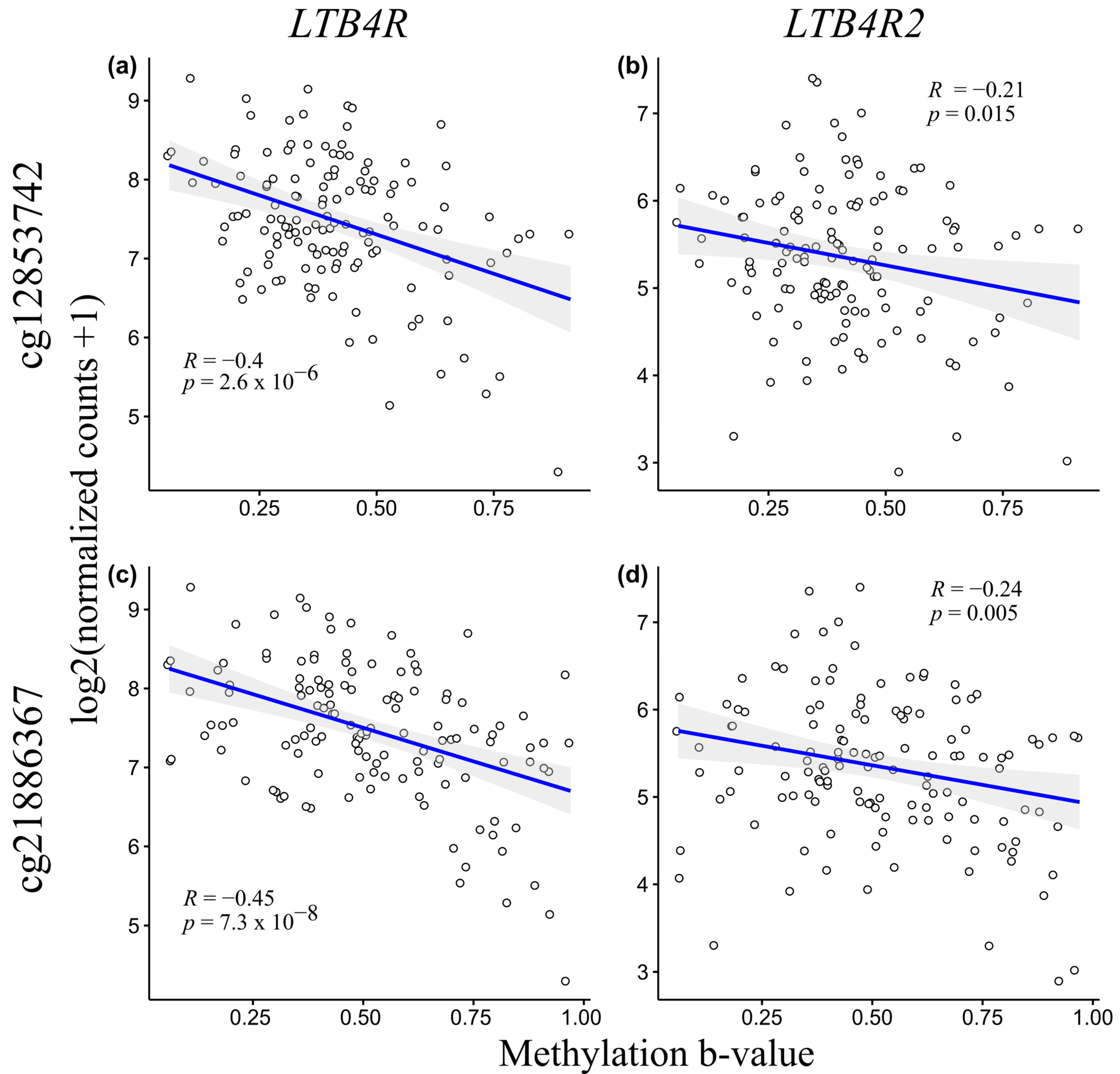
2.3. LTB4R and LTB4R2 CpG Methylation Levels Negatively Correlate with mRNA Expression
2.4. Leukotriene Receptor Inhibitors as Potential New Therapeutic Agents for TNBC
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. XmaI-RRBS
4.3. Data Collecting
4.4. Bisulfite Sanger Sequencing
4.5. Statistical Analysis and Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prat, A.; Parker, J.S.; Fan, C.; Perou, C.M. PAM50 Assay and the Three-Gene Model for Identifying the Major and Clinically Relevant Molecular Subtypes of Breast Cancer. Breast Cancer Res. Treat. 2012, 135, 301. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176. [Google Scholar] [CrossRef]
- Tray, N.; Adams, S.; Esteva, F.J. Antibody-Drug Conjugates in Triple Negative Breast Cancer. Future Oncol. 2018, 14, 2651–2661. [Google Scholar] [CrossRef] [PubMed]
- Sakach, E.; Sacks, R.; Kalinsky, K. Trop-2 as a Therapeutic Target in Breast Cancer. Cancers 2022, 14, 5936. [Google Scholar] [CrossRef]
- El Guerrab, A.; Bamdad, M.; Bignon, Y.J.; Penault-Llorca, F.; Aubel, C. Co-Targeting EGFR and MTOR with Gefitinib and Everolimus in Triple-Negative Breast Cancer Cells. Sci. Rep. 2020, 10, 6367. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Dong, C.; Cao, Y.Z.; Ma, B.L. Dual Target of EGFR and MTOR Suppresses Triple-Negative Breast Cancer Cell Growth by Regulating the Phosphorylation of MTOR Downstream Proteins. Breast Cancer Targets Ther. 2023, 15, 11–24. [Google Scholar] [CrossRef]
- Barchiesi, G.; Roberto, M.; Verrico, M.; Vici, P.; Tomao, S.; Tomao, F. Emerging Role of PARP Inhibitors in Metastatic Triple Negative Breast Cancer. Current Scenario and Future Perspectives. Front. Oncol. 2021, 11, 769280. [Google Scholar] [CrossRef] [PubMed]
- Muvarak, N.E.; Chowdhury, K.; Xia, L.; Robert, C.; Choi, E.Y.; Cai, Y.; Bellani, M.; Zou, Y.; Singh, Z.N.; Duong, V.H.; et al. Enhancing the Cytotoxic Effects of PARP Inhibitors with DNA Demethylating Agents—A Potential Therapy for Cancer. Cancer Cell 2016, 30, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Terranova-Barberio, M.; Thomas, S.; Ali, N.; Pawlowska, N.; Park, J.; Krings, G.; Rosenblum, M.D.; Budillon, A.; Munster, P.N. HDAC Inhibition Potentiates Immunotherapy in Triple Negative Breast Cancer. Oncotarget 2017, 8, 114156–114172. [Google Scholar] [CrossRef]
- Smith, E.R.; Huang, M.; Schlumbrecht, M.P.; George, S.H.L.; Xu, X.X. Rationale for Combination of Paclitaxel and CDK4/6 Inhibitor in Ovarian Cancer Therapy—Non-Mitotic Mechanisms of Paclitaxel. Front. Oncol. 2022, 12, 907520. [Google Scholar] [CrossRef]
- Yao, W.; Jiang, M.; Zhang, M.; Zhang, H.; Liang, X. TTK: A Promising Target in Malignant Tumors. J. Cell. Signal. 2021, 2, 212–220. [Google Scholar] [CrossRef]
- Li, Y.; Zhan, Z.; Yin, X.; Fu, S.; Deng, X. Targeted Therapeutic Strategies for Triple-Negative Breast Cancer. Front. Oncol. 2021, 11, 731535. [Google Scholar] [CrossRef]
- Johnson, A.M.; Kleczko, E.K.; Nemenoff, R.A. Eicosanoids in Cancer: New Roles in Immunoregulation. Front. Pharmacol. 2020, 11, 595498. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.Y.; Pidgeon, G.P. Cross-Talk between Cancer Cells and the Tumour Microenvironment: The Role of the 5-Lipoxygenase Pathway. Int. J. Mol. Sci. 2017, 18, 236. [Google Scholar] [CrossRef]
- Kim, H.; Park, G.S.; Lee, J.E.; Kim, J.H. A Leukotriene B4 Receptor-2 Is Associated with Paclitaxel Resistance in MCF-7/DOX Breast Cancer Cells. Br. J. Cancer 2013, 109, 351. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choi, J.A.; Park, G.S.; Kim, J.H. BLT2 Up-Regulates Interleukin-8 Production and Promotes the Invasiveness of Breast Cancer Cells. PLoS ONE 2012, 7, e49186. [Google Scholar] [CrossRef]
- Kim, K.-M.; Lee, J.; Park, S.H.; Heo, Y.J.; Jang, J.R.; Kim, S.; Park, J.O.; Kang, W.K.; Lee, D.; Han, S.-U.; et al. Reproduction of Gastric Cancer Prognostic Score by Real-Time Quantitative Polymerase Chain Reaction Assay in an Independent Cohort. Precis. Future Med. 2017, 2, 27–32. [Google Scholar] [CrossRef]
- Bhatt, L.; Roinestad, K.; Van, T.; Springman, E.B. Recent Advances in Clinical Development of Leukotriene B4 Pathway Drugs. Semin. Immunol. 2017, 33, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.A.; Lee, J.W.; Kim, H.; Kim, E.Y.; Seo, J.M.; Ko, J.; Kim, J.H. Pro-Survival of Estrogen Receptor-Negative Breast Cancer Cells Is Regulated by a BLT2-Reactive Oxygen Species-Linked Signaling Pathway. Carcinogenesis 2010, 31, 543–551. [Google Scholar] [CrossRef]
- Tanas, A.S.; Sigin, V.O.; Kalinkin, A.I.; Litviakov, N.V.; Slonimskaya, E.M.; Ibragimova, M.K.; Ignatova, E.O.; Simonova, O.A.; Kuznetsova, E.B.; Kekeeva, T.V.; et al. Genome-Wide Methylotyping Resolves Breast Cancer Epigenetic Heterogeneity and Suggests Novel Therapeutic Perspectives. Epigenomics 2019, 11, 605–617. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, G.; Zhang, L.; Wang, Q.; Li, H.; Han, Y.; Xie, L.; Yan, Z.; Li, Y.; An, Y.; et al. Comprehensive Review of Web Servers and Bioinformatics Tools for Cancer Prognosis Analysis. Front. Oncol. 2020, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Luo, H.; Krawczyk, M.; Wei, W.; Wang, W.; Wang, J.; Flagg, K.; Hou, J.; Zhang, H.; Yi, S.; et al. DNA Methylation Markers for Diagnosis and Prognosis of Common Cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 7414–7419. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Golovastova, M.O.; Tsoy, L.V.; Bocharnikova, A.V.; Korolev, D.O.; Gancharova, O.S.; Alekseeva, E.A.; Kuznetsova, E.B.; Savvateeva, L.V.; Skorikova, E.E.; Strelnikov, V.V.; et al. The Cancer-Retina Antigen Recoverin as a Potential Biomarker for Renal Tumors. Tumour Biol. 2016, 37, 9899–9907. [Google Scholar] [CrossRef] [PubMed]
- Baldin, A.V.; Grishina, A.N.; Korolev, D.O.; Kuznetsova, E.B.; Golovastova, M.O.; Kalpinskiy, A.S.; Alekseev, B.Y.; Kaprin, A.D.; Zinchenko, D.V.; Savvateeva, L.V.; et al. Autoantibody against Arrestin-1 as a Potential Biomarker of Renal Cell Carcinoma. Biochimie 2019, 157, 26–37. [Google Scholar] [CrossRef]
- Wolf, I.; O’Kelly, J.; Rubinek, T.; Tong, M.; Nguyen, A.; Lin, B.T.; Tai, H.H.; Karlan, B.Y.; Koeffler, H.P. 15-Hydroxyprostaglandin Dehydrogenase Is a Tumor Suppressor of Human Breast Cancer. Cancer Res. 2006, 66, 7818–7823. [Google Scholar] [CrossRef]
- Han, J.; Puri, R.K. Analysis of the Cancer Genome Atlas (TCGA) Database Identifies an Inverse Relationship between Interleukin-13 Receptor A1 and A2 Gene Expression and Poor Prognosis and Drug Resistance in Subjects with Glioblastoma Multiforme. J. Neurooncol. 2018, 136, 463–474. [Google Scholar] [CrossRef]
- Györffy, B.; Bottai, G.; Fleischer, T.; Munkácsy, G.; Budczies, J.; Paladini, L.; Børresen-Dale, A.L.; Kristensen, V.N.; Santarpia, L. Aberrant DNA Methylation Impacts Gene Expression and Prognosis in Breast Cancer Subtypes. Int. J. Cancer 2016, 138, 87–97. [Google Scholar] [CrossRef]
- Wisastra, R.; Dekker, F.J. Inflammation, Cancer and Oxidative Lipoxygenase Activity Are Intimately Linked. Cancers 2014, 6, 1500–1521. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Yan, X.; Ma, J.; Chen, Z. LTB4R Promotes the Occurrence and Progression of Clear Cell Renal Cell Carcinoma (CcRCC) by Regulating the AKT/MTOR Signaling Pathway. Cells 2022, 11, 3606. [Google Scholar] [CrossRef]
- Long, S.; Ji, S.; Xiao, K.; Xue, P.; Zhu, S. Prognostic and Immunological Value of LTB4R in Pan-Cancer. Math. Biosci. Eng. 2021, 18, 9336–9356. [Google Scholar] [CrossRef]
- Weng, M.S.; Chang, J.H.; Hung, W.Y.; Yang, Y.C.; Chien, M.H. The Interplay of Reactive Oxygen Species and the Epidermal Growth Factor Receptor in Tumor Progression and Drug Resistance. J. Exp. Clin. Cancer Res. 2018, 37, 61. [Google Scholar] [CrossRef]
- Malla, R.R.; Surepalli, N.; Farran, B.; Malhotra, S.V.; Nagaraju, G.P. Reactive Oxygen Species (ROS): Critical Roles in Breast Tumor Microenvironment. Crit. Rev. Oncol. Hematol. 2021, 160, 103285. [Google Scholar] [CrossRef]
- Park, J.I.; Jang, J.H.; Park, G.S.; Chung, Y.; You, H.J.; Kim, J.H. BLT2, a Leukotriene B4 Receptor 2, as a Novel Prognostic Biomarker of Triple-Negative Breast Cancer. BMB Rep. 2018, 51, 373–377. [Google Scholar] [CrossRef]
- Jeon, W.K.; Choi, J.; Park, S.J.; Jo, E.J.; Lee, Y.K.; Lim, S.; Kim, J.H.; Letterio, J.J.; Liu, F.; Kim, S.J.; et al. The Proinflammatory LTB4/BLT1 Signal Axis Confers Resistance to TGF-Β1-Induced Growth Inhibition by Targeting Smad3 Linker Region. Oncotarget 2015, 6, 41650–41666. [Google Scholar] [CrossRef][Green Version]
- Tian, W.; Jiang, X.; Kim, D.; Guan, T.; Nicolls, M.R.; Rockson, S.G. Leukotrienes in Tumor-Associated Inflammation. Front. Pharmacol. 2020, 11, 563120. [Google Scholar] [CrossRef]
- Godinho-Pereira, J.; Lopes, M.D.; Garcia, A.R.; Botelho, H.M.; Malhó, R.; Figueira, I.; Brito, M.A. A Drug Screening Reveals Minocycline Hydrochloride as a Therapeutic Option to Prevent Breast Cancer Cells Extravasation across the Blood–Brain Barrier. Biomedicines 2022, 10, 1988. [Google Scholar] [CrossRef]
- Himmel, L.E.; Lustberg, M.B.; DeVries, A.C.; Poi, M.; Chen, C.S.; Kulp, S.K. Minocycline, a Putative Neuroprotectant, Co-Administered with Doxorubicin-Cyclophosphamide Chemotherapy in a Xenograft Model of Triple-Negative Breast Cancer. Exp. Toxicol. Pathol. 2016, 68, 505–515. [Google Scholar] [CrossRef]
- Pirouzpanah, M.B.; Sabzichi, M.; Pirouzpanah, S.; Chavoshi, H.; Samadi, N. Silibilin-Induces Apoptosis in Breast Cancer Cells by Modulating P53, P21, Bak and Bcl-Xl Pathways. Asian Pac. J. Cancer Prev. 2015, 16, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Molavi, O.; Narimani, F.; Asiaee, F.; Sharifi, S.; Tarhriz, V.; Shayanfar, A.; Hejazi, M.; Lai, R. Silibinin Sensitizes Chemo-Resistant Breast Cancer Cells to Chemotherapy. Pharm. Biol. 2017, 55, 729–739. [Google Scholar] [CrossRef]
- Lucas, J.; Hsieh, T.C.; Halicka, H.D.; Darzynkiewicz, Z.; Wu, J.M. Upregulation of PD-L1 Expression by Resveratrol and Piceatannol in Breast and Colorectal Cancer Cells Occurs via HDAC3/P300-mediated NF-κB Signaling. Available online: https://www.spandidos-publications.com/10.3892/ijo.2018.4512 (accessed on 1 November 2023).
- Zhang, Y.; Wang, J.; Hu, T.; Wang, H.; Long, M.; Liang, B. Adverse Events of PD-1 or PD-L1 Inhibitors in Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. Life 2022, 12, 1990. [Google Scholar] [CrossRef]
- Sahin, I.; Eturi, A.; De Souza, A.; Pamarthy, S.; Tavora, F.; Giles, F.J.; Carneiro, B.A. Glycogen Synthase Kinase-3 Beta Inhibitors as Novel Cancer Treatments and Modulators of Antitumor Immune Responses. Cancer Biol. Ther. 2019, 20, 1047–1056. [Google Scholar] [CrossRef]
- He, R.; Du, S.; Lei, T.; Xie, X.; Wang, Y. Glycogen Synthase Kinase 3β in Tumorigenesis and Oncotherapy (Review). Available online: https://www.spandidos-publications.com/10.3892/or.2020.7817 (accessed on 1 November 2023).
- Vijay, G.V.; Zhao, N.; Den Hollander, P.; Toneff, M.J.; Joseph, R.; Pietila, M.; Taube, J.H.; Sarkar, T.R.; Ramirez-Pena, E.; Werden, S.J.; et al. GSK3β Regulates Epithelial-Mesenchymal Transition and Cancer Stem Cell Properties in Triple-Negative Breast Cancer. Breast Cancer Res. 2019, 21, 37. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt Signal Transduction for Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Liu, H.; Gruber, C.W.; Alewood, P.F.; Möller, A.; Muttenthaler, M. The Oxytocin Receptor Signalling System and Breast Cancer: A Critical Review. Oncogene 2020, 39, 5917–5932. [Google Scholar] [CrossRef]
- Chen, J.; Tang, G. PIM-1 Kinase: A Potential Biomarker of Triple-Negative Breast Cancer. Onco Targets Ther. 2019, 12, 6267. [Google Scholar] [CrossRef]
- Hijazi, M.A.; Gessner, A.; El-Najjar, N. Repurposing of Chronically Used Drugs in Cancer Therapy: A Chance to Grasp. Cancers 2023, 15, 3199. [Google Scholar] [CrossRef]
- Behrouzi, B.; Zokaasadi, M.; Mohagheghi, M.A.; Emami, H.; Sadighi, S. The Effect of Metformin on Survival Outcomes of Non-Metastatic Breast Cancer Patients with Type 2 Diabetes. Asian Pac. J. Cancer Prev. 2021, 22, 611. [Google Scholar] [CrossRef]
- Suknuntha, K.; Yubolphan, R.; Krueaprasertkul, K.; Srihirun, S.; Sibmooh, N.; Vivithanaporn, P. Leukotriene Receptor Antagonists Inhibit Mitogenic Activity in Triple Negative Breast Cancer Cells. Asian Pac. J. Cancer Prev. 2018, 19, 833. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Bassil, C.F.; Murphy, S.K. Bisulfite Sequencing of Cloned Alleles. Methods Mol. Biol. 2013, 1049, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Tanas, A.S.; Borisova, M.E.; Kuznetsova, E.B.; Rudenko, V.V.; Karandasheva, K.O.; Nemtsova, M.V.; Izhevskaya, V.L.; Simonova, O.A.; Larin, S.S.; Zaletaev, D.V.; et al. Rapid and Affordable Genome-Wide Bisulfite DNA Sequencing by XmaI-Reduced Representation Bisulfite Sequencing. Epigenomics 2017, 9, 833–847. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A Flexible Aligner and Methylation Caller for Bisulfite-Seq Applications. Bioinformatics 2011, 27, 1571. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]


| Drug | MOA | R | SE |
|---|---|---|---|
| PICEATANNOL | SYK inhibitor | 0.99 | 3.60 × 10−5 |
| MITIGLINIDE | Insulin secretagogue | 0.99 | 0.0002 |
| 1-AZAKENPAULLONE | Glycogen synthase kinase inhibitor | 0.99 | 0.001 |
| CARBETOCIN | Oxytocin receptor agonist | 0.99 | 0.001 |
| PIM-1-INHIBITOR-2 | PIM kinase inhibitor | 0.99 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalinkin, A.I.; Sigin, V.O.; Kuznetsova, E.B.; Ignatova, E.O.; Vinogradov, I.I.; Vinogradov, M.I.; Vinogradov, I.Y.; Zaletaev, D.V.; Nemtsova, M.V.; Kutsev, S.I.; et al. Epigenomic Profiling Advises Therapeutic Potential of Leukotriene Receptor Inhibitors for a Subset of Triple-Negative Breast Tumors. Int. J. Mol. Sci. 2023, 24, 17343. https://doi.org/10.3390/ijms242417343
Kalinkin AI, Sigin VO, Kuznetsova EB, Ignatova EO, Vinogradov II, Vinogradov MI, Vinogradov IY, Zaletaev DV, Nemtsova MV, Kutsev SI, et al. Epigenomic Profiling Advises Therapeutic Potential of Leukotriene Receptor Inhibitors for a Subset of Triple-Negative Breast Tumors. International Journal of Molecular Sciences. 2023; 24(24):17343. https://doi.org/10.3390/ijms242417343
Chicago/Turabian StyleKalinkin, Alexey I., Vladimir O. Sigin, Ekaterina B. Kuznetsova, Ekaterina O. Ignatova, Ilya I. Vinogradov, Maxim I. Vinogradov, Igor Y. Vinogradov, Dmitry V. Zaletaev, Marina V. Nemtsova, Sergey I. Kutsev, and et al. 2023. "Epigenomic Profiling Advises Therapeutic Potential of Leukotriene Receptor Inhibitors for a Subset of Triple-Negative Breast Tumors" International Journal of Molecular Sciences 24, no. 24: 17343. https://doi.org/10.3390/ijms242417343
APA StyleKalinkin, A. I., Sigin, V. O., Kuznetsova, E. B., Ignatova, E. O., Vinogradov, I. I., Vinogradov, M. I., Vinogradov, I. Y., Zaletaev, D. V., Nemtsova, M. V., Kutsev, S. I., Tanas, A. S., & Strelnikov, V. V. (2023). Epigenomic Profiling Advises Therapeutic Potential of Leukotriene Receptor Inhibitors for a Subset of Triple-Negative Breast Tumors. International Journal of Molecular Sciences, 24(24), 17343. https://doi.org/10.3390/ijms242417343






