Effective Antiviral Application of Antisense in Plants by Exploiting Accessible Sites in the Target RNA
Abstract
:1. Introduction
2. Results
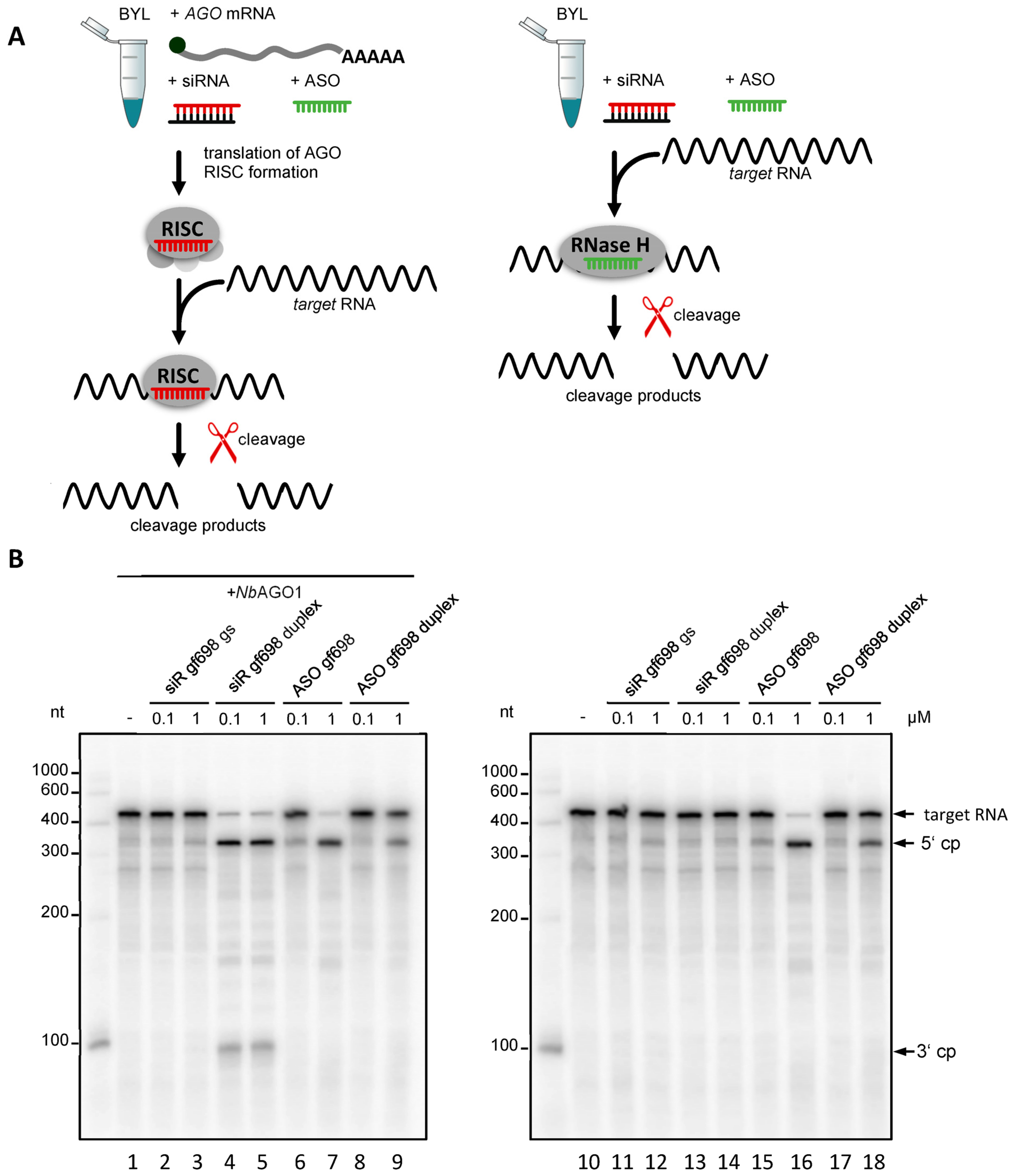
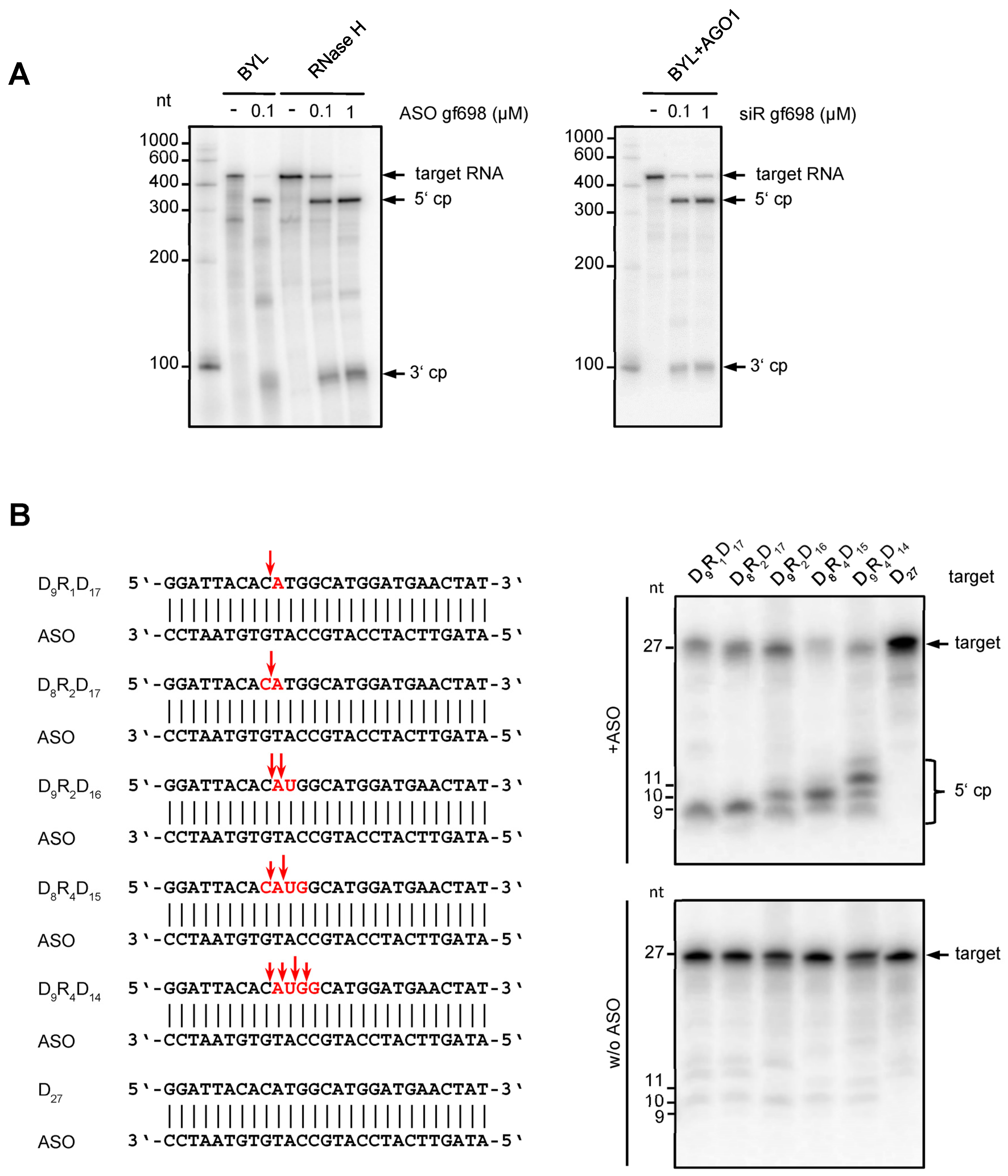
2.1. Nicotiana tabacum Cells Contain Different Types of RNase H Activities
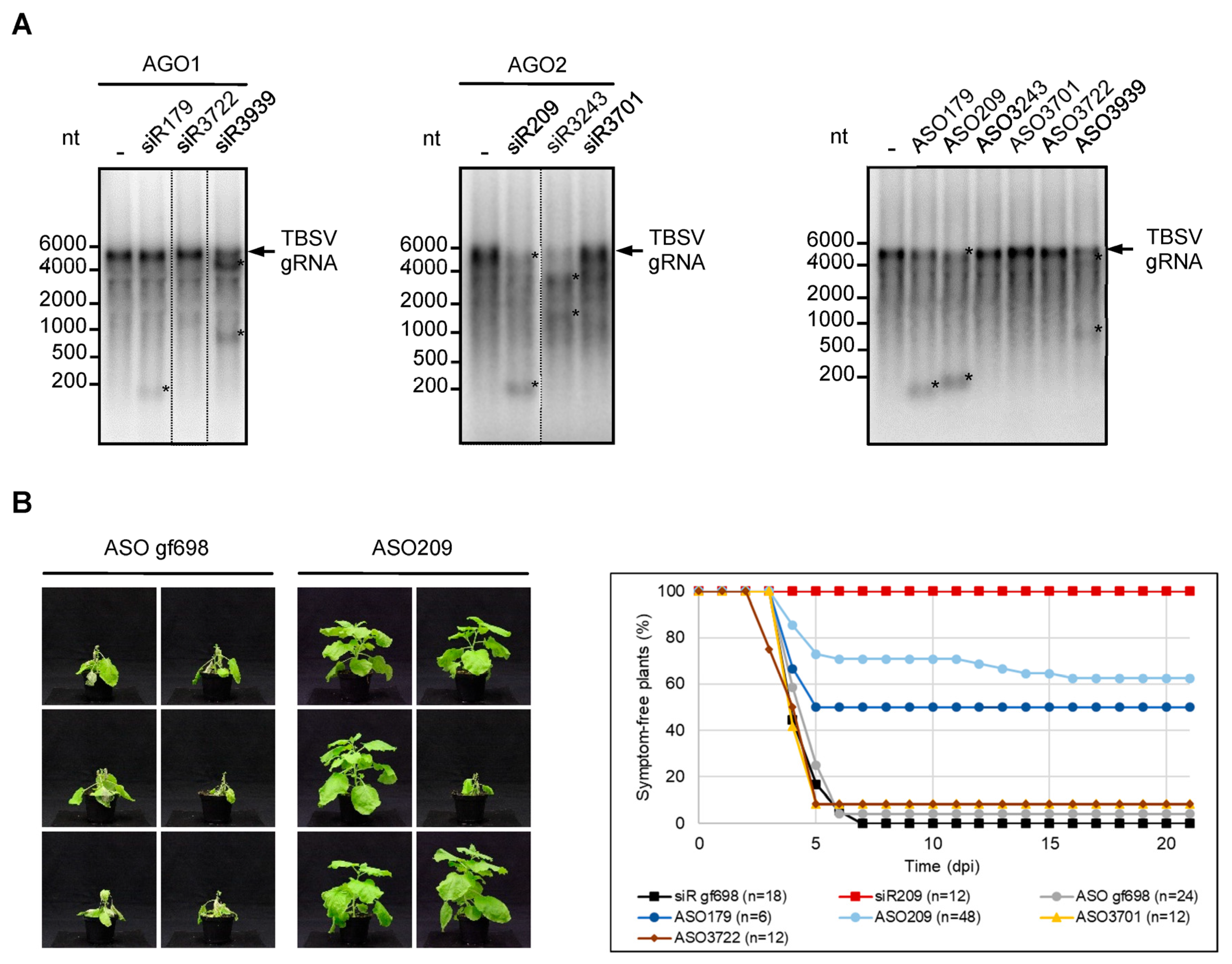
2.2. ASOs Derived from Antiviral esiRNAs Provide Antiviral Protection
2.3. Chemical Modification Increases the Antiviral Effect of eASO
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Preparation of BYL
4.2. In Vitro Transcription
4.3. Oligonucleotides and siRNAs
4.4. In Vitro Cleavage Assays
4.5. 3′-Rapid Amplification of cDNA Ends (3′-RACE)
4.6. Stability of ASOs and siRNAs in BYL
4.7. Plant Treatment, TBSV Challenge, and Analysis of Infection Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Guo, Z.; Li, Y.; Ding, S.-W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant. Res. 2017, 130, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.-S.; Bouteiller, N.; Elmayan, T.; Vaucheret, H. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2015, 81, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Iwasaki, S.; Watanabe, T.; Utsumi, M.; Watanabe, Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008, 49, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Setten, R.L.; Rossi, J.J.; Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Friedrich, M.; Aigner, A. Therapeutic siRNA: State-of-the-Art and Future Perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Corey, D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Liang, X.-H.; Bakre, B.F.; Crooke, R.M. Antisense technology: A review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef] [PubMed]
- Moelling, K.; Broecker, F.; Russo, G.; Sunagawa, S. RNase H as gene modifier, driver of evolution and antiviral defense. Front. Microbiol. 2017, 8, 1745. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef]
- Bevilacqua, P.C.; Ritchey, L.E.; Su, Z.; Assmann, S.Z. Genome-wide analysis of RNA secondary structure. Ann. Rev. Gen. 2016, 50, 235–266. [Google Scholar] [CrossRef] [PubMed]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Shao, Y.; Chan, C.Y.; Maliyekkel, A.; Lawrence, C.E.; Roninson, I.B.; Ding, Y. Effect of target secondary structure on RNAi efficiency. RNA 2007, 13, 1631–1640. [Google Scholar] [CrossRef]
- Tafer, H.; Ameres, S.L.; Obernosterer, G.; Gebeshuber, C.A.; Schroeder, R.; Martinez, J.; Hofacker, I.L. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008, 26, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Gago-Zachert, S.; Schuck, J.; Weinholdt, C.; Knoblich, M.; Große, I.; Pantaleo, V.; Gursinsky, T.; Behrens, S.-E. Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro. Nucleic Acids Res. 2019, 47, 9343–9357. [Google Scholar] [CrossRef]
- Vickers, T.A.; Crooke, S.T. The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acids Res. 2015, 43, 8955–8963. [Google Scholar] [CrossRef]
- Lucks, J.B.; Mortimer, S.A.; Trapnell, C.; Luo, S.; Aviran, S.; Schroth, G.P.; Pachter, L.; Doudna, J.A.; Arkin, A.P. Multiplexed RNA structure characterization with selective 2-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. USA 2011, 108, 11063–11068. [Google Scholar] [CrossRef]
- Spitale, R.C.; Incarnato, D. Probing the dynamic RNA structurome and its functions. Nat. Rev. Genet. 2023, 24, 178–196. [Google Scholar] [CrossRef]
- Lück, S.; Kreszies, T.; Strickert, M.; Schweizer, P.; Kuhlmann, M.; Douchkov, D. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 2019, 10, 1023. [Google Scholar] [CrossRef]
- Sciabola, S.; Xi, H.; Cruz, D.; Cao, Q.; Lawrence, C.; Zhang, T.; Rotstein, S.; Hughes, J.D.; Caffrey, D.R.; Stanton, R. PFRED: A computational platform for siRNA and antisense oligonucleotides design. PLoS ONE 2021, 16, e0238753. [Google Scholar] [CrossRef]
- Eastman, P.; Shi, J.; Ramsundar, B.; Pande, V.S. Solving the RNA design problem with reinforcement learning. PLOS Comput. Biol. 2018, 14, e1006176. [Google Scholar] [CrossRef]
- Carbonell, A.; López, C.; Daròs, J.-A. Fast-forward identification of highly effective artificial small RNAs against different Tomato spotted wilt virus isolates. Mol. Plant Microbe Interact. 2019, 32, 142–156. [Google Scholar] [CrossRef]
- Komoda, K.; Naito, S.; Ishikawa, M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. USA 2004, 101, 1863–1867. [Google Scholar] [CrossRef] [PubMed]
- Gursinsky, T.; Schulz, B.; Behrens, S.-E. Replication of Tomato bushy stunt virus RNA in a plant in vitro system. Virology 2009, 390, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Schuck, J.; Gursinsky, T.; Pantaleo, V.; Burgyán, J.; Behrens, S.-E. AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 2013, 41, 5090–5103. [Google Scholar] [CrossRef]
- Iki, T.; Yoshikawa, M.; Nishikiori, M.; Jaudal, M.C.; Matsumoto-Yokoyama, E.; Mitsuhara, I.; Meshi, T.; Ishikawa, M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 2010, 39, 282–291. [Google Scholar]
- Gursinsky, T.; Pirovano, W.; Gambino, G.; Friedrich, S.; Behrens, S.-E.; Pantaleo, V. Homeologs of the Nicotiana benthamiana antiviral ARGONAUTE1 show different susceptibilities to microRNA168-mediated control. Plant Physiol. 2015, 168, 938–952. [Google Scholar] [CrossRef] [PubMed]
- Pertermann, R.; Tamilarasan, S.; Gursinsky, T.; Gambino, G.; Schuck, J.; Weinholdt, C.; Lilie, H.; Grosse, I.; Golbik, R.P.; Pantaleo, V.; et al. A viral suppressor modulates the plant immune response early in infection by regulating microRNA activity. mBio 2018, 9, e00419-18. [Google Scholar] [CrossRef]
- Eder, P.S.; Walder, R.Y.; Walder, J.A. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie 1999, 75, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Kuciński, J.; Chamera, S.; Kmera, A.; Rowley, M.J.; Fujii, S.; Khurana, P.; Nowotny, M.; Wierzbicki, A.T. Evolutionary History and Activity of RNase H1-Like Proteins in Arabidopsis thaliana. Plant Cell Physiol. 2020, 61, 1107–1119. [Google Scholar] [CrossRef]
- Majorek, K.A.; Dunin-Horkawicz, S.; Steczkiewicz, K.; Muszewska, A.; Nowotny, M.; Ginalski, K.; Bujnicki, J.M. The RNase H-like superfamily: New members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014, 42, 4160–4179. [Google Scholar] [CrossRef]
- Chon, H.; Sparks, J.L.; Rychlik, M.; Nowotny, M.; Burgers, P.M.; Crouch, R.J.; Cerritelli, S.M. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013, 41, 3130–3143. [Google Scholar] [CrossRef]
- Jeong, H.-S.; Backlund, P.S.; Chen, H.-C.; Karavanov, A.A.; Crouch, R.J. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004, 32, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, Y.; Scholthof, H.B. Tomato bushy stunt virus. A resilient model system to study virus-plant interactions. Mol. Plant Pathol. 2005, 6, 491–502. [Google Scholar] [CrossRef]
- Campbell, J.M.; Bacon, T.A.; Wickstrom, E. Oligodeoxynucleoside phosphorothioate stability in subcellular extracts, culture media, sera and cerebrospinal fluid. J. Biochem. Bioph. Meth. 1990, 20, 259–267. [Google Scholar] [CrossRef]
- Voloudakis, A.E.; Holeva, M.; Sarin, P.L.; Bamford, D.H.; Vargas, M.; Poranen, M.M.; Tenllado, F. Efficient double-stranded RNA production methods for utilization in plant virus control. In Plant Virology Protocols. Methods in Molecular Biology; Uyeda, I., Masuta, C., Eds.; Humana Press: New York, NY, USA, 2014; pp. 255–274. [Google Scholar]
- Šponer, J.; Bussi, G.; Krepl, M.; Banáš, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurečka, P.; et al. RNA structural dynamics as captured by molecular simulations: A comprehensive overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Grigull, J.; Ore, M.O.; Morin, S.; White, K.A. Global organization of a positive-strand RNA virus genome. PLOS Pathog. 2013, 9, e1003363. [Google Scholar] [CrossRef] [PubMed]
- Miozzi, L.; Gambino, G.; Burgyan, J.; Pantaleo, V. Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol. Plant Pathol. 2013, 14, 30–43. [Google Scholar] [CrossRef]
- Cox, D.B.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Dinç, E.; Tóth, S.Z.; Schansker, G.; Ayaydin, F.; Kovács, L.; Dudits, D.; Garab, G.; Bottka, S. Synthetic antisense oligodeoxynucleotides to transiently suppress different nucleus- and chloroplast-encoded proteins of higher plant chloroplasts. Plant Physiol. 2011, 157, 1628–1641. [Google Scholar] [CrossRef]
- Kawai-Toyooka, H.; Kuramoto, C.; Orui, K.; Motoyama, K.; Kikuchi, K.; Kanegae, T.; Wada, M. DNA interference. A simple and efficient gene-silencing system for high-throughput functional analysis in the fern Adiantum. Plant Cell Physiol. 2004, 45, 1648–1657. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.; Wang, L.; Yang, L.-B.; Zhang, L.; Peng, X.; Sun, M.-X. Antisense oligodeoxynucleotide inhibition as an alternative and convenient method for gene function analysis in pollen tubes. PLoS ONE 2013, 8, e59112. [Google Scholar] [CrossRef]
- Nowak, M.; Wyszko, E.; Fedoruk-Wyszomirska, A.; Pospieszny, H.; Barciszewska, M.Z.; Barciszewski, J. A new and efficient method for inhibition of RNA viruses by DNA interference. FEBS J. 2009, 276, 4372–4380. [Google Scholar] [CrossRef] [PubMed]
- Krasnodębski, C.; Sawula, A.; Kazmierczak, U.; Zuk, M. Oligo—Not only for silencing: Overlooked potential for multidirectional action in plants. Int. J. Mol. Sci. 2023, 24, 4466. [Google Scholar] [CrossRef] [PubMed]
- Wdowikowska, A.; Janicka, M. Antisense oligonucleotide technology as a research tool in plant biology. Funct. Plant Biol. 2022, 49, 1–12. [Google Scholar] [CrossRef]
- Lima, W.; Wu, H.; Crooke, S.T. The RNase H mechanism. In Antisense Drug Technology—Principles, Strategies, and Applications, 2nd ed.; Crooke, S.T., Ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 47–74. [Google Scholar]
- Vickers, T.A.; Koo, S.; Bennett, C.F.; Crooke, S.T.; Dean, N.M.; Baker, B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H- dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003, 278, 7108–7118. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Laikova, K.V.; Repetskaya, A.I.; Kenyo, I.M.; Gorlov, M.V.; Kasich, I.N.; Krasnodubets, A.M.; Gal’chinsky, N.V.; Fomochkina, I.I.; Zaitsev, A.S.; et al. A half-century history of applications of aqntisense oligonucleotides in medicine, agriculture and forestry: We should continue the journey. Molecules 2018, 23, 1302. [Google Scholar] [CrossRef] [PubMed]
- Hearne, P.Q.; Knorr, D.A.; Hillman, B.I.; Morris, T.J. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 1990, 177, 141–151. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruber, C.; Gursinsky, T.; Gago-Zachert, S.; Pantaleo, V.; Behrens, S.-E. Effective Antiviral Application of Antisense in Plants by Exploiting Accessible Sites in the Target RNA. Int. J. Mol. Sci. 2023, 24, 17153. https://doi.org/10.3390/ijms242417153
Gruber C, Gursinsky T, Gago-Zachert S, Pantaleo V, Behrens S-E. Effective Antiviral Application of Antisense in Plants by Exploiting Accessible Sites in the Target RNA. International Journal of Molecular Sciences. 2023; 24(24):17153. https://doi.org/10.3390/ijms242417153
Chicago/Turabian StyleGruber, Cornelia, Torsten Gursinsky, Selma Gago-Zachert, Vitantonio Pantaleo, and Sven-Erik Behrens. 2023. "Effective Antiviral Application of Antisense in Plants by Exploiting Accessible Sites in the Target RNA" International Journal of Molecular Sciences 24, no. 24: 17153. https://doi.org/10.3390/ijms242417153
APA StyleGruber, C., Gursinsky, T., Gago-Zachert, S., Pantaleo, V., & Behrens, S.-E. (2023). Effective Antiviral Application of Antisense in Plants by Exploiting Accessible Sites in the Target RNA. International Journal of Molecular Sciences, 24(24), 17153. https://doi.org/10.3390/ijms242417153







