VEGFR and DPP-IV as Markers of Severe COVID-19 and Predictors of ICU Admission
Abstract
:1. Introduction
2. Results
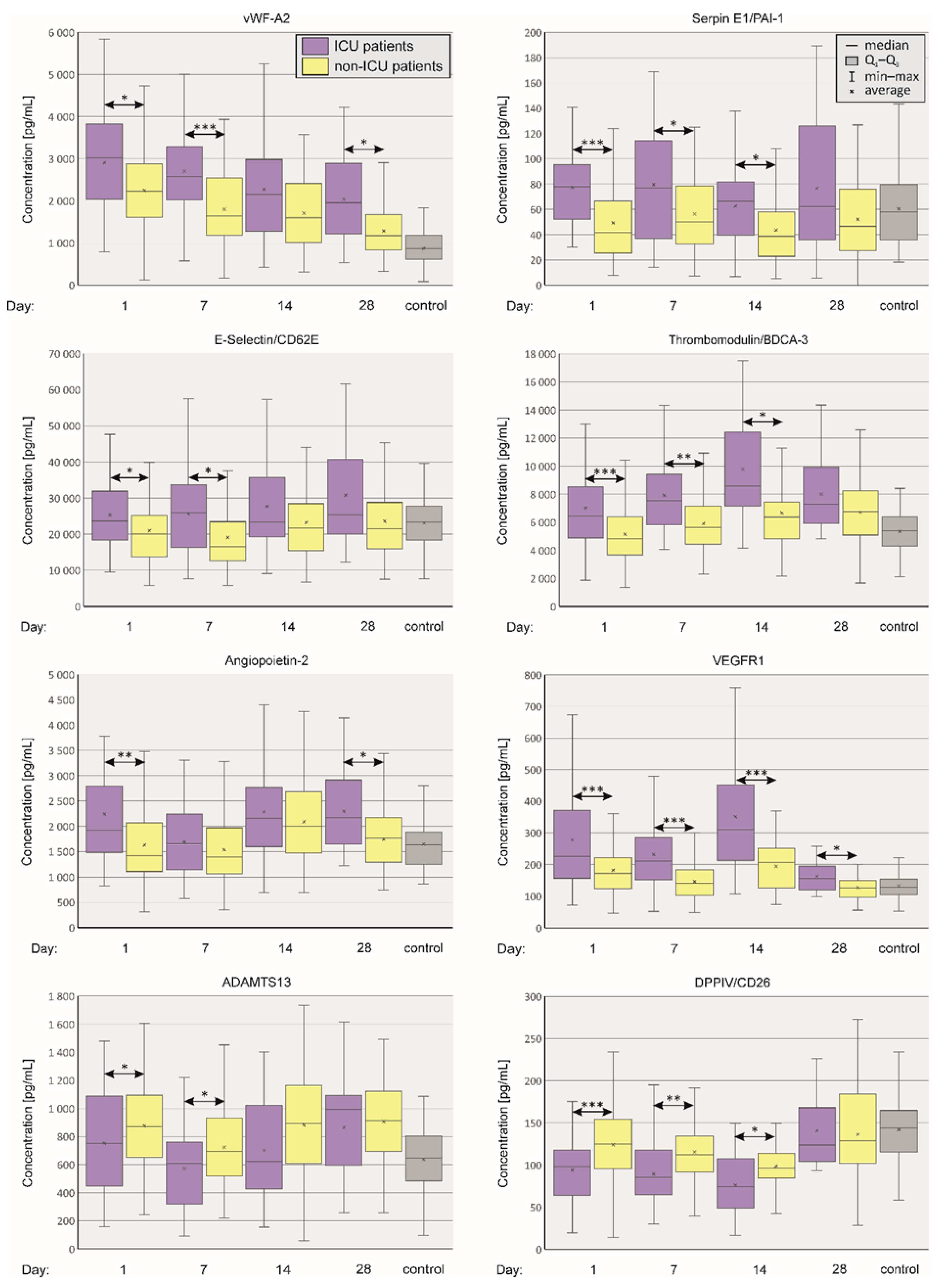
2.1. Comparison of Plasma Concentrations of Selected Vasoactive Agents between ICU and Non-ICU Patients
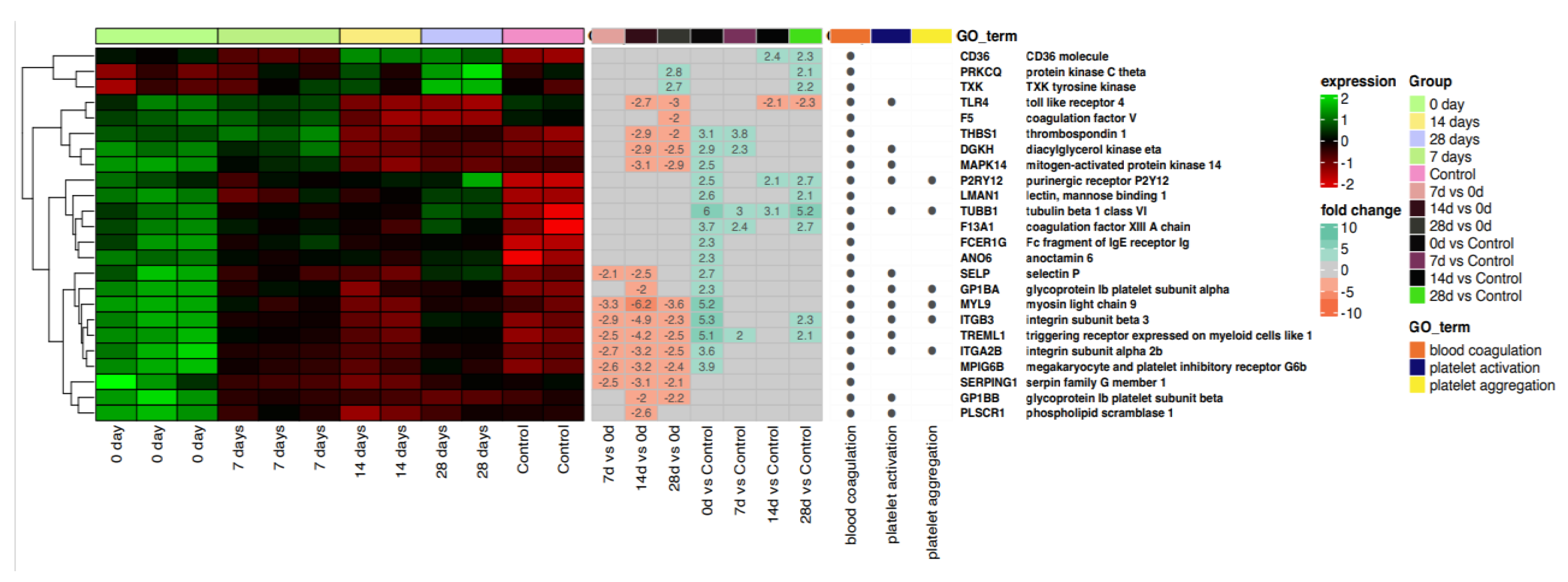
2.2. Blood Coagulation, Platelet Activation and Aggregation
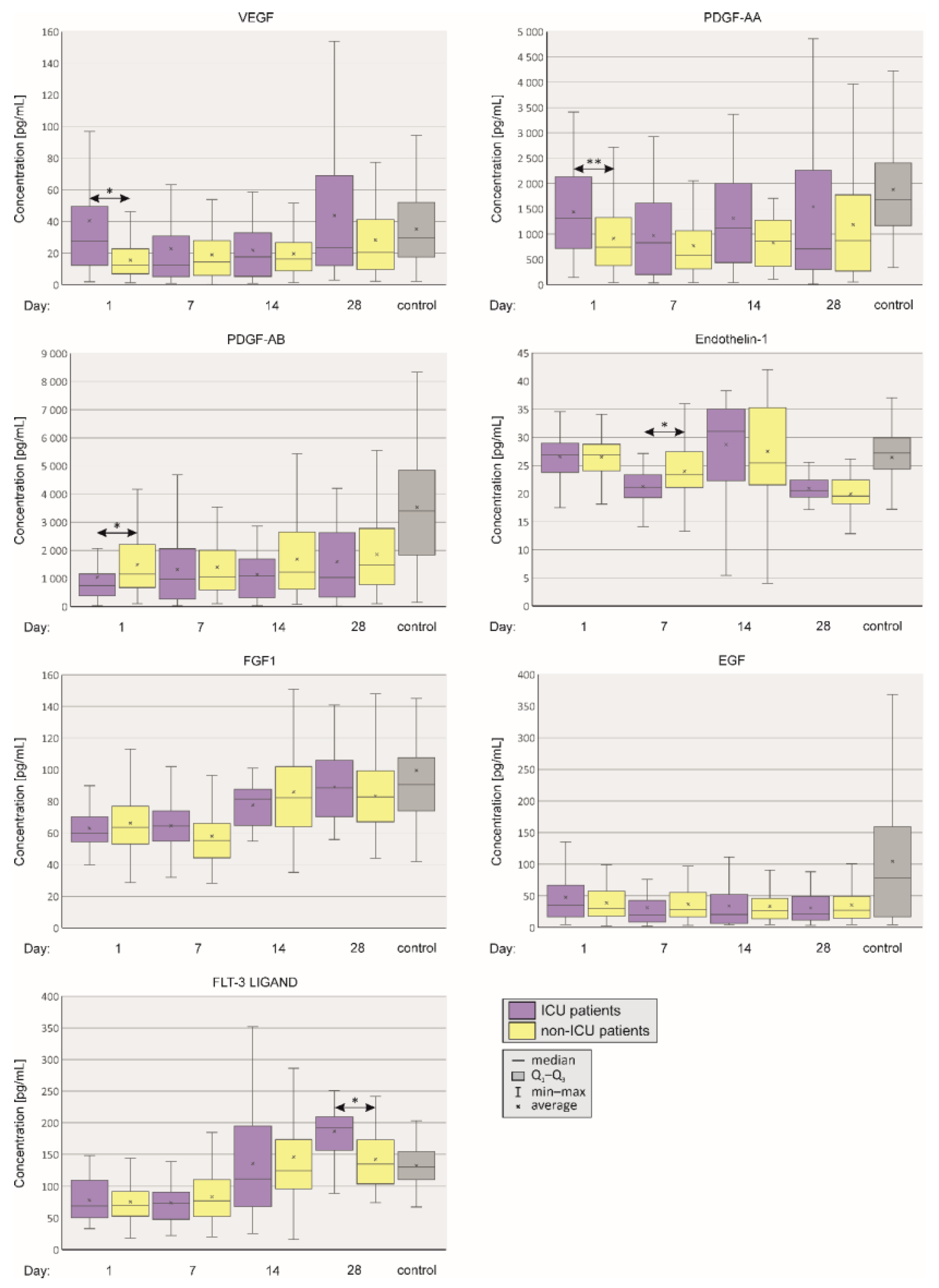
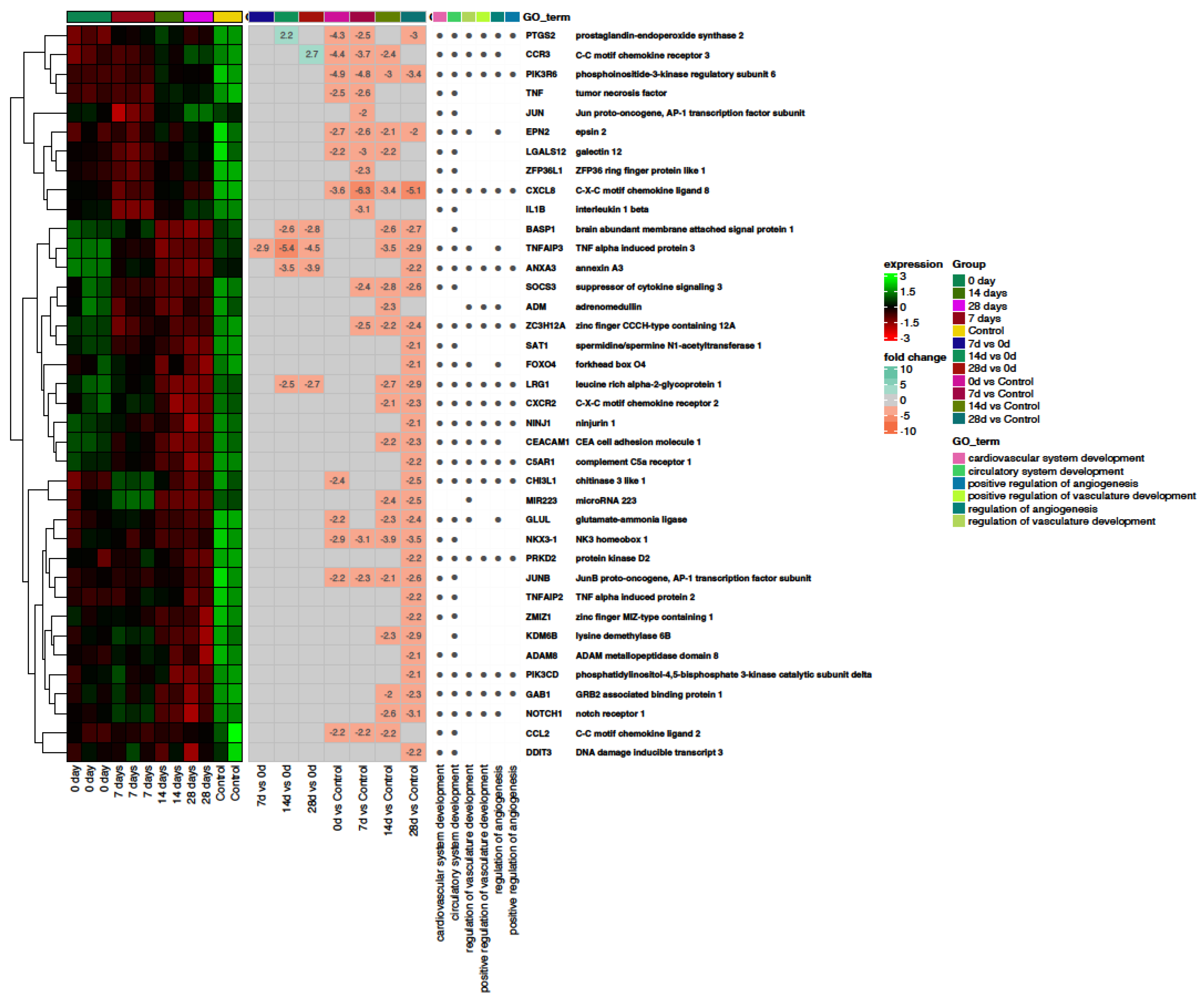
2.3. Angiogenesis
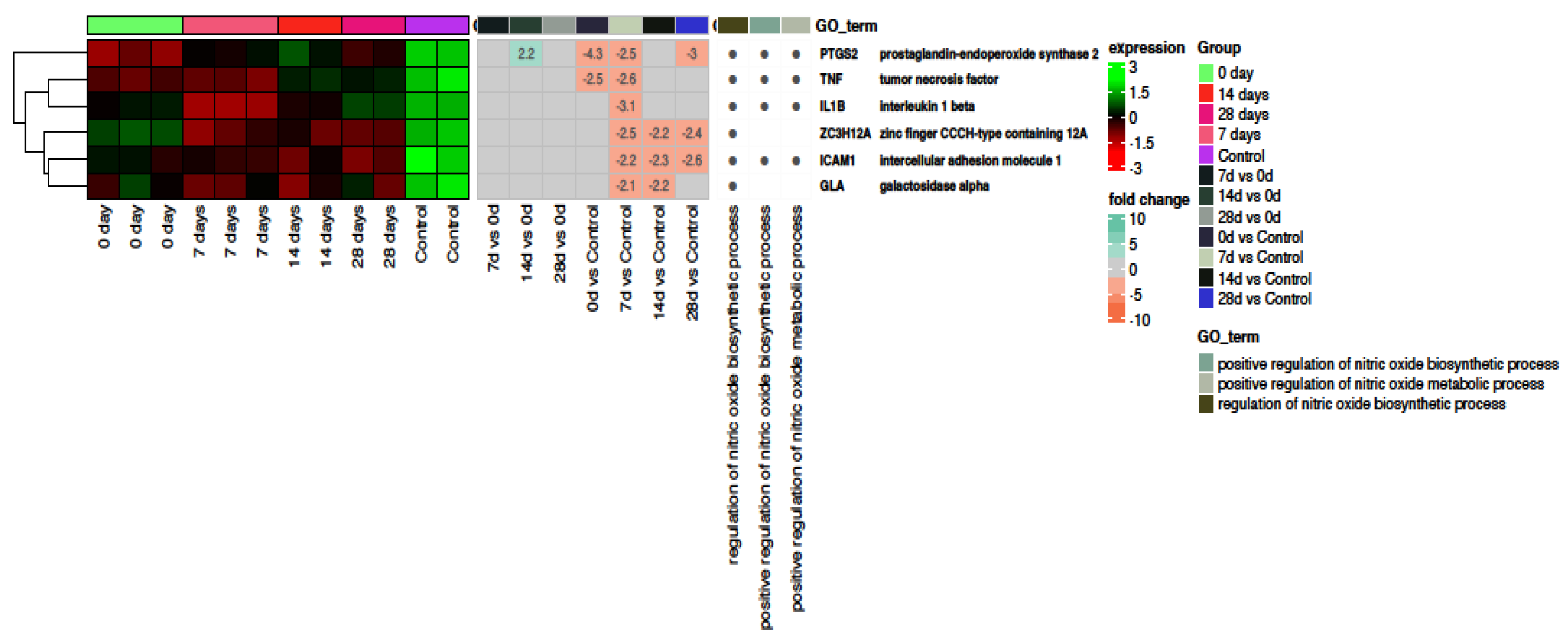
2.4. Nitric Oxide
3. Discussion
4. Materials and Methods
4.1. Study Group
4.2. General Health Questionnaire
4.3. Plasma Collection
4.4. RNA Isolation
4.5. qRT–PCR Assays for Detecting SARS-CoV-2 RNA
4.6. Affymetrix GeneChip Microarray Expression Study
4.7. Assignment of Differentially Expressed Genes to Relevant Gene Ontology Biological Process (GO BP) Terms
4.8. Luminex Assay
4.9. Statistical Analysis
5. Conclusions
6. Strengths of the Study
7. Study Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Butt, A.A.; Dargham, S.R.; Chemaitelly, H.; Al Khal, A.; Tang, P.; Hasan, M.R.; Coyle, P.V.; Thomas, A.G.; Borham, A.M.; Concepcion, E.G.; et al. Severity of Illness in Persons Infected With the SARS-CoV-2 Delta Variant vs Beta Variant in Qatar. JAMA Intern. Med. 2022, 182, 197. [Google Scholar] [CrossRef]
- Hendaus, M.A.; Jomha, F.A. Delta variant of COVID-19: A simple explanation. Qatar Med. J. 2021, 2021, 49. [Google Scholar] [CrossRef] [PubMed]
- Meo, S.A.; Meo, A.S.; Al-Jassir, F.F.; Klonoff, D.C. Omicron SARS-CoV-2 new variant: Global prevalence and biological and clinical characteristics. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 8012–8018. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, Z.; Wang, Z.; Li, K.; Tian, Y.; Lu, W.; Hong, J.; Peng, X.; Shi, J.; Zhang, Z.; et al. Clinical characteristics of 1139 mild cases of the SARS-CoV-2 Omicron variant infected patients in Shanghai. J. Med. Virol. 2023, 95, e28224. [Google Scholar] [CrossRef] [PubMed]
- Parums, D.V. Editorial: A Rapid Global Increase in COVID-19 is Due to the Emergence of the EG.5 (Eris) Subvariant of Omicron SARS-CoV-2. Med. Sci. Monit. 2023, 29, e942244. [Google Scholar] [CrossRef]
- Da Rosa Mesquita, R.; Francelino Silva Junior, L.C.; Santos Santana, F.M.; De Oliveira, T.F.; Campos Alcântara, R.; Monteiro Arnozo, G.; Filho, E.R.d.S.; dos Santos, A.G.G.; da Cunha, E.J.O.; de Aquino, S.H.S.; et al. Clinical manifestations of COVID-19 in the general population: Systematic review. Wien. Klin. Wochenschr. 2021, 133, 377–382. [Google Scholar] [CrossRef]
- Lee, Y.; Min, P.; Lee, S.; Kim, S.-W. Prevalence and Duration of Acute Loss of Smell or Taste in COVID-19 Patients. J. Korean Med. Sci. 2020, 35, e174. [Google Scholar] [CrossRef]
- Parasher, A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad. Med. J. 2021, 97, 312–320. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, J.; Dai, Z.; Deng, H.; Li, X.; Huang, Q.; Wu, Y.; Sun, L.; Xu, Y. Prevalence and severity of corona virus disease 2019 (COVID-19): A systematic review and meta-analysis. J. Clin. Virol. 2020, 127, 104371. [Google Scholar] [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Raveendran, A.V.; Jayadevan, R.; Sashidharan, S. Long COVID: An overview. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 869–875. [Google Scholar] [CrossRef]
- Montazersaheb, S.; Khatibi, S.M.H.; Hejazi, M.S.; Tarhriz, V.; Farjami, A.; Sorbeni, F.G.; Farahzadi, R.; Ghasemnejad, T. COVID-19 infection: An overview on cytokine storm and related interventions. Virol. J. 2022, 19, 92. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Liu, F.; Li, L.; Xu, M.; Wu, J.; Luo, D.; Zhu, Y.; Li, B.; Song, X.; Zhou, X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020, 127, 104370. [Google Scholar] [CrossRef]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef]
- Luo, P.; Liu, Y.; Qiu, L.; Liu, X.; Liu, D.; Li, J. Tocilizumab treatment in COVID-19: A single center experience. J. Med. Virol. 2020, 92, 814–818. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, Z.; Li, J.-W.; Zhao, H.; Wang, G.-Q. Cytokine release syndrome in severe COVID-19: Interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents 2020, 55, 105954. [Google Scholar] [CrossRef]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X.; et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef]
- Zhang, X.; Song, K.; Tong, F.; Fei, M.; Guo, H.; Lu, Z.; Wang, J.; Zheng, C. First case of COVID-19 in a patient with multiple myeloma successfully treated with tocilizumab. Blood Adv. 2020, 4, 1307–1310. [Google Scholar] [CrossRef]
- Joffre, J.; Rodriguez, L.; Matthay, Z.A.; Lloyd, E.; Fields, A.T.; Bainton, R.J.; Kurien, P.; Sil, A.; Calfee, C.S.; Woodruff, P.G.; et al. COVID-19–associated Lung Microvascular Endotheliopathy: A “From the Bench” Perspective. Am. J. Respir. Crit. Care Med. 2022, 206, 961–972. [Google Scholar] [CrossRef]
- Jin, Y.; Ji, W.; Yang, H.; Chen, S.; Zhang, W.; Duan, G. Endothelial activation and dysfunction in COVID-19: From basic mechanisms to potential therapeutic approaches. Signal Transduct. Target. Ther. 2020, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Pius-Sadowska, E.; Niedźwiedź, A.; Kulig, P.; Baumert, B.; Sobuś, A.; Rogińska, D.; Łuczkowska, K.; Ulańczyk, Z.; Wnęk, S.; Karolak, I.; et al. CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course. Int. J. Mol. Sci. 2022, 23, 11338. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Perez, R.; Brandão, M.; Pazdernik, M.; Kresoja, K.-P.; Carpenito, M.; Maeda, S.; Casado-Arroyo, R.; Muscoli, S.; Pöss, J.; Fontes-Carvalho, R.; et al. Cardiovascular disease and COVID-19, a deadly combination: A review about direct and indirect impact of a pandemic. World J. Clin. Cases 2022, 10, 9556–9572. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Rathore, S.S.; Khan, H.; Karale, S.; Chawla, Y.; Iqbal, K.; Bhurwal, A.; Tekin, A.; Jain, N.; Mehra, I.; et al. Association of Obesity With COVID-19 Severity and Mortality: An Updated Systemic Review, Meta-Analysis, and Meta-Regression. Front. Endocrinol. 2022, 13, 780872. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Mathioudakis, A.G.; Higham, A. Chronic obstructive pulmonary disease and COVID-19: Interrelationships. Curr. Opin. Pulm. Med. 2022, 28, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.; Daniels, L.B.; DeFilippis, A.P.; Hamburg, N.M.; Khan, Y.; Keith, R.J.; Kumar, R.S.; Strokes, A.C.; Robertson, R.M.; Bhatnagar, A. Smoking is associated with increased risk of cardiovascular events, disease severity, and mortality among patients hospitalized for SARS-CoV-2 infections. Santulli G, editor. PLoS ONE 2022, 17, e0270763. [Google Scholar] [CrossRef]
- Statsenko, Y.; Al Zahmi, F.; Habuza, T.; Almansoori, T.M.; Smetanina, D.; Simiyu, G.L.; Gorkom, K.N.-V.; Ljubisavljevic, M.; Awawdeh, R.; Elshekhali, H.; et al. Impact of Age and Sex on COVID-19 Severity Assessed From Radiologic and Clinical Findings. Front. Cell. Infect. Microbiol. 2022, 11, 777070. [Google Scholar] [CrossRef]
- Monari, C.; Sagnelli, C.; Maggi, P.; Sangiovanni, V.; Numis, F.G.; Gentile, I.; Masullo, A.; Rescigno, C.; Calabria, G.; Megna, A.S.; et al. More Severe COVID-19 in Patients with Active Cancer: Results of a Multicenter Cohort Study. Front. Oncol. 2021, 11, 662746. [Google Scholar] [CrossRef]
- Sienko, J.; Marczak, I.; Kotowski, M.; Bogacz, A.; Tejchman, K.; Sienko, M.; Kotfis, K. Association of ACE2 Gene Variants with the Severity of COVID-19 Disease—A Prospective Observational Study. Int. J. Environ. Res. Public Health 2022, 19, 12622. [Google Scholar] [CrossRef]
- Zheng, X.L. ADAMTS13 and von Willebrand Factor in Thrombotic Thrombocytopenic Purpura. Annu. Rev. Med. 2015, 66, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Seibert, F.S.; Blazquez-Navarro, A.; Hölzer, B.; Doevelaar, A.A.; Nusshag, C.; Merle, U.; Morath, C.; Zgoura, P.; Dittmer, R.; Schneppenheim, S.; et al. Effect of plasma exchange on COVID-19 associated excess of von Willebrand factor and inflammation in critically ill patients. Sci. Rep. 2022, 12, 4801. [Google Scholar] [CrossRef] [PubMed]
- Helms, J.; Tacquard, C.; Severac, F.; Leonard-Lorant, I.; Ohana, M.; Delabranche, X.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Gandet, F.F.; et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: A multicenter prospective cohort study. Intensive Care Med. 2020, 46, 1089–1098. [Google Scholar] [CrossRef]
- Panigada, M.; Bottino, N.; Tagliabue, P.; Grasselli, G.; Novembrino, C.; Chantarangkul, V.; Pesenti, A.; Peyvandi, F.; Tripodi, A. Hypercoagulability of COVID-19 patients in intensive care unit: A report of thromboelastography findings and other parameters of hemostasis. J. Thromb. Haemost. 2020, 18, 1738–1742. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.W.; Van Wijk, X.M.R.; Pham, H.P.; Marin, M.J. Role of von Willebrand Factor in COVID-19 Associated Coagulopathy. J. Appl. Lab. Med. 2021, 6, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Escher, R.; Breakey, N.; Lämmle, B. ADAMTS13 activity, von Willebrand factor, factor VIII and D-dimers in COVID-19 inpatients. Thromb. Res. 2020, 192, 174–175. [Google Scholar] [CrossRef] [PubMed]
- Sillen, M.; Declerck, P.J. A Narrative Review on Plasminogen Activator Inhibitor-1 and Its (Patho)Physiological Role: To Target or Not to Target? Int. J. Mol. Sci. 2021, 22, 2721. [Google Scholar] [CrossRef]
- Baycan, O.F.; Barman, H.A.; Bolen, F.; Atici, A.; Erman, H.; Korkmaz, R.; Calim, M.; Atalay, B.; Aciksari, G.; Cekmen, M.B.; et al. Plasminogen Activator Inhibitor-1 Levels as an Indicator of Severity and Mortality for COVID-19. North. Clin. Istanb. 2023, 10, 1–9. [Google Scholar] [CrossRef]
- Conway, E.M. Thrombomodulin and its role in inflammation. Semin. Immunopathol. 2012, 34, 107–125. [Google Scholar] [CrossRef]
- Melincovici, C.S.; Boşca, A.B.; Şuşman, S.; Mărginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular endothelial growth factor (VEGF)—Key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. Rev. Roum. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar]
- Raisky, O.; Nykänen, A.I.; Krebs, R.; Hollmén, M.; Keränen, M.A.I.; Tikkanen, J.M.; Sihvola, R.; Alhonen, L.; Salven, P.; Wu, Y.; et al. VEGFR-1 and -2 Regulate Inflammation, Myocardial Angiogenesis, and Arteriosclerosis in Chronically Rejecting Cardiac Allografts. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 819–825. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.-P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Birnhuber, A.; Fließer, E.; Gorkiewicz, G.; Zacharias, M.; Seeliger, B.; David, S.; Welte, T.; Schmidt, J.; Olschewski, H.; Wygrecka, M.; et al. Between inflammation and thrombosis: Endothelial cells in COVID-19. Eur. Respir. J. 2021, 58, 2100377. [Google Scholar] [CrossRef]
- Miggiolaro, A.F.R.S.; Da Silva, F.P.G.; Wiedmer, D.B.; Godoy, T.M.; Borges, N.H.; Piper, G.W.; Oricil, A.G.G.; Klein, C.K.; Hlatchuk, E.C.; Dagostini, J.C.H.; et al. COVID-19 and Pulmonary Angiogenesis: The Possible Role of Hypoxia and Hyperinflammation in the Overexpression of Proteins Involved in Alveolar Vascular Dysfunction. Viruses 2023, 15, 706. [Google Scholar] [CrossRef]
- Sayin Kocakap, D.B.; Kaygusuz, S.; Aksoy, E.; ŞahïN, Ö.; Baççioğlu, A.; EkïCï, A.; Kalpaklıoğlu, A.F.; Ekici, M.S.; Gül, S.; Kaçmaz, B.; et al. Adverse effect of VEGFR-2 (rs1870377) polymorphism on the clinical course of COVID-19 in females and males in an age-dependent manner. Microbes Infect. 2023, 105188. [Google Scholar] [CrossRef] [PubMed]
- Flatt, P.R. Dipeptidyl peptidase IV (DPP IV) and related molecules in type 2 diabetes. Front. Biosci. 2008, 13, 3648. [Google Scholar] [CrossRef] [PubMed]
- Green, B.D.; Flatt, P.R.; Bailey, C.J. Dipeptidyl peptidase IV (DPP IV) inhibitors: A newly emerging drug class for the treatment of type 2 diabetes. Diabetes Vasc. Dis. Res. 2006, 3, 159–165. [Google Scholar] [CrossRef]
- Alomair, B.M.; Al-kuraishy Hayder, M.; Al-Buhadily, A.K.; Al-Gareeb, A.I.; De Waard, M.; Elekhnawy, E.; Batiha, G.E.-S. Is sitagliptin effective for SARS-CoV-2 infection: False or true prophecy? Inflammopharmacology 2022, 30, 2411–2415. [Google Scholar] [CrossRef]
- Lamers, D.; Famulla, S.; Wronkowitz, N.; Hartwig, S.; Lehr, S.; Ouwens, D.M.; Eckardt, K.; Kaufman, J.M.; Ryden, M.; Müller, S.; et al. Dipeptidyl Peptidase 4 Is a Novel Adipokine Potentially Linking Obesity to the Metabolic Syndrome. Diabetes 2011, 60, 1917–1925. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Jiang, S.; Lu, L.; Du, L.; Debnath, A.K. Putative conformations of the receptor-binding domain in S protein of hCoV-EMC in complex with its receptor dipeptidyl peptidase-4. J. Infect. 2013, 67, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Raha, A.A.; Chakraborty, S.; Henderson, J.; Mukaetova-Ladinska, E.; Zaman, S.; Trowsdale, J.; Raha-Chowdhury, R. Investigation of CD26, a potential SARS-CoV-2 receptor, as a biomarker of age and pathology. Biosci. Rep. 2020, 40, BSR20203092. [Google Scholar] [CrossRef] [PubMed]
- Schlicht, K.; Rohmann, N.; Geisler, C.; Hollstein, T.; Knappe, C.; Hartmann, K.; Schwarz, J.; Tran, F.; Schunk, D.; Junker, R.; et al. Circulating levels of soluble Dipeptidylpeptidase-4 are reduced in human subjects hospitalized for severe COVID-19 infections. Int. J. Obes. 2020, 44, 2335–2338. [Google Scholar] [CrossRef] [PubMed]
- Bardaweel, S.K.; Hajjo, R.; Sabbah, D.A. Sitagliptin: A potential drug for the treatment of COVID-19? Acta Pharm. 2021, 71, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Solerte, S.B.; D’addio, F.; Trevisan, R.; Lovati, E.; Rossi, A.; Pastore, I.; Dell’acqua, M.; Ippolito, E.; Scaranna, C.; Bellante, R.; et al. Sitagliptin Treatment at the Time of Hospitalization Was Associated With Reduced Mortality in Patients With Type 2 Diabetes and COVID-19: A Multicenter, Case-Control, Retrospective, Observational Study. Diabetes Care 2020, 43, 2999–3006. [Google Scholar] [CrossRef] [PubMed]
- Al-kuraishy Hayder, M.; Al-Gareeb, A.I.; Qusty, N.; Alexiou, A.; Batiha, G.E.-S. Impact of Sitagliptin on Non-diabetic COVID-19 Patients. Curr. Mol. Pharmacol. 2022, 15, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Abuhasira, R.; Ayalon-Dangur, I.; Zaslavsky, N.; Koren, R.; Keller, M.; Dicker, D.; Grossman, A. A Randomized Clinical Trial of Linagliptin vs. Standard of Care in Patients Hospitalized With Diabetes and COVID-19. Front. Endocrinol. 2021, 12, 794382. [Google Scholar] [CrossRef]
- Kokic Males, V. Letter to the editor in response to the article “COVID-19 and diabetes: Can DPP4 inhibition play a role?”. Diabetes Res. Clin. Pract. 2020, 163, 108163. [Google Scholar] [CrossRef]
- Bouhanick, B.; Cracowski, J.-L.; Faillie, J.-L. DPP-4 inhibitors and severe course of illness in patients with COVID-19. Therapies 2021, 76, 359–360. [Google Scholar] [CrossRef]
- Narayanan, N.; Naik, D.; Sahoo, J.; Kamalanathan, S. Dipeptidyl peptidase 4 inhibitors in COVID-19: Beyond glycemic control. World J. Virol. 2022, 11, 399–410. [Google Scholar] [CrossRef]
- Klok, F.A.; Kruip, M.J.H.A.; Van Der Meer, N.J.M.; Arbous, M.S.; Gommers, D.A.M.P.J.; Kant, K.M.; Kaptein, F.H.J.; van Paassen, J.; Stals, M.A.M.; Huisman, M.V.; et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 2020, 191, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Middeldorp, S.; Coppens, M.; Van Haaps, T.F.; Foppen, M.; Vlaar, A.P.; Müller, M.C.A.; Bouman, C.C.S.; Beenen, L.F.M.; Kootte, R.S.; Heijmans, J.; et al. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1995–2002. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mies, B.; Gómez-Rojo, M.; Carretero-Barrio, I.; Bardi, T.; Benito, A.; García-Cosío, M.; Caballero, Á.; de Pablo, R.; Galán, J.C.; Pestaña, D.; et al. Pulmonary vascular proliferation in patients with severe COVID-19: An autopsy study. Thorax 2021, 76, 1044–1046. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.R.; Huckaby, L.V.; Shiva, S.S.; Zuckerbraun, B.S. Nitric Oxide and Endothelial Dysfunction. Crit. Care Clin. 2020, 36, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Huang, Y.L.; Ma, S.H.; Yeh, C.T.; Chiou, S.Y.; Chen, L.K.; Liao, C.L. Inhibition of Japanese encephalitis virus infection by nitric oxide: Antiviral effect of nitric oxide on RNA virus replication. J. Virol. 1997, 71, 5227–5235. [Google Scholar] [CrossRef] [PubMed]
- Akaberi, D.; Krambrich, J.; Ling, J.; Luni, C.; Hedenstierna, G.; Järhult, J.D.; Lennerstrand, J.; Lundkvist, Å. Mitigation of the replication of SARS-CoV-2 by nitric oxide in vitro. Redox Biol. 2020, 37, 101734. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, R.A.; Berra, L.; Gladwin, M.T. Home Nitric Oxide Therapy for COVID-19. Am. J. Respir. Crit. Care Med. 2020, 202, 16–20. [Google Scholar] [CrossRef]
- Gelzo, M.; Scialò, F.; Cacciapuoti, S.; Pinchera, B.; De Rosa, A.; Cernera, G.; Comegna, M.; Tripodi, L.; Moriello, N.S.; Mormile, M.; et al. Inducible Nitric Oxide Synthase (iNOS): Why a Different Production in COVID-19 Patients of the Two Waves? Viruses 2022, 14, 534. [Google Scholar] [CrossRef]
- Stelcer, E.; Komarowska, H.; Jopek, K.; Żok, A.; Iżycki, D.; Malińska, A.; Szczepaniak, B.; Komekbai, Z.; Karczewski, M.; Wierzbicki, T.; et al. Biological response of adrenal carcinoma and melanoma cells to mitotane treatment. Oncol. Lett. 2022, 23, 120. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Kleszcz, R.; Studzińska-Sroka, E.; Łukaszyk, A.; Szoszkiewicz, A.; Stelcer, E.; Jopek, K.; Rucinski, M.; Cielecka-Piontek, J.; Krajka-Kuźniak, V. Lichen Secondary Metabolites Inhibit the Wnt/β-Catenin Pathway in Glioblastoma Cells and Improve the Anticancer Effects of Temozolomide. Cells 2022, 11, 1084. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Fresno, C.; Fernández, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 16 October 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pius-Sadowska, E.; Kulig, P.; Niedźwiedź, A.; Baumert, B.; Łuczkowska, K.; Rogińska, D.; Sobuś, A.; Ulańczyk, Z.; Kawa, M.; Paczkowska, E.; et al. VEGFR and DPP-IV as Markers of Severe COVID-19 and Predictors of ICU Admission. Int. J. Mol. Sci. 2023, 24, 17003. https://doi.org/10.3390/ijms242317003
Pius-Sadowska E, Kulig P, Niedźwiedź A, Baumert B, Łuczkowska K, Rogińska D, Sobuś A, Ulańczyk Z, Kawa M, Paczkowska E, et al. VEGFR and DPP-IV as Markers of Severe COVID-19 and Predictors of ICU Admission. International Journal of Molecular Sciences. 2023; 24(23):17003. https://doi.org/10.3390/ijms242317003
Chicago/Turabian StylePius-Sadowska, Ewa, Piotr Kulig, Anna Niedźwiedź, Bartłomiej Baumert, Karolina Łuczkowska, Dorota Rogińska, Anna Sobuś, Zofia Ulańczyk, Miłosz Kawa, Edyta Paczkowska, and et al. 2023. "VEGFR and DPP-IV as Markers of Severe COVID-19 and Predictors of ICU Admission" International Journal of Molecular Sciences 24, no. 23: 17003. https://doi.org/10.3390/ijms242317003
APA StylePius-Sadowska, E., Kulig, P., Niedźwiedź, A., Baumert, B., Łuczkowska, K., Rogińska, D., Sobuś, A., Ulańczyk, Z., Kawa, M., Paczkowska, E., Parczewski, M., Machalińska, A., & Machaliński, B. (2023). VEGFR and DPP-IV as Markers of Severe COVID-19 and Predictors of ICU Admission. International Journal of Molecular Sciences, 24(23), 17003. https://doi.org/10.3390/ijms242317003





