SMALL PLANT AND ORGAN 1 (SPO1) Encoding a Cellulose Synthase-like Protein D4 (OsCSLD4) Is an Important Regulator for Plant Architecture and Organ Size in Rice
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Characterization of the spo1 Mutant
2.2. Map-Based Cloning Revealed That SPO1 Gene Encodes Cellulose Synthase-like D4
2.3. The Tissue-Specific and Stressed-Induced Expression Patterns of SPO1
2.4. SPO1 Regulates Leaf Shape, Plant Height, and Grain Size through Affecting Cell Division and/or Cell Expansion
2.5. Hormone Contents Were Significantly Altered in the spo1 Mutant
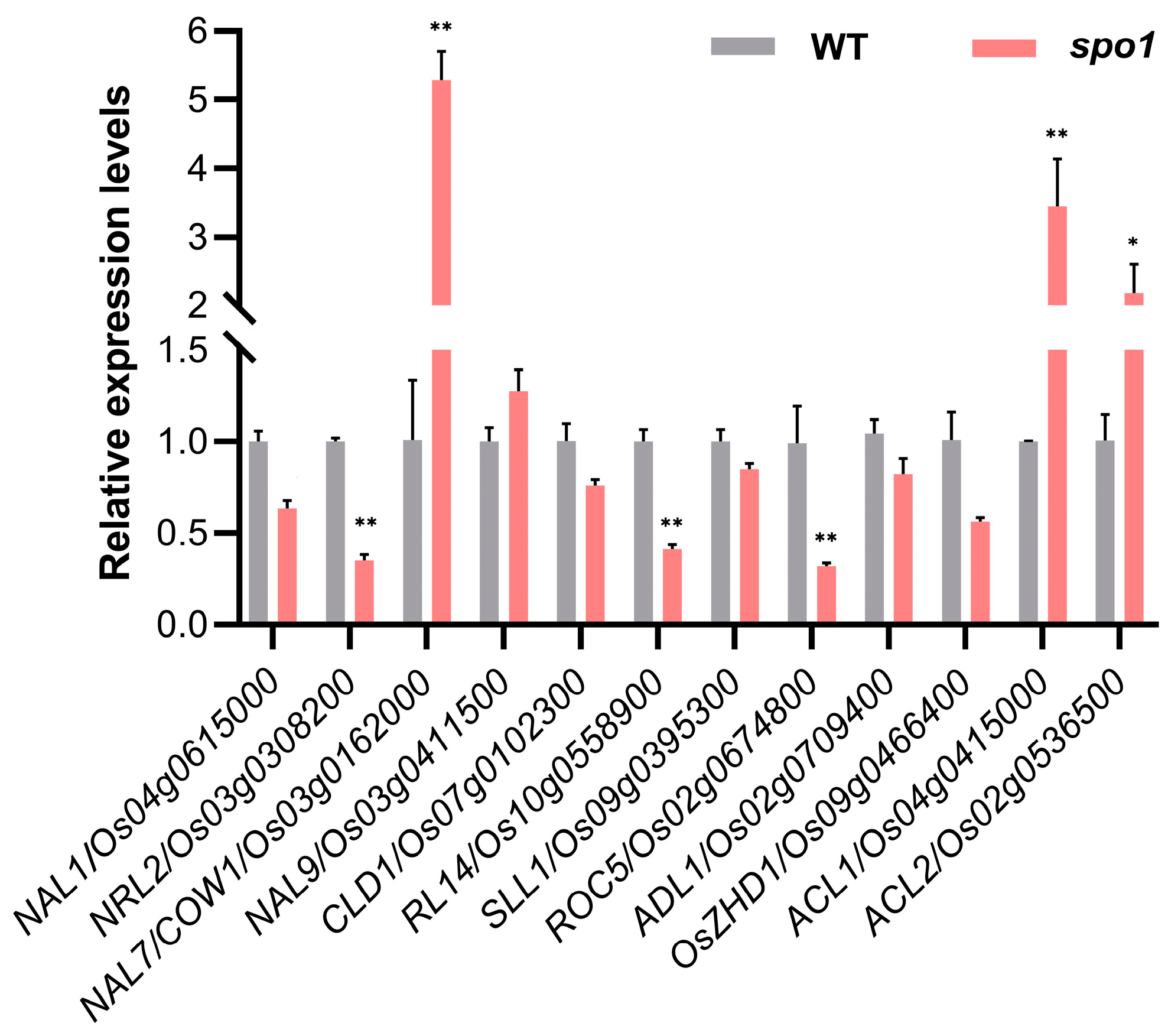
2.6. Loss-of-Function of SPO1 Affects the Expression of Leaf-Shape-Related Genes
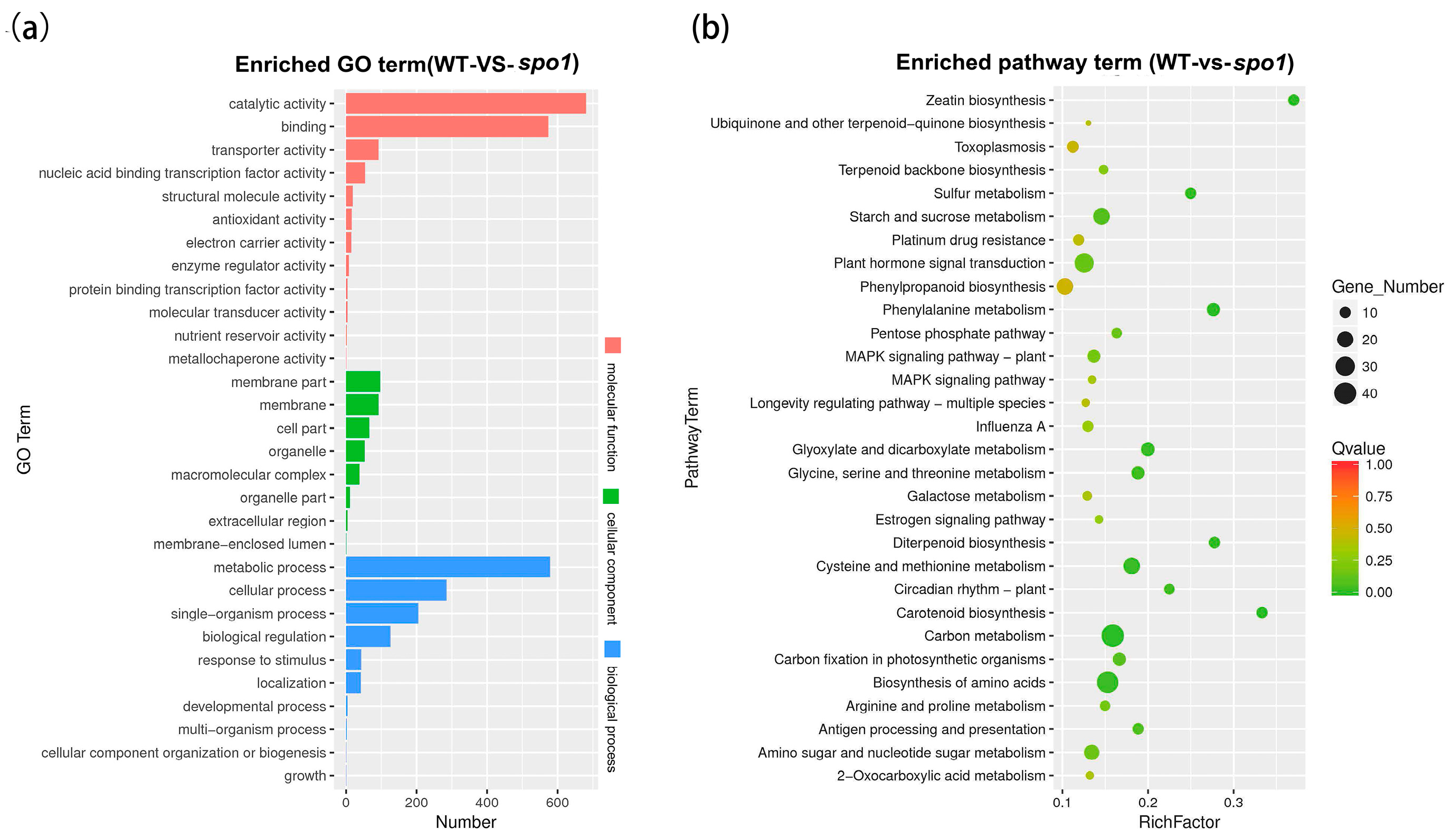
2.7. Transcriptomic Analysis Revealed SPO1 Plays an Important Role in Hormone, Cell Cycle, and Cell Wall Formation Pathways
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. The Mapping of the SPO1 Gene
4.3. Vector Construction and Plant Transformation
4.4. Microscopy Analysis
4.5. RNA Extraction and Gene Expression Analysis
4.6. Measurement of Endogenous Hormone Content
4.7. RNA-seq Analysis
4.8. Measurements
4.9. Phylogenetic Analysis and Protein Structure Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xing, Y.; Zhang, Q. Genetic and Molecular Bases of Rice Yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Genes controlling plant architecture. Curr. Opin. Biotechnol. 2006, 17, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Breuninger, H.; Lenhard, M. Control of Tissue and Organ Growth in Plants. In Plant Development; Elsevier: Amsterdam, The Netherlands, 2010; pp. 185–220. [Google Scholar]
- Li, W.; Yang, Z.; Yao, J.; Li, J.; Song, W.; Yang, X. Cellulose synthase-like D1 controls organ size in maize. BMC Plant Biol. 2018, 18, 239. [Google Scholar] [CrossRef] [PubMed]
- Niinemets, Ü.; Portsmuth, A.; Tobias, M. Leaf size modifies support biomass distribution among stems, petioles and mid-ribs in temperate plants. New Phytol. 2006, 171, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Braybrook, S.A.; Kuhlemeier, C. How a Plant Builds Leaves. Plant Cell 2010, 22, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X.; et al. Mutation of the Rice Narrow leaf1 Gene, Which Encodes a Novel Protein, Affects Vein Patterning and Polar Auxin Transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Yoo, S.C.; Zhang, H.; Pandeya, D.; Koh, H.J.; Hwang, J.Y.; Kim, G.T.; Paek, N.C. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013, 198, 1071–1084. [Google Scholar] [CrossRef]
- Li, W.; Wu, C.; Hu, G.; Xing, L.; Qian, W.; Si, H.; Sun, Z.; Wang, X.; Fu, Y.; Liu, W. Characterization and fine mapping of a novel rice narrow leaf mutant nal9. J. Integr. Plant Biol. 2013, 55, 1016–1025. [Google Scholar] [CrossRef]
- Wu, Y.; Luo, L.; Chen, L.; Tao, X.; Huang, M.; Wang, H.; Chen, Z.; Xiao, W. Chromosome mapping, molecular cloning and expression analysis of a novel gene response for leaf width in rice. Biochem. Biophys. Res. Commun. 2016, 480, 394–401. [Google Scholar] [CrossRef]
- Zhao, S.; Zhao, L.; Liu, F.; Wu, Y.; Zhu, Z.; Sun, C.; Tan, L. NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J. Integr. Plant Biol. 2016, 58, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.-D.; Um, T.Y.; Yang, W.T.; Park, T.H.; Hong, S.Y.; Kim, K.M.; Chung, Y.S.; Yun, D.J.; Kim, D.H. Characterization of dwarf and narrow leaf (dnl-4) mutant in rice. Plant Signal Behav. 2020, 16, 1849490. [Google Scholar] [CrossRef]
- Uzair, M.; Long, H.; Zafar, S.A.; Patil, S.B.; Chun, Y.; Li, L.; Fang, J.; Zhao, J.; Peng, L.; Yuan, S.; et al. Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol. 2021, 186, 497–518. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Xiao, W.; Zhou, Y.; Shen, W.; Ye, L.; Yu, P.; Yu, G.; Duan, Q.; Zhang, X.; He, Z.; et al. The APC/CTAD1-WIDE LEAF 1-NARROW LEAF 1 pathway controls leaf width in rice. Plant Cell 2022, 34, 4313–4328. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yan, S.; Jiang, S.; Bai, L.; Liu, Y.; Peng, S.; Chen, R.; Liu, Q.; Xiao, Y.; Kang, H. Identification of a Rice Leaf Width Gene Narrow Leaf 22 (NAL22) through Genome-Wide Association Study and Gene Editing Technology. Int. J. Mol. Sci. 2023, 24, 4073. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.H.; Xu, Q.; Zhu, X.D.; Qian, Q.; Xue, H.W. SHALLOT-LIKE1 Is a KANADI Transcription Factor That Modulates Rice Leaf Rolling by Regulating Leaf Abaxial Cell Development. Plant Cell 2009, 21, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Wang, X.Q.; An, L.S.; Zhang, J.L. Overexpression of ACL1 (abaxially curled leaf 1) increased Bulliform cells and induced Abaxial curling of leaf blades in rice. Mol. Plant 2010, 3, 807–817. [Google Scholar] [CrossRef]
- Zou, L.P.; Sun, X.H.; Zhang, Z.G.; Liu, P.; Wu, J.X.; Tian, C.J.; Qiu, J.L.; Lu, T.G. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 2011, 156, 1589–1602. [Google Scholar] [CrossRef]
- Xiang, J.J.; Zhang, G.H.; Qian, Q.; Xue, H.W. SEMI-ROLLED LEAF1 Encodes a Putative Glycosylphosphatidylinositol-Anchored Protein and Modulates Rice Leaf Rolling by Regulating the Formation of Bulliform Cells. Plant Physiol. 2012, 159, 1488–1500. [Google Scholar] [CrossRef]
- Fang, L.; Zhao, F.; Cong, Y.; Sang, X.; Du, Q.; Wang, D.; Li, Y.; Ling, Y.; Yang, Z.; He, G. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 2012, 10, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.; Long, Q.; Huang, J.; Wang, Y.; Zhou, K.; Zheng, M.; Sun, J.; Chen, H.; Chen, S.; et al. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 2014, 239, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Shen, A.; Xiong, W.; Sun, Q.L.; Luo, Q.; Song, T.; Li, Z.L.; Luan, W.J. Overexpression of OsHox32 Results in Pleiotropic Effects on Plant Type Architecture and Leaf Development in Rice. Rice 2016, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Q.; Li, W.Q.; Miao, H.; Gan, P.F.; Qiao, L.; Chang, Y.L.; Shi, C.H.; Chen, K.M. REL2, A Gene Encoding An Unknown Function Protein which Contains DUF630 and DUF632 Domains Controls Leaf Rolling in Rice. Rice 2016, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Fang, J.; Lou, L.; Zhao, J.; Yuan, S.; Yin, L.; Sun, W.; Peng, L.; Guo, B.; Li, X. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE 2015, 10, e0118169. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wen, H.; Teotia, S.; Du, Y.; Zhang, J.; Li, J.; Sun, H.; Tang, G.; Peng, T.; Zhao, Q. Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol. 2017, 17, 215. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Cui, Y.; Hu, H.; Xu, Q.; Rao, Y.; Yu, X.; Zhang, Y.; Wang, Y.; Peng, Y.; Zeng, D.; et al. AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J. 2019, 100, 813–824. [Google Scholar] [CrossRef]
- Li, W.Q.; Zhang, M.J.; Gan, P.F.; Qiao, L.; Yang, S.Q.; Miao, H.; Wang, G.F.; Zhang, M.M.; Liu, W.T.; Li, H.F.; et al. CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 2017, 92, 904–923. [Google Scholar] [CrossRef]
- Lin, L.; Zhao, Y.; Liu, F.; Chen, Q.; Qi, J. Narrow leaf 1 (NAL1) regulates leaf shape by affecting cell expansion in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2019, 516, 957–962. [Google Scholar] [CrossRef]
- Wu, Y.; Fu, Y.; Zhao, S.; Gu, P.; Zhu, Z.; Sun, C.; Tan, L. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 2016, 14, 377–386. [Google Scholar] [CrossRef]
- Zhang, Y.; Gan, L.; Zhang, Y.; Huang, B.; Wan, B.; Li, J.; Tong, L.; Zhou, X.; Wei, Z.; Li, Y.; et al. OsCBL5–CIPK1–PP23 module enhances rice grain size and weight through the gibberellin pathway. Plant J. 2023, 115, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Busov, V.B.; Brunner, A.M.; Strauss, S.H. Genes for control of plant stature and form. New Phytol. 2008, 177, 589–607. [Google Scholar] [CrossRef]
- Xu, P.; Ali, A.; Han, B.; Wu, X. Current Advances in Molecular Basis and Mechanisms Regulating Leaf Morphology in Rice. Front. Plant Sci. 2018, 9, 1528. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Rieu, I.; Steber, C.M. Gibberellin Metabolism and Signaling. Vitam. Horm. 2005, 72, 289–338. [Google Scholar]
- Perrot-Rechenmann, C. Cellular Responses to Auxin: Division versus Expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Chen, W.; Cheng, Z.; Liu, L.; Wang, M.; You, X.; Wang, J.; Zhang, F.; Zhou, C.; Zhang, Z.; Zhang, H.; et al. Small Grain and Dwarf 2, encoding an HD-Zip II family transcription factor, regulates plant development by modulating gibberellin biosynthesis in rice. Plant Sci. 2019, 288, 110208. [Google Scholar] [CrossRef]
- Sazuka, T.; Kamiya, N.; Nishimura, T.; Ohmae, K.; Sato, Y.; Imamura, K.; Nagato, Y.; Koshiba, T.; Nagamura, Y.; Ashikari, M.; et al. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 2009, 60, 227–241. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Inukai, Y.; Kitano, H.; Fujioka, S. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 2013, 73, 676–688. [Google Scholar] [CrossRef]
- Shan, C.; Mei, Z.; Duan, J.; Chen, H.; Feng, H.; Cai, W. OsGA2ox5, a Gibberellin Metabolism Enzyme, Is Involved in Plant Growth, the Root Gravity Response and Salt Stress. PLoS ONE 2014, 9, e87110. [Google Scholar] [CrossRef]
- Shi, C.L.; Dong, N.Q.; Guo, T.; Ye, W.W.; Shan, J.X.; Lin, H.X. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef] [PubMed]
- Sims, K.; Abedi-Samakush, F.; Szulc, N.; Macias Honti, M.G.; Mattsson, J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. Int. J. Mol. Sci. 2021, 22, 4089. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tong, H.; Xiao, Y.; Che, R.; Xu, F.; Hu, B.; Liang, C.; Chu, J.; Li, J.; Chu, C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11102–11107. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Yoshida, H.; Aya, K.; Kawamura, M.; Hayashi, M.; Hobo, T.; Sato-Izawa, K.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING Form a Complex to Integrate Auxin and Brassinosteroid Signaling in Rice. Mol. Plant 2017, 10, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Lv, B.; Ding, T.; Bai, M.; Ding, Z. Auxin-BR Interaction Regulates Plant Growth and Development. Front. Plant Sci. 2018, 8, 2256. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, P.; Zhang, X.; Li, X.; Yan, X.; Fu, D.; Wu, G. The genetic and molecular basis of crop height based on a rice model. Planta 2017, 247, 1–26. [Google Scholar] [CrossRef]
- Duan, E.; Lin, Q.; Wang, Y.; Ren, Y.; Xu, H.; Zhang, Y.; Wang, Y.; Teng, X.; Dong, H.; Wang, Y.; et al. The transcriptional hub SHORT INTERNODES1 integrates hormone signals to orchestrate rice growth and development. Plant Cell 2023, 35, 2871–2886. [Google Scholar] [CrossRef]
- Song, Y.; You, J.; Xiong, L. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 2009, 70, 297–309. [Google Scholar] [CrossRef]
- Keegstra, K. Plant Cell Walls. Plant Physiol. 2010, 154, 483–486. [Google Scholar] [CrossRef]
- Farrokhi, N.; Burton, R.A.; Brownfield, L.; Hrmova, M.; Wilson, S.M.; Bacic, A.; Fincher, G.B. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 2005, 4, 145–167. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.T.; Kirienko, D.H.; Sylvester, A.W.; Peter, G.F.; McCarty, D.R.; Koch, K.E. Cellulose Synthase-Like D1 Is Integral to Normal Cell Division, Expansion, and Leaf Development in Maize. Plant Physiol. 2012, 158, 708–724. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lee, B.-H.; Dellinger, M.; Cui, X.; Zhang, C.; Wu, S.; Nothnagel, E.A.; Zhu, J.-K. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [CrossRef]
- Li, M.; Xiong, G.; Li, R.; Cui, J.; Tang, D.; Zhang, B.; Pauly, M.; Cheng, Z.; Zhou, Y. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, L.; Zeng, D.; Gao, Z.; Guo, L.; Fang, Y.; Zhang, G.; Dong, G.; Yan, M.; Liu, J.; et al. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010, 73, 283–292. [Google Scholar] [CrossRef]
- Wu, C.; Fu, Y.; Hu, G.; Si, H.; Cheng, S.; Liu, W. Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 2010, 232, 313–324. [Google Scholar] [CrossRef]
- Luan, W.; Liu, Y.; Zhang, F.; Song, Y.; Wang, Z.; Peng, Y.; Sun, Z. OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol. J. 2011, 9, 513–524. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Eiguchi, M.; Hibara, K.I.; Ito, J.I.; Nagato, Y. Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J. Exp. Bot. 2013, 64, 2049–2061. [Google Scholar] [CrossRef]
- Ding, Z.; Lin, Z.; Li, Q.; Wu, H.; Xiang, C.; Wang, J. DNL1, encodes cellulose synthase-like D4, is a major QTL for plant height and leaf width in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2015, 457, 133–140. [Google Scholar] [CrossRef]
- Shi, L.; Wei, X.J.; Adedze, Y.M.N.; Sheng, Z.H.; Tang, S.Q.; Hu, P.S.; Wang, J.L. Characterization and gene cloning of the rice (Oryza sativa L.) dwarf and narrow-leaf mutant dnl3. Genet Mol. Res. 2016, 15, gmr.15038731. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Z.; Wang, Y.; Wang, J.; Xiao, M.; Liu, H.; Quan, R.; Zhang, H.; Huang, R.; Zhu, L.; et al. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 2021, 20, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hu, Y.; Du, A.; Yu, L.; Fu, X.; Wu, C.; Lu, L.; Liu, Y.; Wang, S.; Huang, W.; et al. Cell Wall Matrix Polysaccharides Contribute to Salt–Alkali Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 15019. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhao, H.; Wang, Z.; Xu, J.; Liu, Y.; Wang, J.; Chen, M.; Liu, X.; Zhang, Z.; Cen, J.; et al. Leaf Morphology Genes SRL1 and RENL1 Co-Regulate Cellulose Synthesis and Affect Plant drought Tolerance. Rice Sci. 2023, 31. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, A.; Ozawa, M.; Nagasaki, H.; Kato, M.; Noda, Y.; Yamaguchi, T.; Nosaka, M.; Shimizu-Sato, S.; Nagasaki, A.; Maekawa, M.; et al. Two WUSCHEL-related homeobox Genes, narrow leaf2 and narrow leaf3, Control Leaf Width in Rice. Plant Cell Physiol. 2013, 54, 779–792. [Google Scholar] [CrossRef]
- Vanstraelen, M.; Benková, E. Hormonal Interactions in the Regulation of Plant Development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef]
- Sakamoto, T.; Inukai, Y. Characterization of a Tos17 Insertion Mutant of Rice Auxin Signal Transcription Factor Gene, OsARF24. Am. J. Plant Sci. 2013, 04, 84–91. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Hsing, Y.I.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef]
- Sasaki, A.; Itoh, H.; Gomi, K.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Jeong, D.H.; An, G.; Kitano, H.; Ashikari, M.; et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 2003, 299, 1896–1898. [Google Scholar] [CrossRef]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. slender Rice, a Constitutive Gibberellin Response Mutant, Is Caused by a Null Mutation of the SLR1 Gene, an Ortholog of the Height-Regulating Gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef]
- Itoh, H.; Ueguchi-Tanaka, M.; Sentoku, N.; Kitano, H.; Matsuoka, M.; Kobayashi, M. Cloning and functional analysis of two gibberellin 3b -hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 8909–8914. [Google Scholar] [CrossRef] [PubMed]
- Spielmeyer, W.; Ellis, M.; Chandler, P.M. Semidwarf (sd-1), ‘‘green revolution’’ rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Lin, Q.; Zhou, C.; Ren, Y.; Liu, X.; Miao, R.; Jing, R.; Mou, C.; Nguyen, T.; Zhu, X.; et al. Small grain and semi-dwarf 3, a WRKY transcription factor, negatively regulates plant height and grain size by stabilizing SLR1 expression in rice. Plant Mol. Biol. 2020, 104, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhiguo, E.; Zhang, D.; Yun, Q.; Zhou, Y.; Niu, B.; Chen, C. OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiol. 2021, 185, 934–950. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1947–1952. [Google Scholar] [CrossRef]
- Ma, L.; Sang, X.; Zhang, T.; Yu, Z.; Li, Y.; Zhao, F.; Wang, Z.; Wang, Y.; Yu, P.; Wang, N.; et al. ABNORMAL VASCULAR BUNDLES regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol. 2016, 213, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, J.; Qian, Q.; Xu, Y.; Zhang, C.; Xiao, J.; Du, C.; Luo, W.; Zou, G.; Chen, M.; et al. Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 2011, 23, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, S.; Park, G.; Cho, H.; Choi, D.; Umeda, M.; Choi, Y.; Hwang, D.; Hwang, I. CYTOKININ-RESPONSIVE GROWTH REGULATOR regulates cell expansion and cytokinin-mediated cell cycle progression. Plant Physiol. 2021, 186, 1734–1746. [Google Scholar] [CrossRef]
- Rong, C.; Liu, Y.; Chang, Z.; Liu, Z.; Ding, Y.; Ding, C.; Zhang, J. Cytokinin oxidase/dehydrogenase family genes exhibit functional divergence and overlap in rice growth and development, especially in control of tillering. J. Exp. Bot. 2022, 73, 3552–3568. [Google Scholar] [CrossRef]
- Hirose, N.; Makita, N.; Kojima, M.; Kamada-Nobusada, T.; Sakakibara, H. Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 2007, 48, 523–539. [Google Scholar] [CrossRef]
- Depuydt, S.; Hardtke, C.S. Hormone Signalling Crosstalk in Plant Growth Regulation. Curr. Biol. 2011, 21, R365–R373. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Zhang, B.; Wang, N.; Zhou, Y.; Xiong, L. Increased Leaf Angle1, a Raf-Like MAPKKK That Interacts with a Nuclear Protein Family, Regulates Mechanical Tissue Formation in the Lamina Joint of Rice. Plant Cell 2011, 23, 4334–4347. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, M.; Qiao, L.; Chen, Y.; Zhang, D.; Jing, X.; Gan, P.; Huang, Y.; Gao, J.; Liu, W.; et al. Characterization of wavy root 1, an agravitropism allele, reveals the functions of OsPIN2 in fine regulation of auxin transport and distribution and in ABA biosynthesis and response in rice (Oryza sativa L.). Crop J. 2022, 10, 980–992. [Google Scholar] [CrossRef]
- Chen, H.; Cheng, Z.; Ma, X.; Wu, H.; Liu, Y.; Zhou, K.; Chen, Y.; Ma, W.; Bi, J.; Zhang, X.; et al. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep. 2013, 32, 1855–1867. [Google Scholar] [CrossRef]
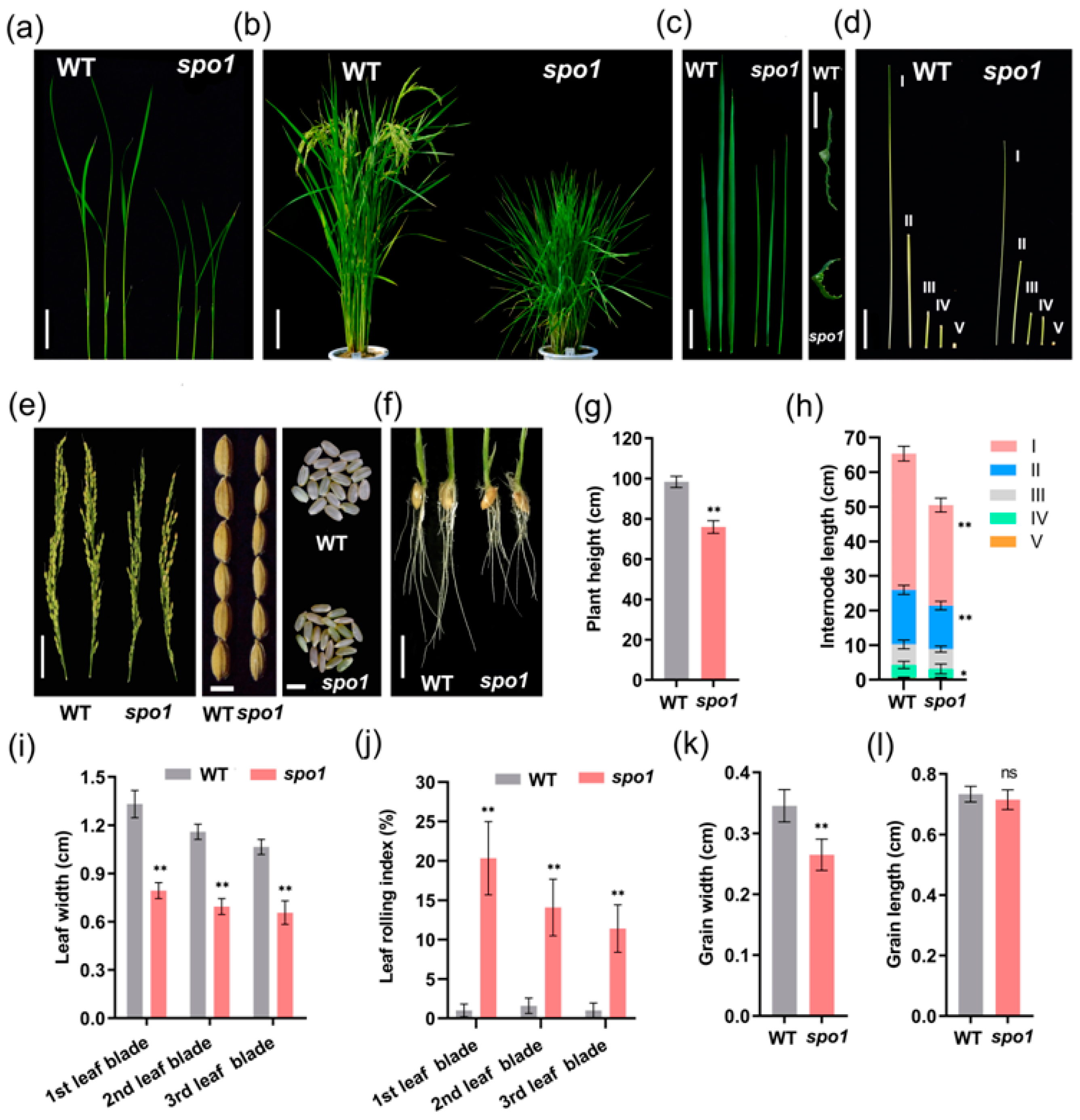
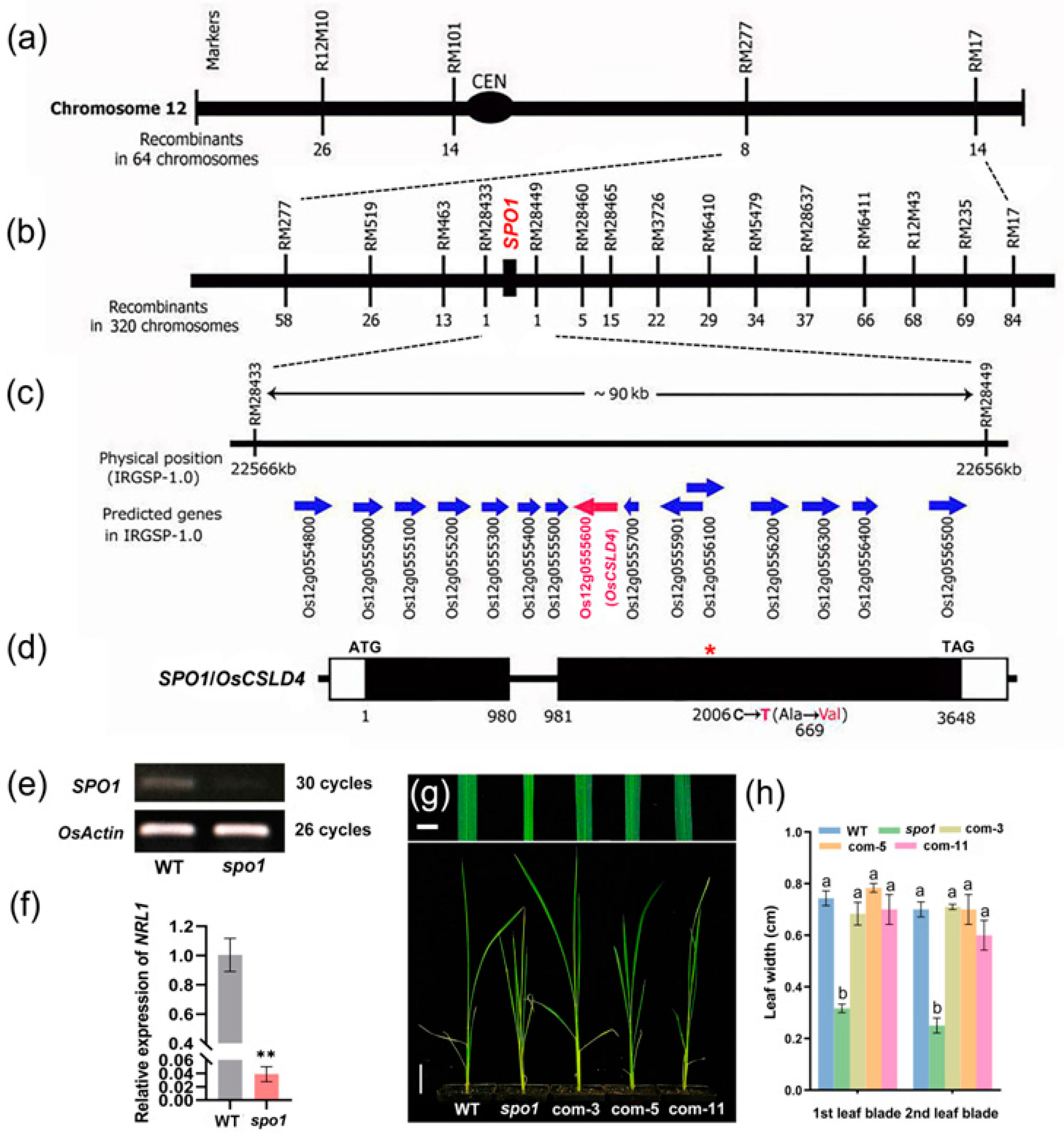
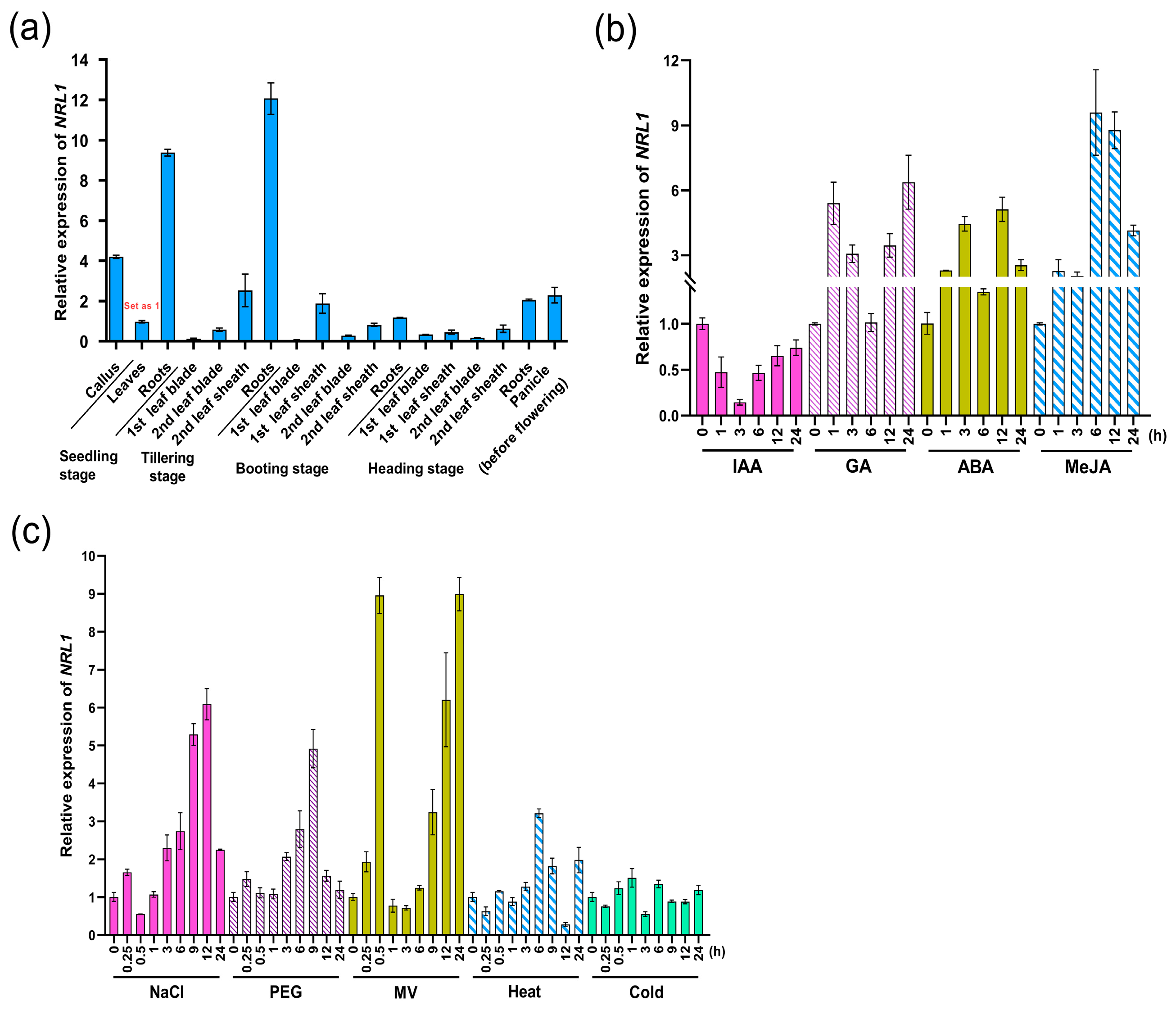
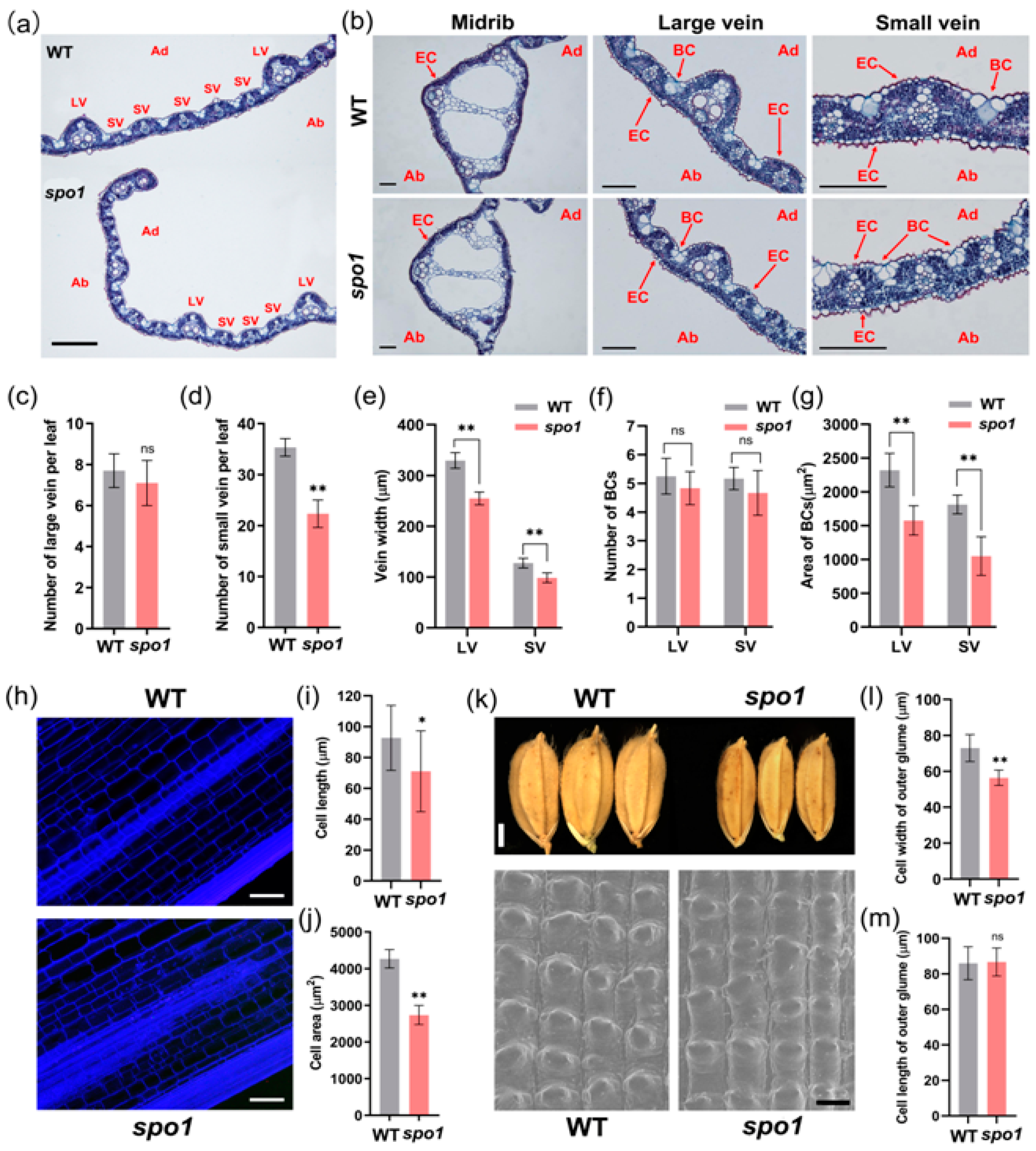
| Traits | WT | spo1 |
|---|---|---|
| Flag leaf length (cm) | 33.38 ± 3.46 | 30.63 ± 4.44 |
| Second leaf length (cm) | 42.67 ± 1.94 | 37.81 ± 1.25 ** |
| Third leaf length (cm) | 42.13 ± 1.64 | 33.44 ± 2.44 ** |
| Tillering number | 12 ± 2.23 | 25 ± 3.91 ** |
| Leaf angle of flag leaf blade (°) | 9.38 ± 1.77 | 18.5 ± 7.63 * |
| Leaf angle of second leaf blade (°) | 9.25 ± 1.39 | 18.75 ± 7.03 ** |
| Leaf angle of third leaf blade (°) | 12.38 ± 2.33 | 25.13 ± 9.26 ** |
| Fertility rate (%) | 82.55 ± 6.92 | 55.29 ± 7.09 ** |
| 1000-grain weight (g) | 25.08 ± 0.24 | 16.53 ± 0.22 ** |
| Panicle length (cm) | 23.26 ± 0.73 | 21.14 ± 1.15 ** |
| Number of adventive roots | 14 ± 2.15 | 12 ± 2.28 * |
| Primary root length (cm) | 8.72 ± 0.83 | 5.87 ± 0.86 ** |
| Plant Hormone | WT (ng/g FW) | spo1 (ng/g FW) |
|---|---|---|
| IAA | 67.854 ± 5.086 | 37.220 ± 1.185 ** |
| ABA | 114.697 ± 7.687 | 90.670 ± 6.971 ** |
| GA3 | 0.861 ± 0.136 | 1.118 ± 0.054 ** |
| GA4 | 0.905 ± 0.051 | 1.316 ± 0.097 ** |
| ZR | 2.652 ± 0.075 | 3.042 ± 0.132 ** |
| dhZR | 0.814 ± 0.035 | 1.002 ± 0.035 ** |
| Gene ID | Gene Description | log2 Fold Change | p-Value | Regulation |
|---|---|---|---|---|
| Genes involved in auxin pathway | ||||
| LOC_Os04g56850 | Auxin response factor, OsARF11 | 2.56219 | 2.90 × 10−6 | Up |
| LOC_Os01g59110 | Similar to indole-3-acetate beta-glucosyl transferase | 2.0965 | 1.48 × 10−18 | Up |
| LOC_Os01g69070 | Similar to Efflux carrier of polar auxin transport, OsPIN5a | 1.69253 | 7.39 × 10−10 | Up |
| LOC_Os03g08850 | Auxin receptor; auxin-signaling F-Box (AFB) gene, OsAFB6 | 1.42947 | 6.45 × 10−10 | Up |
| LOC_Os02g07110 | OsSAUR6-Auxin-responsive SAUR gene family member | 1.39592 | 1.42 × 10−5 | Up |
| LOC_Os01g07500 | Tryptophan amino transferase, FIB; OsTAR2; OsTAA1 | 1.34337 | 2.07 × 10−7 | Up |
| LOC_Os10g05690 | AUX1/LAX gene; auxin carrier; auxin influx transporter, OsAUX4 | −2.65314 | 1.54 × 10−6 | Down |
| LOC_Os05g37470 | AUX1/LAX gene; auxin carrier; auxin influx transporter OsAUX3; qGL5 | −1.42293 | 0.00015 | Down |
| LOC_Os02g56120 | OsIAA9-Auxin-responsive Aux/IAA gene family member | −1.57906 | 0.00380 | Down |
| LOC_Os01g53880 | Auxin-responsive Aux/IAA gene family member, OsIAA6 | −1.4906 | 1.74 × 10−6 | Down |
| LOC_Os07g25540 | IAA synthetic pathway gene, OsYUCCA6 | −1.32218 | 0.00781 | Down |
| Genes involved in GA pathway | ||||
| LOC_Os07g01340 | Gibberellin 2-oxidase gene, OsGA2ox5 | 2.34471 | 4.40 × 10−11 | Up |
| LOC_Os01g08220 | Gibberellin 3β-hydroxylase, GA metabolism, d18; OsGA3ox2 | 1.38532 | 4.77 × 10−5 | Up |
| LOC_Os01g66100 | Semidwarf-1; gibberellin 20-oxidase gene, sd1, OsGA20ox2 | 1.30841 | 0.00011 | Up |
| LOC_Os03g21400 | Cytochrome P450 gene; gibberellin 13-oxidase, GA homeostasis, CYP714B2 | 1.27219 | 1.42 × 10−11 | Up |
| LOC_Os07g44900 | Gibberellin receptor GID1L2 | 1.48675 | 1.57 × 10−10 | Up |
| LOC_Os09g28230 | Gibberellin receptor GID1L2 | 1.28099 | 1.08 × 10−12 | Up |
| Genes involved in CK pathway | ||||
| LOC_Os05g08480 | UDP-glucuronosyl/UDP-glucosyltransferase family protein, cytokinin-O-glucosyltransferase 1 | 3.16484 | 2.15 × 10−22 | Up |
| LOC_Os07g30610 | UDP-glucuronosyl/UDP-glucosyltransferase family protein, cytokinin-O-glucosyltransferase 2 | 1.29078 | 3.37 × 10−7 | Up |
| LOC_Os01g71310 | Cytokinin oxidase/dehydrogenase family gene, OsCKX4; REN1 | 2.1701 | 6.74 × 10−12 | Up |
| LOC_Os10g34230 | Cytokinin oxidase/dehydrogenase, OsCKX3 | 1.19109 | 0.00044 | Up |
| LOC_Os04g57720 | A-type response regulator gene, OsRR6 | 1.19145 | 6.26 × 10−5 | Up |
| LOC_Os03g50860 | Cytokinin receptor family, OHK4 | 1.12033 | 9.44 × 10−8 | Up |
| Genes related to cell cycle processes | ||||
| LOC_Os07g30240 | Meiotic recombination; MutS-homolog family gene; ZMM protein, OsMSH4 | 2.89183 | 2.12 × 10−12 | Up |
| LOC_Os12g04980 | Homologous pairing aberration in rice meiosis; meiosis-specific DNA recombinase, OsDMC1A; DMC1A | 1.72769 | 0.00218 | Up |
| LOC_Os04g53680 | Rice cyclin gene, CycP1;1 | 1.44644 | 9.56 × 10−9 | Up |
| LOC_Os01g13260 | Rice cyclin gene, CycA1;1 | −1.24606 | 0.00589 | Down |
| LOC_Os05g33040 | P-type cyclin gene, OsCYCP1;1; CycP3;1 | −1.27283 | 4.56 × 10−5 | Down |
| LOC_Os05g41880 | Meiotic reciprocal recombination; MutS-homolog gene; ZMM protein, OsMSH5 | −1.23514 | 0.00798 | Down |
| LOC_Os09g38768 | Cell cycle control protein | −1.16037 | 0.00016 | Down |
| Genes related to cell wall formation | ||||
| LOC_Os08g01330 | NAC Transcription Factor, affects the synthesis of cellulose in the secondary wall, OsSWN3; NAC31 | 2.32202 | 1.28 × 10−6 | Up |
| LOC_Os10g29470 | Cinnamyl alcohol dehydrogenase 3, participates in lignin biosynthesis, OsCAD3 | 1.16085 | 2.48 × 10−9 | Up |
| LOC_Os04g50770 | R2R3-type MYB family transcription factor, regulation of cellulose biosynthesis during secondary cell wall formation | −2.49826 | 1.01 × 10−7 | Down |
| LOC_Os06g04090 | NAC transcription factor, regulation of secondary wall biosynthesis by affects the content of lignin, OsSWN1 | −1.25174 | 0.00072 | Down |
| LOC_Os10g40960 | 2OG-Fe (II) oxygenase family protein, regulating the formation of secondary cell walls by affecting their components, RL14 | −1.09748 | 3.86 × 10−6 | Down |
| LOC_Os08g02300 | NAC Transcription Factor, OsSWN2; NAC29, affects cellulose synthesis | −1.05195 | 0.00827 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, L.; Wu, Q.; Yuan, L.; Huang, X.; Yang, Y.; Li, Q.; Shahzad, N.; Li, H.; Li, W. SMALL PLANT AND ORGAN 1 (SPO1) Encoding a Cellulose Synthase-like Protein D4 (OsCSLD4) Is an Important Regulator for Plant Architecture and Organ Size in Rice. Int. J. Mol. Sci. 2023, 24, 16974. https://doi.org/10.3390/ijms242316974
Qiao L, Wu Q, Yuan L, Huang X, Yang Y, Li Q, Shahzad N, Li H, Li W. SMALL PLANT AND ORGAN 1 (SPO1) Encoding a Cellulose Synthase-like Protein D4 (OsCSLD4) Is an Important Regulator for Plant Architecture and Organ Size in Rice. International Journal of Molecular Sciences. 2023; 24(23):16974. https://doi.org/10.3390/ijms242316974
Chicago/Turabian StyleQiao, Lei, Qilong Wu, Liuzhen Yuan, Xudong Huang, Yutao Yang, Qinying Li, Nida Shahzad, Haifeng Li, and Wenqiang Li. 2023. "SMALL PLANT AND ORGAN 1 (SPO1) Encoding a Cellulose Synthase-like Protein D4 (OsCSLD4) Is an Important Regulator for Plant Architecture and Organ Size in Rice" International Journal of Molecular Sciences 24, no. 23: 16974. https://doi.org/10.3390/ijms242316974
APA StyleQiao, L., Wu, Q., Yuan, L., Huang, X., Yang, Y., Li, Q., Shahzad, N., Li, H., & Li, W. (2023). SMALL PLANT AND ORGAN 1 (SPO1) Encoding a Cellulose Synthase-like Protein D4 (OsCSLD4) Is an Important Regulator for Plant Architecture and Organ Size in Rice. International Journal of Molecular Sciences, 24(23), 16974. https://doi.org/10.3390/ijms242316974






