Abstract
Many tumors are characterized by marked extracellular acidosis due to increased glycolytic metabolism, which affects gene expression and thereby tumor biological behavior. At the same time, acidosis leads to altered expression of several microRNAs (Mir7, Mir183, Mir203, Mir215). The aim of this study was to analyze whether the acidosis-induced changes in cytokines and tumor-related genes are mediated via pH-sensitive microRNAs. Therefore, the expression of Il6, Nos2, Ccl2, Spp1, Tnf, Acat2, Aox1, Crem, Gls2, Per3, Pink1, Txnip, and Ypel3 was examined in acidosis upon simultaneous transfection with microRNA mimics or antagomirs in two tumor lines in vitro and in vivo. In addition, it was investigated whether microRNA expression in acidosis is affected via known pH-sensitive signaling pathways (MAPK, PKC, PI3K), via ROS, or via altered intracellular Ca2+ concentration. pH-dependent microRNAs were shown to play only a minor role in modulating gene expression. Individual genes (e.g., Ccl2, Txnip, Ypel3) appear to be affected by Mir183, Mir203, or Mir215 in acidosis, but these effects are cell line-specific. When examining whether acid-dependent signaling affects microRNA expression, it was found that Mir203 was modulated by MAPK and ROS, Mir7 was affected by PKC, and Mir215 was dependent on the intracellular Ca2+ concentration. Mir183 could be increased by ROS scavenging. These correlations could possibly result in new therapeutic approaches for acidotic tumors.
1. Introduction
In addition to the pronounced oxygen deficiency in tumor tissues, extracellular acidification plays an important role as a metabolic stress factor for tumor cells and also for normal cells (e.g., fibroblasts, immune cells), which are part of the tumor tissue. On the one hand, tumor acidosis is the result of insufficient O2 supply to the tissue (due to inadequate tumor perfusion), but on the other hand, it may also be caused by the phenomenon of aerobic glycolysis, which has been a well-known phenomenon in tumor cells for many years. It has already been shown in various studies, that the acidic extracellular environment can lead to functional changes in tumor cells and thereby influence the malignant behavior of tumors. Thus, experimental studies have shown that a lowered pH leads to changes in cell–cell and cell–matrix adhesion [1,2,3], increases the migratory activity of tumor cells [4,5], and thereby promotes metastasis [6]. The expression of numerous proteins is affected by acidosis [3,7]. For example, epithelial–mesenchymal transition (EMT) is promoted [8], but also markers of cell proliferation and cell death are affected. It has also been shown that the activity of drug transporters responsible for the development of multidrug resistance is enhanced under acidotic conditions [9]. Finally, the secretion of inflammatory cytokines and the phagocytosis activity of monocytes and macrophages are also altered in acidosis [10].
These numerous effects of acidosis lead directly to the question of the mechanism by which extracellular pH affects gene expression and functional parameters. In studying various signaling pathways, it has already been shown that the MAP kinases p38 and ERK1/2 are activated by acidosis [11,12] and the PI3K/Akt and PKC (protein kinase C) pathways are affected [9,13]. Also, the formation of reactive O2 species (ROS) is stimulated by extracellular acidosis [11] and there is a decrease in intracellular Ca2+ concentration [9]. However, the exact mechanism by which pH triggers these changes in signaling cascades is not yet clear.
In particular, other mechanisms influencing gene expression must also be considered. For the milieu factor hypoxia, it is known that O2 deficiency in tumors can lead to altered expression of non-coding RNA (e.g., microRNA). For example, hypoxia leads to an increase in Mir210, or decreased expression of Mir34 via the activation of HIF-1α. In light of this, it has been shown that extracellular acidosis can also lead to altered expression of various microRNAs. In a previous study the expression of numerous microRNAs was analyzed by microRNA sequencing and qPCR, and it was found that several microRNAs were affected by the low extracellular pH [14]. The most strongly regulated microRNAs were Mir7, Mir96, Mir133b, Mir141, Mir7, Mir144, Mir183, Mir200c, Mir203, and Mir215. However, validating the results in another tumor cell line led to different results. Only four microRNAs were uniformly regulated by acidosis in different tumor lines. Mir7 was up-regulated and Mir183, Mir203, and Mir215 were down-regulated [14]. The pH-dependent regulation of Mir7 and Mir203 was also confirmed in human tumor cell lines (Supplementary Figure S1). For this reason, the present study analyzed the impact of these four pH-dependent microRNAs on gene expression.
MicroRNAs are an important regulator of post-transcriptional gene regulation. MicroRNAs are short (~22-nucleotide) single-strand non-coding RNAs. They are formed in a multi-step process from pri-microRNAs which are cleaved, exported from the nucleus, integrated in a complex with DICER1, and finally leading to single-strand mature microRNAs [15]. The mature microRNA can interact with mRNA of different genes to inhibit translation. Inhibition acts by binding the microRNA sequence to the respective complementary strand of the mRNA. If the complementary is perfect the mRNA is eliminated. If the complementary is imperfect the translation is only repressed [16]. In oncology the inhibition of translation can have different effects. If the microRNA target is an oncogene, a reduced microRNA expression will lead to a pro-oncogenic effect, whereas a higher microRNA level will act antitumorigenic. If the microRNA target is a tumor suppressor the effect of an altered microRNA expression will be reversed [17]. The target genes affect every step of tumor progression or repression. For instance, Mir203, which was shown to be acidosis-dependent, can modulate the expression of ABCE1 which in turn affects EMT or cancer progression [18,19].
Using bioinformatic techniques various genes can be predicted as targets of the abovementioned pH-dependent microRNAs, which could be associated with the functional changes described above. Therefore, the question arises of whether tumor acidosis could mediate the observed effects on inflammatory cytokine expression or on genes involved in proliferation or metastasis via pH-dependent microRNAs.
Against this background, the present study aimed to analyze whether the altered expression of the inflammatory genes Il6 (interleukin 6), Nos2 (nitric oxide synthase 2), Ccl2 (C-C motif chemokine ligand 2), Spp1 (secreted phosphoprotein 1), and Tnf (tumor necrosis factor alpha) induced by extracellular acidosis, as well as the tumor-relevant genes Acat2 (acetyl-CoA acetyltransferase 2), Aox1 (aldehyde oxidase 1), Crem (cAMP responsive element modulator), Gls2 (glutaminase 2), Per3 (period circadian regulator 3), Pink1 (PTEN induced kinase 1), Txnip (thioredoxin interacting protein), and Ypel3 (yippee like 3) can be regulated by the abovementioned pH-dependent microRNAs. The investigations were carried out both in vitro and in vivo (in experimental tumors). For this purpose, tumor cells were exposed to extracellular acidosis and expression was compared to control conditions. At the same time, the cells were transfected either with the microRNAs themselves (mimics) or with their inhibitors (antagomirs) to mimic or antagonize the conditions under acidosis.
The hypothesis described so far assumes that acidosis alters the expression of microRNAs and that these modulate gene expression either directly or via other signaling pathways (e.g., MAP kinases, PI3K). However, there is also evidence that signaling pathways (PKC, MAPK) themselves can affect the expression of microRNAs. Furthermore, because it is currently unclear how extracellular acidosis leads to altered expression of Mir7, Mir183, Mir203, and Mir215, we also investigated whether the previously described signaling pathways (MAP kinases, PI3K/Akt, PKC), increased ROS formation, or decreased intracellular Ca2+ concentration, to alter microRNA expression. To do so, cells were exposed to acidosis and incubated with inhibitors of the respective signaling pathways, and the expression of pH-sensitive microRNAs was subsequently determined.
2. Results
2.1. Genes of Inflammatory Mediators
Figure 1 shows the impact of acidosis with and without modification of the microRNA expression in both tumor cell lines in vitro. Acidosis per se had a strong impact on almost all genes. Only Nos2 (iNOS) was slightly affected by low extracellular pH. Il6 and Ccl2 showed a disparate behavior to acidosis in the two different tumor cell lines studied. Additional transfection with microRNA mimics or inhibitors showed mostly no effect. Only Mir215 in AT1 cells led to changes in mRNA expression. For Il6 and Spp1 at normal pH the inhibition of Mir215 led to an increased expression, and for Ccl2 the acidosis-induced increase could be counteracted by Mir215 overexpression reducing the Ccl2 expression to the control level. These results may indicate that Mir215 plays a role in regulating the expression of these inflammatory genes and that their altered expression during acidosis might—at least partially—be mediated by changes in Mir215 expression.
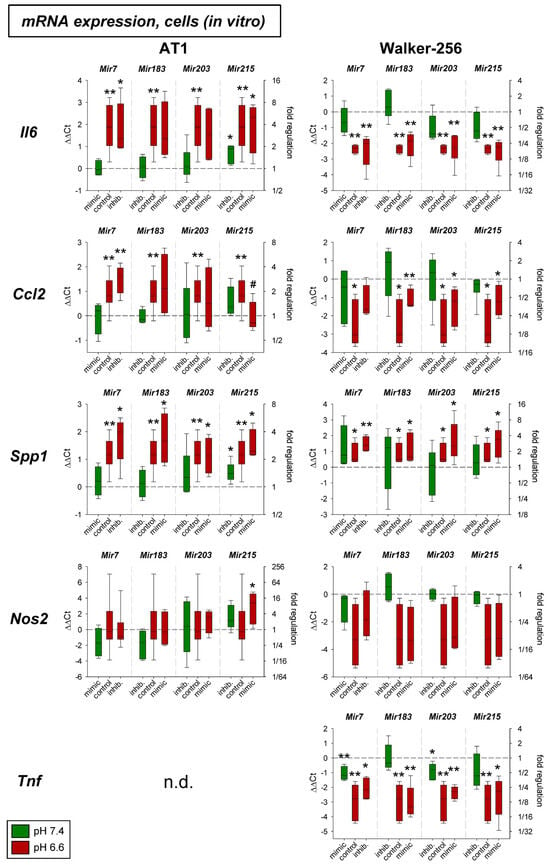
Figure 1.
mRNA expression of inflammatory genes in AT1 and Walker-256 carcinoma cells after 24 h at pH 7.4 or pH 6.6 in combination with overexpression (mimic) or inhibition (inhib.) of pH-dependent microRNAs. n = 4–16, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control; (#) p < 0.05 vs. pH 6.6 control; n.d. not detectable.
Figure 2 shows the results of the experiments in experimental AT1 tumors in vivo. Here neither acidosis alone nor additional overexpression or inhibition of the microRNAs had a systematic impact on the expression of the inflammatory genes analyzed.
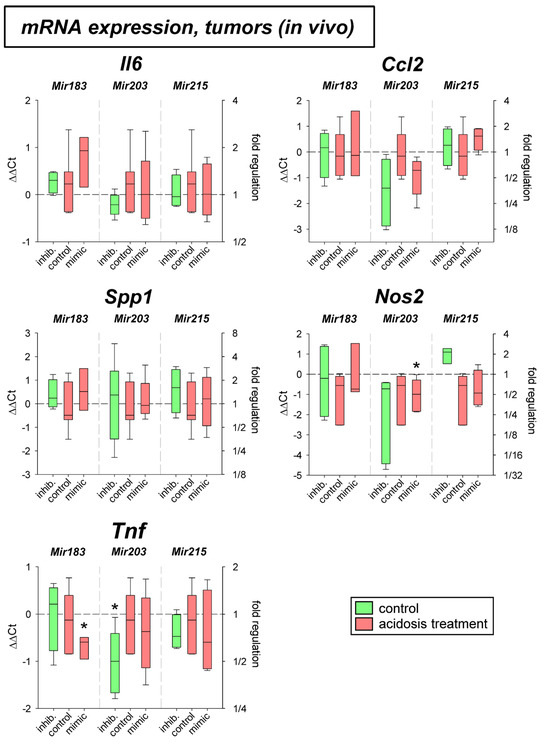
Figure 2.
mRNA expression of inflammatory genes in AT1 tumors in vivo after 24 h under control or acidotic conditions in combination with overexpression (mimic) or inhibition (inhib.) of pH-dependent microRNAs. n = 3–7, (*) p < 0.05 vs. pH 7.4 control.
2.2. Genes Related to Tumor Progression
In a previous study the mRNA expression of different genes under acidic conditions was analyzed and it was found that several genes were up- or down-regulated [3]. For eight of these genes (Crem, Gls2, Per3, Txnip, Acat2, Aox1, Pink1, Ypel3), it was tested whether for the pH-dependent regulation the pH-sensitive microRNAs are relevant. Figure 3 shows mRNA expression of Crem, Gls2, Per3, and Txnip in cells exposed to acidic pH with concomitant overexpression and inhibition of the four microRNAs analyzed. Gls2, Per3, and Txnip were up-regulated at low pH and Crem was unaffected in AT1 cells but down-regulated in Walker-256 cells. The additional transfection with microRNA mimics or inhibitors had almost no impact. For these genes the protein expression was also analyzed (Figure 4). In this group of tumor-relevant proteins, only TXNIP was significantly upregulated under acidotic conditions in both cell lines. The transfection with Mir7, Mir183, and Mir203 had almost no impact. However, in Walker-256 cells the transfection with Mir215 mimics under acidosis reduced the elevated TXNIP expression to the control level.
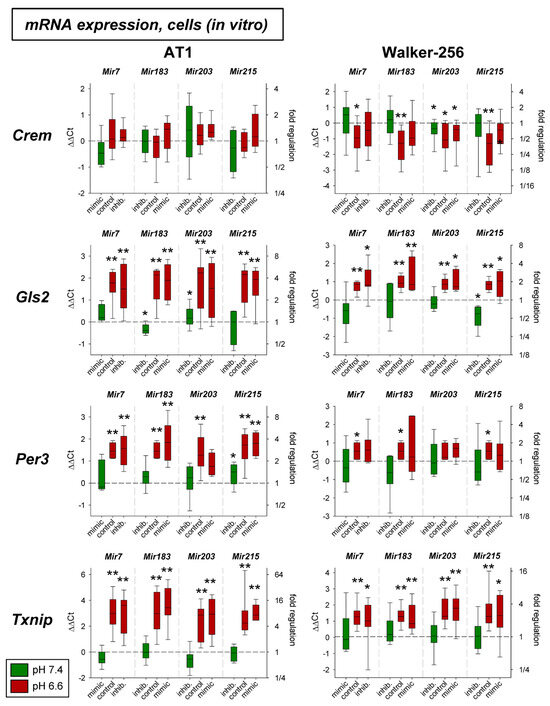
Figure 3.
mRNA expression of tumor-associated genes in AT1 and Walker-256 carcinoma cells after 24 h at pH 7.4 or pH 6.6 in combination with overexpression (mimic) or inhibition (inhib.) of pH-dependent microRNAs. n = 4–20, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control.
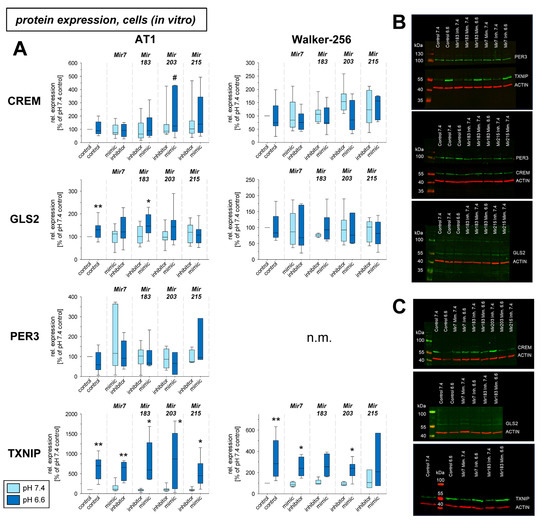
Figure 4.
(A) Protein expression of tumor-associated genes in AT1 and Walker-256 carcinoma cells after 24 h at pH 7.4 or pH 6.6 in combination with overexpression (mimic) or inhibition of pH-dependent microRNAs. Example Western blots of (B) AT1 and (C) Walker-256 cells. n = 2–24, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control; (#) p < 0.05 vs. pH 6.6 control; n.m. not measured.
Under in vivo conditions in experimental tumors Per3 and Txnip mRNA was elevated by the acidosis treatment (Figure 5), whereas Gls2 expression was significantly lowered. The transfection with microRNAs showed an effect, only for Mir215 mimics at low pH. Here, the reduced Gls2 expression was slightly elevated and the Txnip expression was further increased even above the level of acidotic control. The protein expression in vivo showed, however, a different picture (Figure 6). In experimental tumors the overexpression of Mir183 under acidic conditions led to a significant increase of CREM and GLS2 protein. The overexpression of Mir203 at low pH, however, reduced the acidosis-induced elevated TXNIP expression to the control level. Finally, overexpression of Mir215 increased the PER3 expression during the acidosis treatment.
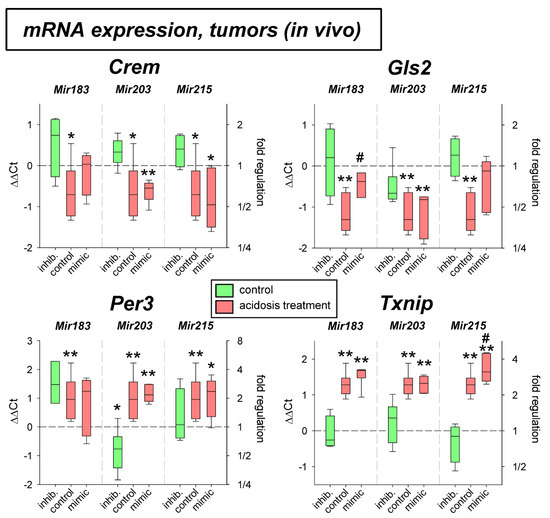
Figure 5.
mRNA expression of tumor-associated genes in AT1 tumors in vivo after 24 h under control or acidotic conditions in combination with overexpression (mimic) or inhibition (inhib.) of pH-dependent microRNAs. n = 3–9, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control, (#) p < 0.05 vs. pH 6.6 control (without transfection at the respective pH).
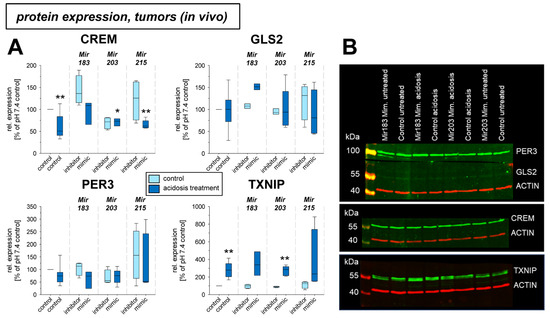
Figure 6.
(A) Protein expression of tumor-associated genes in AT1 tumors in vivo after 24 h under control or acidotic conditions in combination with overexpression (mimic) or inhibition of pH-dependent microRNAs. (B) Example Western blots. n = 2–14, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control.
In addition, the expression of four other genes (Acat2, Aox1, Pink1, Ypel3) was tested solely on the mRNA level. As shown in Figure 7 the expression of Aox1, Pink1, and Ypel3 was up-regulated by acidosis in both cell lines, whereas Acat2 was down-regulated, however, only in AT1 cells. The additional transfection with mimics or antagomirs of the different microRNAs had almost no impact. Only the overexpression of Mir215 under acidic conditions led to a significant reduction of Ypel3.
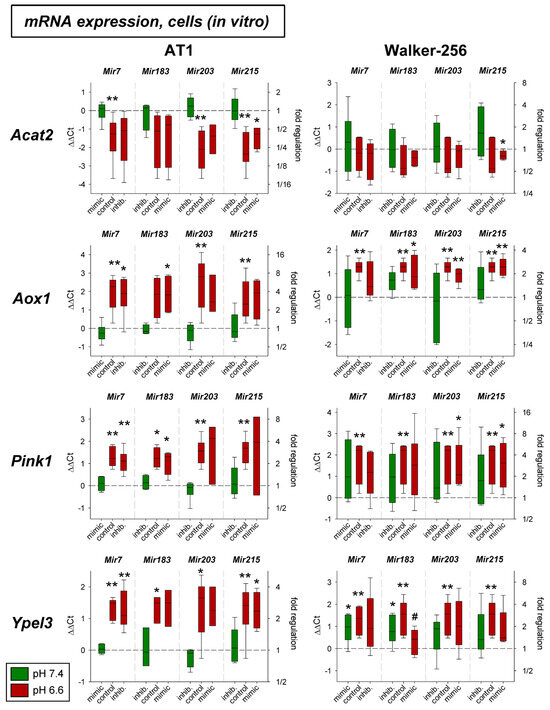
Figure 7.
mRNA expression of tumor-associated genes in AT1 and Walker-256 carcinoma cells after 24 h at pH 7.4 or pH 6.6 in combination with overexpression (mimic) or inhibition (inhib.) of pH-dependent microRNAs. n = 3–7, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control, (#) p < 0.05 vs. pH 6.6 control (without transfection at the respective pH).
The effect of acidosis alone (up-regulation of Aox1, Pink1, and Ypel3 and down-regulation of Acat2) was seen in AT1 tumors in vivo (Figure 8). In these tumors, however, none of the microRNAs showed any impact on mRNA expression.
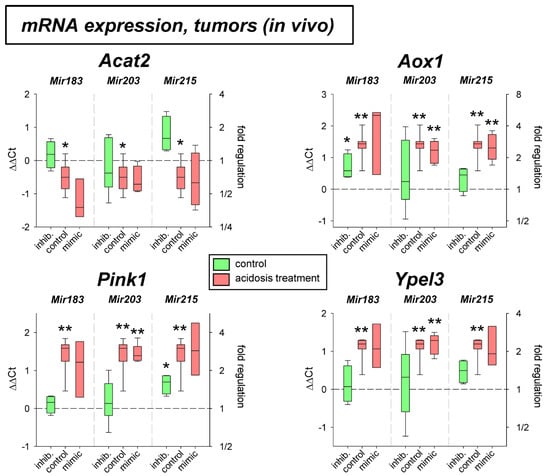
Figure 8.
mRNA expression of tumor-associated genes in AT1 tumors in vivo after 24 h under control or acidotic conditions in combination with overexpression (mimic) or inhibition (inhib.) of pH-dependent microRNAs. n = 3–9, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control.
2.3. microRNA Expression and Signaling Pathways
In the third part of the study, it was tested whether signaling pathways known to be activated by acidosis [9,11] may be responsible for the acidosis-induced change in microRNA expression. Here, we analyzed PKC, MAP kinase, and Akt/mTOR signaling as well as the role of reactive oxygen species (ROS) and low intracellular Ca2+ (both already shown to be modulated by low extracellular pH [9,11]). To analyze the role of the signaling cascades the pathways were blocked by chemical inhibitors (PKC by BIM 1 µM; MAPK by U0126 10 μM + SB203580 10 μM; PI3K by Wortmannin 0.1 µM). ROS generation was reduced by Tiron (1 mM) and low intracellular Ca2+ was induced by incubating the cells in Ca2+-free medium (Ca2+: ~5 µM) [9]. With these methods the signaling pathways were blocked during extracellular acidosis and the expression of the four pH-dependent microRNAs was measured. As Figure 9 shows, inhibition of the PKC reduced the acidosis effect on Mir7 (increased by acidosis) and Mir215 (reduced expression). Inhibition of the MAPKs had a strong effect on the expression of the Mir203, which was down-regulated under acidic conditions but was highly up-regulated with the combination of low pH and MAPK inhibition. Inhibition of the PI3K/Akt pathway reversed the effect of acidosis on Mir215 expression resulting in a strong up-regulation. ROS scavenging led to a significant increase in the expression of Mir183 and Mir203 under low pH conditions. Finally, the induction of low intracellular Ca2+ reduced the expression of the Mir7 (opposite to an extracellular acidosis) as well as the Mir183, which corresponds to the effect of extracellular acidosis.
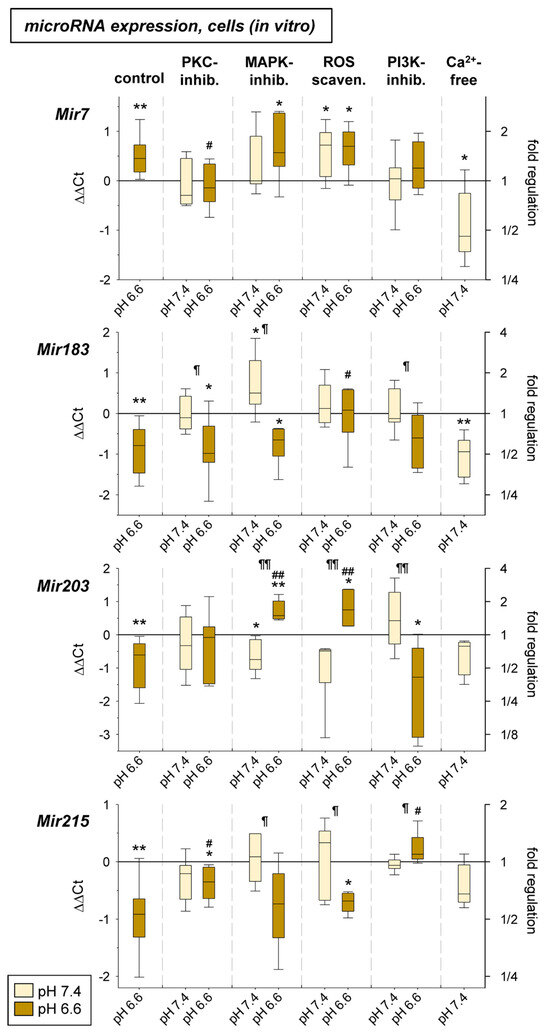
Figure 9.
microRNA expression in AT1 carcinoma cells after 24 h at pH 7.4 or pH 6.6 in combination with an inhibition of different signaling pathways (PKC: BIM bisindolylmaleimide 1 µM; MAPK: U0126 10 μM + SB203580 10 μM; ROS: Tiron 1 mM; PI3K: Wortmannin 0.1 µM; Ca2+: ~5 µM). n = 4–17, (*) p < 0.05, (**) p < 0.01 vs. pH 7.4 control, (#) p < 0.05, (##) p < 0.01 vs. pH 6.6 control, (¶) p < 0.05, (¶¶) p < 0.01 pH 6.6 vs. pH 7.4 (both with inhibition).
3. Discussion
In the present study, we investigated whether the alteration of gene expression of various inflammatory mediators and tumor-relevant proteins caused by extracellular acidosis may be mediated by pH-sensitive microRNAs. For this purpose, cells were transfected with mimics or inhibitors of the corresponding microRNAs. For Mir7, which is overexpressed under acidosis, mimic transfection at normal pH should mimic the acidosis situation and inhibitor transfection at low pH should antagonize the acidosis effect. For Mir183, Mir203, and Mir215, which are decreased in acidosis, the opposite transfection was chosen.
3.1. Inflammatory Mediators
In cell culture experiments, an increase in the expression of almost all mediators was observed under acidic control conditions in both cell lines (Figure 1). These results are in good agreement with previous experiments in tumor and normal cell lines [10,20,21]. On the other hand, measurements in experimental tumors in vivo could not confirm these acidosis effects. In artificially acidified tumors, there was no increase in mRNA of the mediators studied (Figure 2). Such a difference has also been observed by other authors. It was shown that different acid anions had different effects on cytokine secretion. For example, in RAW 264.7 cells acidification with HCl led to an increase in iNOS expression, whereas acidification with lactic acid led to a decrease in expression [22]. Also, the interaction of normal and tumor cells in tissue in vivo may influence expression [23].
The influence of pH-dependent microRNAs on expression showed almost no effect. Only overexpression of Mir215 led to a significant decrease in Ccl2 expression in AT1 cells under acidotic conditions (Figure 1). Here, transfection of the cells with the Mir215 mimic reduced Ccl2 expression to approximately the control level at pH 7.4. These results may suggest that Mir215 expression has a relevant effect on Ccl2 expression under acidosis. Although the Ccl2 gene does not have a target region for Mir215, so that direct influence by microRNA is unlikely, indirect effects via other signaling pathways must also be considered. For example, Mir215 is known to affect the PI3K pathway [24,25], which in turn can then modulate Ccl2. Also, the interference of Mir215 expression with the PI3K pathway observed in the present work (Figure 9) may be indicative of functional coupling of Mir215 with pH-dependent signaling pathways.
3.2. Genes Related to Tumor Progression
Concerning genes regulated by pH-dependent microRNAs the bioinformatic software miRWalk version 2.0 [26] was used to predict possible target genes. MiRWalk2.0 combines the information of possible miRNA binding sites within the complete sequence of a gene from several miRNA-target prediction databases (e.g., miRBase Targetscan, miRTarBase). The list of possible target genes of each pH-dependent miRNA was then analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [27,28] to determine further possible functions of these genes. The analysis revealed that genes regulated by Mir7 play a role for transcription control, cell morphogenesis, and nutrient metabolism. Additionally, this microRNA affects the activation of several signaling cascades (e.g., ERK1/2). These processes might affect cell proliferation [29,30,31] and several processes of malignant progression including multidrug resistance. In other studies, it was also shown that overexpression of Mir7 (as found under acidic conditions) can increase tumor cell migration [31,32] and tumor cell invasion [33], both important steps of metastasis formation. Genes regulated by Mir183 are involved in DNA damage repair relevant for sensitivity to non-surgical treatment modalities. Mir183 also has an impact on proliferation by regulating different genes such as low-density lipoprotein receptor-related protein 6 (LRP6) or Forkhead box O1 (FoxO1) [34,35,36]. Also, several genes relevant for cell–cell and cell–matrix adhesion are under control of this microRNA which has an impact on metastatic spread and invasiveness of tumors [37,38,39]. Mir203 is strongly related to growth regulation and morphogenesis which could have a stimulating effect on tumor growth [40,41,42]. This microRNA affects cell adhesion (positively cell–cell adhesion, negatively cell–matrix adhesion) which could have an impact on different steps of metastasis formation [42,43,44,45]. Mir215 seems to be involved in stress response (similar to response to hypoxia) and to autophagy. This microRNA was also demonstrated to suppress tumor proliferation [46,47,48] which is of high importance since acidosis leads to a reduced Mir215 expression. This microRNA was also demonstrated to modulate tumor cell migration and invasiveness [49,50]. Surprisingly for three of the four pH-dependent microRNAs a regulation of genes related to biological rhythms was predicted (e.g., Per3 period circadian regulator 3). However, the role of these biological rhythm associated genes for oncology remains currently unclear.
Analyzing the signaling pathways which are under control of the pH-dependent microRNAs shows that the mTOR pathway is regulated by Mir7 and Mir183. Akt signaling was described as being affected by Mit215 [51]. MAP kinases are modulated by Mir183, whereas Mir203 has an impact on IP3 signaling and the Ras/Raf pathway. All these signaling cascades are known to affect the malignant behavior of tumors.
In the present study, in addition to the inflammatory mediators described above, eight genes were investigated for which there was evidence from previous studies that (1) their expression might be affected by extracellular acidosis [3], and that (2) they are important for tumor malignant behavior, such as migration (e.g., Txnip, Per3, Ypel3), proliferation (e.g., Crem, Gls2, Per3, Ypel3), adhesion (e.g., Pink1, Txnip), metastasis (e.g., Pink1, Txnip), apoptosis, and necrosis (e.g., Aox1, Acat2, Crem, Per3, Txnip). For four of these genes (Crem, Gls2, Per3, Txnip), expression was determined at both the mRNA and protein levels (Figure 3 and Figure 4). Of these four genes, acidosis in vitro increased mRNA expression of Gls2, Per3, and Txnip, but only TXNIP was also shown to increase in protein level. The marked increase in Txnip expression agrees with other studies, that found Txnip increased, in some cases 60-fold, at an extracellular pH of 6.5 [52]. Crem showed no mRNA or protein change in acidosis. In experimental tumors, Per3 and Txnip were upregulated by acidosis, with surprisingly decreased expression of Gls2 by acidosis. The differences between in vitro and in vivo conditions may be due to the fact, that the experimental tumors contain normal tissue cells (e.g., fibroblasts and immune cells) in addition to tumor cells, which may show an opposite response to acidic pH.
In cell culture, there was no effect on mRNA expression by the pH-dependent microRNAs studied. Only for Crem, expression was there found to be reduced acidosis in Walker 256 cells, which led to an increase in Crem expression approximately to the control level (pH 7.4) upon simultaneous transfection with Mir183. A similar effect of Mir183 was also found for Crem expression in AT1 tumors in vivo at both the mRNA (Figure 5) and protein (Figure 6) levels. A link between Crem expression and Mir183 was previously inferred from a bioinformatics analysis of human tumors [53]. However, the correlation between Mir183 and Txnip expression described for neuropathic pain could not be found in the present study in tumor cells. However, at the protein level, Mir215 was shown to affect TXNIP expression in Walker-256 cells (Figure 4). Overexpression of Mir215 reduced Txnip expression, which was increased by acidosis, back to the control level. This may indicate that Mir215 expression, which is decreased in acidosis, may be involved in the increase in the Txnip level. For the TXNIP protein, a clear interference by overexpression of Mir203 was also observed, leading to a strong reduction of the TXNIP level (Figure 6).
For four other genes (Acat2, Aox1, Pink1, Ypel3), the influence of acidosis in combination with altered microRNA expression was investigated at the mRNA level. Both in cell culture and in vivo, lowering the pH resulted in altered expression. Acat2 was decreased expressed, whereas there was an increase in expression for Aox1, Pink1, and Ypel3 (Figure 7 and Figure 8). Decreased Acat expression in acidosis was also described previously for other tumor lines [52].
With regard to the influence of the microRNAs, these four genes showed only minor effects. Only for Ypel3, was overexpression of Mir183 found to reduce the otherwise increased expression back to the control level (pH 7.4) upon acidosis (Figure 7). There is no evidence in the literature for a correlation between Mir183 and Ypel3. Only reduced Mir215 was found to coincide with a decrease in Ypel3 expression [54], but no causal link was established. For Pink1, associations with microRNAs were described in the literature. In endothelial cells, for example, a correlation between Mir7 and Pink1 seems to exist [55]. A causal relationship between the Mir203 and Pink1 was also shown for neurons [55,56]. To what extent these are cell line-specific effects and whether these mechanisms are relevant to tumor cells must currently remain open.
3.3. microRNA Expression and Signaling Pathways
There is ample evidence in the literature that microRNA expression can interfere with various intracellular signaling pathways and by this modulate the biological behavior of tumors. In principle, two different directions of interference are possible: (1) microRNAs modulate the activity of signaling pathways, and (2) activation of cellular signaling affects the expression of microRNAs. Concerning the first direction, several mechanisms have been described. MicroRNAs have been found to modulate the cellular ROS formation (e.g., regulation of ROS formation by NOX4 or modulation of ROS scavenging enzymes as the manganese-dependent superoxide dismutase) [57,58]. ROS themselves can either directly affect cellular process like genomic instability or can activate other signaling cascades like p38 or ERK1/2 MAP kinases [57]. MicroRNAs have also been described to directly affect the expression of several enzymes of the MAPK pathways [59]. Mir203, which was found to be pH-dependent, reduces the expression of the insulin receptor substrates 1 (IRS-1) and by this affects ERK1/2 activity [60]. A similar effect was also described for the Mir145 which also targets IRS-1 and by this can affect Akt phosphorylation [61]. Other studies show that the expression of microRNAs modulate the PI3K/Akt pathway. Mir126 was found to directly target the p85 subunit of PI3K [62] Other mechanisms by which microRNAs can modulate Akt signaling include a reduced expression of the EGF receptor (e.g., by Mir133b) [63] or by a reduced expression of the Akt activator PTEN [64,65]. But also, many other proteins involved in the PI3K/Akt/mTOR way are targeted by various microRNAs [66]. Finally, the JAK/STAT3 way seems to be modulated by microRNAs. Here Mir147 was described to downregulate STAT3 maybe in combination with the long noncoding RNA (lncRNA) MEG3 [67].
On the other hand, activation of different intracellular signaling pathways may modulated the microRNA expression. For instance, ERK1/2 activity modulates phosphorylation of TRBP (transactivation response element RNA-binding protein) which acts as a co-factor of DICER which in turn is a key complex of microRNA maturation [68]. It was also shown that the transcription of pri-microRNA is affected by MAP kinases probably by an alteration of a microRNA-gene-promotor [69]. Guo et al. [70] found that EGF receptor activation increased the expression of Mir145 which was inhibited by blocking ERK1/2 activation. Also, p38, which is activated in an acidic tumor environment, can modulate the expression of microRNAs; however, the underlying mechanism is still unclear [71]. Akt also modulates microRNA expression. Mir21 was shown to be upregulated whereas Mir199a was repressed upon Akt activation [62]. Here indication was found that the microRNAs are post-transcriptionally regulated by different members of the SMAD family. Another possible mechanism by which PI3K/Akt/mTOR signaling can affect microRNA expression could be via Mdm2-dependent ubiquitination of DROSHA, which is an essential component of the microprocessor complex for the formation of pri-microRNA [66]. DROSHA (as well as DICER) can be directly affected by reactive oxygen species and through this can modulate the expression of numerous microRNAs [58].
To analyze the role of signaling pathways on the expression of pH-dependent microRNAs we investigated whether PKC, MAP kinases, PI3K/Akt signaling or other intracellular messengers (ROS, Ca2+ ions), which are known to be altered by acidosis, could have caused the observed changes in microRNA expression. Inhibition of protein kinase C had an inhibitory effect on Mir7 expression under acidotic conditions (Figure 9). In the literature, the influence of PKC on Mir15a and Mir200c has been described so far [72,73], but not for Mir7. Future studies should analyze the mechanism by which activation or inhibition of PKC may lead to modulation of miR expression and whether this mechanism has functional significance for tumor cells. Regarding MAP kinases p38 and ERK1/2, it was shown that upon inhibition, Mir203 was significantly altered during acidosis. Whereas this microRNA was decreased in acidosis, there was a significant increase upon inhibition of MAPK (at lowered pH) (Figure 9). Regulation of MAP kinases by Mir203 was described previously [74], but the reverse process has only been found for Mir7 [69,75]. Clearly, these results need further investigation, especially regarding which of the MAP kinases is responsible for the effect. However, new therapeutic approaches could also arise from such a mechanism, as Mir203 is involved in many functional processes of tumor cells. Regarding PI3K/Akt, it was found that Mir215 was significantly increased in expression by inhibiting the pathway by acidosis (Figure 9). Comparable to the situation of the interaction between MAPK and Mir203, there are findings showing activation of the PI3K/Akt pathway by Mir215 [24,51]. On the other hand, such influence seems to be reciprocal and the PI3K/Akt pathway can modulate the expression of microRNAs [65]. However, there are no findings so far for an interaction between PI3K/Akt and Mir215.
Previous studies demonstrated that extracellular acidosis leads to increased ROS formation [11]. For their part, ROS can affect the expression of various microRNAs at different levels [58]. For this reason, we investigated whether ROS scavenging during acidosis affects the formation of pH-sensitive microRNAs. Here, we found that ROS scavenging resulted in a significant increase in Mir203 (Figure 9). In the literature, evidence for such an influence comes from data of cerebral ischemia/reperfusion experiments, in which there was a reduction of Mir203 expression via a long-non-coding RNA [56]. In addition, it has been shown that extracellular acidosis leads to a reduction of intracellular Ca2+ concentration by about 30–50% [9]. For the current studies, the decrease was mimicked at normal pH by incubation with a Ca2+-free medium. It was found that under these conditions Mir183 was reduced comparably to acidosis (Figure 9). For the interaction between intracellular Ca2+ and microRNAs, both an influence of Ca2+ handling by microRNAs and of microRNA expression by Ca2+ have been described [76,77]. However, no results are yet available for Mir183.
3.4. Role of pH-Dependent microRNAs for Malignant Behavior and Therapeutic Outcome
In the present study, the influence of acidosis-dependent microRNAs on the expression of various cytokines and genes which influence the malignant behavior of tumors was investigated. In recent years, it has been shown that the extracellular pH of tumors can influence numerous functional parameters. For example, increased hematogenous metastasis was described [5], which was linked to an increased migration capacity as well as to an altered adhesion of tumor cells under acidotic conditions [3,5]. However, proliferation behavior and tumor growth were also pH-dependent [3]. Finally, it was shown that the activity of active drug transport was increased under acidotic conditions, which led to an increase in multi-drug resistance [9,78,79]. In addition to the pH-dependent changes in various intracellular signaling cascades already described in the past, the present work postulates a possible alternative mechanism by which the metabolic microenvironment may alter functional properties of tumors. Based on the present measurements, it is possible that acidosis-induced microRNAs directly influence the expression of specific relevant genes or that microRNAs indirectly interact with intracellular signaling cascades (e.g., MAP kinases, Akt/mTOR) and thereby influence the functional properties. However, no functional measurements of proliferation, metastasis, or chemoresistance were carried out in the present study. For this reason, no conclusions can be made about the validity of the hypothesis of the functional significance of microRNAs. For this purpose, extensive studies must be carried out in vitro and in vivo in the future. This study, therefore, only serves to generate a hypothesis.
These further studies should also analyze whether the acidosis-dependent microRNAs are also influenced by other parameters of the metabolic microenvironment. It has been known for some time that, for example, Mir210 is induced by hypoxia and that this can lead to changes in gene expression and tumor cell function [80]. However, it is interesting to note that the pH-dependent microRNAs investigated in the present study (in particular Mir203 and Mir215) are also associated with hypoxia [56,81,82,83,84,85]. However, other metabolic stress factors (e.g., oxidative stress or glucose deprivation) also led to altered microRNA expression, e.g., Mir203 [86,87]. In this respect, it could be possible that the metabolic microenvironment of tumors (hypoxia, acidosis, glucose deprivation, ROS formation, etc.) influences the malignant behavior of tumor cells via common microRNA pathways.
A more precise knowledge of the relationships between the metabolic microenvironment, microRNAs, intracellular signaling pathways, and functional changes in tumor cells could also potentially lead to new therapeutic approaches. For example, pharmacological intervention in intracellular signaling cascades could influence the effect of microRNAs on cell function or modulate the expression of specific microRNAs. It would therefore be conceivable to influence the impact of metabolic milieu factors on proliferation, metastasis, or therapy resistance by interfering with microRNA expression.
4. Materials and Methods
4.1. Cell Lines
All studies were performed with two tumor cell lines of the rat: (a) subline AT1 of the Dunning prostate carcinoma R3327 (CLS # 500121, CLS GmbH, Eppelheim, Germany), and (b) Walker-256 mammary carcinoma (ATCC # CCL-38, LGC Standards GmbH, Wesel, Germany). Both cell lines were cultured with room air containing 5% CO2 in RPMI medium supplemented with 10% fetal calf serum (FCS) and for Walker-256 cells additionally with 10 mM L-glutamine, 20 mM HEPES, 7.5% NaHCO3. For the experiments, cells were incubated under serum starvation for 24 h either in medium buffered with NaHCO3, 10 mM MES (morpholino-ethanesulfonic acid) and 10 mM HEPES, pH adjustment to pH 7.4 or 6.6 with 1 N HCl.
4.2. In Vivo Tumor Models
Solid tumors of AT-1 cells were induced in vivo in male Copenhagen rats (body weight 123–279 g), housed in the animal care facility of the University of Halle. All experiments had previously been approved by the regional animal ethics committee and were conducted in accordance with the German Law for Animal Protection and the UKCCCR Guidelines [88]. Solid tumors were induced by heterotopic injection of cell suspension (6–8 × 106 cells/0.4 mL isotonic saline) subcutaneously into the dorsum of the hind foot. Tumor volumes were determined by measuring the three orthogonal diameters with a caliper and with the formula: V = d1 × d2 × d3 × π/6. Tumors were investigated when they reached a volume of 0.48–1.63 mL.
In order to induce a more pronounced tumor acidosis in vivo, metabolic acidosis was intensified by treating tumor-bearing animals with a combination of inspiratory hypoxia and meta-iodobenzylguanidine (MIBG) which forces glycolytic metabolism [89]. Animals received an MIBG injection (20 mg/kg b.w., i.p., dissolved in isotonic saline) and were then housed in a hypoxic atmosphere containing 10% O2 and 90% N2 for 24 h. This procedure reduced the extracellular pH in AT1 tumors from 7.02 ± 0.04 to 6.48 ± 0.08 [14].
4.3. miRNA Transfection
To test whether four pH-dependent miRNAs (Mir7, Mir183, Mir203, Mir215) have an impact on gene and protein expression the miRNA expression was either increased or decreased by transfecting the cells with respective mimics or inhibitors (antagomirs). For the three miRNAs which were down-regulated under acidosis (Mir183, Mir203, Mir215) two experimental approaches were performed: (1) miR inhibitor at pH 7.4 (to mimic the miR expression at low pH), and (2) miR mimic at pH 6.6 (to antagonize the acidosis miR effect). For the Mir7, which was up-regulated by acidosis, the experiments were reversed ((1) mimic at pH 7.4, and (2) inhibitor at pH 6.6).
Transient transfection was performed with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. In brief, AT1 and Walker-256 cells (0.5–0.7 × 106 cells/mL) were incubated with lipofectamine and the respective miRNA mimics or antagomirs (miRCURY LNA, Qiagen, Hilden, Germany) for 24 h. The list of miRNA sequences used for transfection is shown in Supplementary Table S3. Transfection with unspecific miRNA sequences served as controls (Qiagen). Mimics were used at a final concentration of 1.7 nM and antagomirs at 16.7 nM. After 24 h the lipofectamine containing medium was replaced with media with different pHs, in which the cells were incubated for another 24 h.
For in vivo experiments different transfection procedures were used. For experiments with miR mimics the cells were transfected with the respective miRNA mimic and afterwards these cells were implanted as described above by subcutaneous injection. For inhibitor experiments untransfected tumor cells were implanted. When the tumors reached the desired volume a small amount (20 µL) of lipofectamine + respective miRNA inhibitor was injected intratumorally (under isoflurane anesthesia). The RNA strands are taken up into the cells and modulate the miRNA expression accordingly for at least 48 h.
4.4. mRNA Expression
For mRNA expression analyses, total RNA was isolated from cells or tumor homogenates using TRIzol according to the manufacturer’s instructions. For qPCR measurements 1 µg RNA was subjected to reverse transcription with SuperScript II reverse transcriptase (Thermo Fisher Scientific) and analyzed by qPCR using the Platinum SYBR Green qPCR Supermix (Thermo Fisher Scientific). The obtained data were normalized against 18S or Hprt1 and were related to the respective control. Supplementary Table S1 shows the primers used.
4.5. miRNA Expression
MiRNA expression in cells was assessed by Taqman-qPCR which was performed according to the manufacturer’s instructions (Taqman MicroRNA assay; Applied Biosystems, Waltham, MA, USA), normalized to unspecific snoRNA (RNU64702) or U6 snRNA, respectively, and Ct values were related to controls at pH 7.4. The list of oligonucleotides used for miRNA-PCR is shown in the Supplementary Table S2.
4.6. Western Blot
Western blotting was performed according to standard protocols. In brief, cells were lysed (0.5 M Tris-HCl pH 6.8; 10% SDS; 10% 2-mercaptoethanol; 20% glycerol; 0.01% bromophenol blue), separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred to a nitrocellulose membrane. Subsequently, membranes were incubated with antibodies specific for CREM (#PA5-29927, Invitrogen, Darmstadt, Germany), GLS2 (#PA5-78475, Invitrogen), PER3 (#PA5-40922, Invitrogen), and TXNIP (#14715, Cell Signaling, Danvers MA, USA). The bound primary antibody was visualized by IRDye secondary antibodies (Licor Biosciences, Lincoln, NE, USA) with the imaging system Odyssey (Licor Biosciences, Lincoln, NE, USA). Quantitative analysis was performed with Image Studio Lite Ver. 5.2.5 (Licor Biosciences, Lincoln, NE, USA).
4.7. Statistical Analysis
Results are expressed as box plots with median, 5%, 25%, 75%, and 95% percentile. Differences between groups were assessed by the two-tailed t-test for paired and unpaired samples. The significance level was set at α = 5% for all comparisons without Bonferroni correction.
5. Conclusions
In summary, the current study demonstrates that numerous tumor-related genes and cytokines are altered in their expression by extracellular acidosis in tumor cells both in vitro and in vivo. However, acidosis-dependent microRNAs appear to play only a minor role in this modulation. The data suggest that individual genes are affected by overexpression or inhibition of microRNAs, but these effects were cell line-specific. Nevertheless, the results raise broader questions when indirect effects (e.g., microRNA-induced changes in various signaling pathways) are included. In this context, the results of the last part of the study are of interest, in which it was shown that acidosis-activated signaling pathways (e.g., MAP kinases) can in turn influence microRNA expression. This could possibly also lead to new therapeutic approaches to influence the biological behavior of tumor cells in an acidic environment.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms242316919/s1.
Author Contributions
Conceptualization, O.T. and A.R.; methodology, O.T., A.R. and M.R.; validation, O.T., A.R. and M.R.; formal analysis, O.T., A.R. and M.R.; investigation, M.R. and S.R.; writing—original draft preparation, O.T.; writing—review and editing, O.T., A.R. and M.R.; visualization, O.T.; supervision, O.T.; funding acquisition, O.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Deutsche Forschungsgemeinschaft, grant number grant TH 482/6-1.
Institutional Review Board Statement
The animal experiments had previously been approved by the regional animal ethics committee and the Landesverwaltungsamt Sachsen-Anhalt, Halle (Saale), Germany (ref # 42502-2-1425 MLU, 11 January 2017).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Materials file. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rofstad, E.K.; Mathiesen, B.; Kindem, K.; Galappathi, K. Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006, 66, 6699–6707. [Google Scholar] [CrossRef]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Rauschner, M.; Lange, L.; Hüsing, T.; Reime, S.; Nolze, A.; Maschek, M.; Thews, O.; Riemann, A. Impact of the acidic environment on gene expression and functional parameters of tumors in vitro and in vivo. J. Exp. Clin. Cancer Res. 2021, 40, 10. [Google Scholar] [CrossRef] [PubMed]
- Stock, C.; Gassner, B.; Hauck, C.R.; Arnold, H.; Mally, S.; Eble, J.A.; Dieterich, P.; Schwab, A. Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J. Physiol. 2005, 567, 225–238. [Google Scholar] [CrossRef]
- Riemann, A.; Schneider, B.; Gündel, D.; Stock, C.; Thews, O.; Gekle, M. Acidic priming enhances metastatic potential of cancer cells. Pflügers Arch. 2014, 466, 2127–2138. [Google Scholar] [CrossRef] [PubMed]
- Thews, O.; Riemann, A. Tumor pH and metastasis: A malignant process beyond hypoxia. Cancer Metastasis Rev. 2019, 38, 113–129. [Google Scholar] [CrossRef]
- Yao, J.; Czaplinska, D.; Ialchina, R.; Schnipper, J.; Liu, B.; Sandelin, A.; Pedersen, S.F. Cancer Cell Acid Adaptation Gene Expression Response Is Correlated to Tumor-Specific Tissue Expression Profiles and Patient Survival. Cancers 2020, 12, 2183. [Google Scholar] [CrossRef]
- Audero, M.M.; Carvalho, T.M.A.; Ruffinatti, F.A.; Loeck, T.; Yassine, M.; Chinigo, G.; Folcher, A.; Farfariello, V.; Amadori, S.; Vaghi, C.; et al. Acidic Growth Conditions Promote Epithelial-to-Mesenchymal Transition to Select More Aggressive PDAC Cell Phenotypes In Vitro. Cancers 2023, 15, 2572. [Google Scholar] [CrossRef]
- Thews, O.; Gassner, B.; Kelleher, D.K.; Schwerdt, G.; Gekle, M. Impact of extracellular acidity on the activity of p-glycoprotein and the cytotoxicity of chemotherapeutic drugs. Neoplasia 2006, 8, 143–152. [Google Scholar] [CrossRef]
- Riemann, A.; Wußling, H.; Loppnow, H.; Fu, H.; Reime, S.; Thews, O. Acidosis differently modulates the inflammatory program in monocytes and macrophages. Biochim. Biophys. Acta 2016, 1862, 72–81. [Google Scholar] [CrossRef]
- Riemann, A.; Schneider, B.; Ihling, A.; Nowak, M.; Sauvant, C.; Thews, O.; Gekle, M. Acidic environment leads to ROS-induced MAPK signaling in cancer cells. PLoS ONE 2011, 6, e22445. [Google Scholar] [CrossRef] [PubMed]
- Riemann, A.; Ihling, A.; Thomas, J.; Schneider, B.; Thews, O.; Gekle, M. Acidic environment activates inflammatory programs in fibroblasts via a cAMP-MAPK pathway. Biochim. Biophys. Acta 2015, 1853, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Balgi, A.D.; Diering, G.H.; Donohue, E.; Lam, K.K.; Fonseca, B.D.; Zimmerman, C.; Numata, M.; Roberge, M. Regulation of mTORC1 signaling by pH. PLoS ONE 2011, 6, e21549. [Google Scholar] [CrossRef] [PubMed]
- Riemann, A.; Reime, S.; Thews, O. Acidic extracellular environment affects miRNA expression in tumors in vitro and in vivo. Int. J. Cancer 2019, 144, 1609–1618. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Gao, J.; Jung, M.; Mayoh, C.; Venkat, P.; Hannan, K.M.; Fletcher, J.I.; Kamili, A.; Gifford, A.J.; Kusnadi, E.P.; Pearson, R.B.; et al. Suppression of ABCE1-Mediated mRNA Translation Limits N-MYC-Driven Cancer Progression. Cancer Res. 2020, 80, 3706–3718. [Google Scholar] [CrossRef]
- Inoue, J.; Inazawa, J. Cancer-associated miRNAs and their therapeutic potential. J. Hum. Genet. 2021, 66, 937–945. [Google Scholar] [CrossRef]
- Avnet, S.; Di Pompo, G.; Chano, T.; Errani, C.; Ibrahim-Hashim, A.; Gillies, R.J.; Donati, D.M.; Baldini, N. Cancer-associated mesenchymal stroma fosters the stemness of osteosarcoma cells in response to intratumoral acidosis via NF-κB acivation. Int. J. Cancer 2016, 140, 1331–1345. [Google Scholar] [CrossRef]
- Riemann, A.; Reime, S.; Thews, O. Tumor acidosis and hypoxia differently modulate the inflammatory program: Measurements in vitro and in vivo. Neoplasia 2017, 19, 1033–1042. [Google Scholar] [CrossRef]
- Kellum, J.A.; Song, M.; Li, J. Science review: Extracellular acidosis and the immune response: Clinical and physiologic implications. Crit. Care 2004, 8, 331–336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peiffer, R.; Boumahd, Y.; Gullo, C.; Crake, R.; Letellier, E.; Bellahcene, A.; Peulen, O. Cancer-Associated Fibroblast Diversity Shapes Tumor Metabolism in Pancreatic Cancer. Cancers 2022, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, J.; Liu, Q.; Gong, Y.; Zhang, Y.; Zheng, Y.; Yu, D.; Zhang, Z. Mir-215-5p induces autophagy by targeting PI3K and activating ROS-mediated MAPK pathways in cardiomyocytes of chicken. J. Inorg. Biochem. 2019, 193, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Y.; Huang, C.; Lin, S. miR-215-5p is an anticancer gene in multiple myeloma by targeting RUNX1 and deactivating the PI3K/AKT/mTOR pathway. J. Cell. Biochem. 2020, 121, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, Z.; Huang, J.; Huang, S.; Li, Y.; Yu, S.; Yu, S.; Liu, X. miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int. J. Oncol. 2014, 44, 1571–1580. [Google Scholar] [CrossRef]
- Wang, F.; Qiang, Y.; Zhu, L.; Jiang, Y.; Wang, Y.; Shao, X.; Yin, L.; Chen, J.; Chen, Z. MicroRNA-7 downregulates the oncogene VDAC1 to influence hepatocellular carcinoma proliferation and metastasis. Tumour Biol. 2016, 37, 10235–10246. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, M.; Wang, G. Upregulation of microRNA-7 contributes to inhibition of the growth and metastasis of osteosarcoma cells through the inhibition of IGF1R. J. Cell. Physiol. 2019, 234, 22195–22206. [Google Scholar] [CrossRef]
- Yin, C.Y.; Kong, W.; Jiang, J.; Xu, H.; Zhao, W. miR-7-5p inhibits cell migration and invasion in glioblastoma through targeting SATB1. Oncol. Lett. 2019, 17, 1819–1825. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.X.; Bradbury, R.; Flamini, V.; Wu, B.; Jordan, N.; Jiang, W.G. MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br. J. Cancer 2017, 117, 89–101. [Google Scholar] [CrossRef]
- Ueno, K.; Hirata, H.; Shahryari, V.; Deng, G.; Tanaka, Y.; Tabatabai, Z.L.; Hinoda, Y.; Dahiya, R. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br. J. Cancer 2013, 108, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, X.; Li, Z.; Liu, H.; Teng, Y. MicroRNA-183 suppresses retinoblastoma cell growth, invasion and migration by targeting LRP6. FEBS J. 2014, 281, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Quan, H.; Wang, S.; Li, X.; Che, X. MiR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol. 2015, 36, 8121–8126. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, Y.; Qi, P.; Chen, Y.; Xu, P.; Yang, X.; Jin, X.; Tian, X. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol. Oncol. 2016, 141, 166–174. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Zheng, J.Y.; Liu, J.; Huang, C.L.; Zhang, W.; Zeng, Y. miR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed. Pharmacother. 2015, 70, 151–157. [Google Scholar] [CrossRef]
- Zhu, J.; Feng, Y.; Ke, Z.; Yang, Z.; Zhou, J.; Huang, X.; Wang, L. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am. J. Pathol. 2012, 180, 2440–2451. [Google Scholar] [CrossRef]
- Chen, T.; Xu, C.; Chen, J.; Ding, C.; Xu, Z.; Li, C.; Zhao, J. MicroRNA-203 inhibits cellular proliferation and invasion by targeting Bmi1 in non-small cell lung cancer. Oncol. Lett. 2015, 9, 2639–2646. [Google Scholar] [CrossRef][Green Version]
- Tian, L.; Li, M.; Ge, J.; Guo, Y.; Sun, Y.; Liu, M.; Xiao, H. MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumours. Tumour Biol. 2014, 35, 5953–5963. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zhou, Y.; Wang, C.; Zhang, S.; Pan, Y.; Wang, Y.; Yan, X.; Zhang, J.; Zhang, C.Y.; et al. miR-203 suppresses the proliferation and migration and promotes the apoptosis of lung cancer cells by targeting SRC. PLoS ONE 2014, 9, e105570. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Park, S.I.; McCauley, L.K.; Wang, C.Y. Signaling between transforming growth factor ß (TGF-ß) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 2013, 288, 10241–10253. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Bian, C.; Wang, H.; Huang, S.; Wu, D. MiR-203 down-regulates Rap1A and suppresses cell proliferation, adhesion and invasion in prostate cancer. J. Exp. Clin. Cancer Res. 2015, 34, 8. [Google Scholar] [CrossRef]
- Zhang, K.; Dai, L.; Zhang, B.; Xu, X.; Shi, J.; Fu, L.; Chen, X.; Li, J.; Bai, Y. miR-203 is a direct transcriptional target of E2F1 and causes G1 arrest in esophageal cancer cells. J. Cell. Physiol. 2015, 230, 903–910. [Google Scholar] [CrossRef]
- Cai, X.; Peng, D.; Wei, H.; Yang, X.; Huang, Q.; Lin, Z.; Xu, W.; Qian, M.; Yang, C.; Liu, T.; et al. miR-215 suppresses proliferation and migration of non-small cell lung cancer cells. Oncol. Lett. 2017, 13, 2349–2353. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khella, H.W.; Bakhet, M.; Allo, G.; Jewett, M.A.; Girgis, A.H.; Latif, A.; Girgis, H.; Von Both, I.; Bjarnason, G.A.; Yousef, G.M. miR-192, miR-194 and miR-215: A convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013, 34, 2231–2239. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Merhautova, J.; Machackova, T.; Gutierrez-Garcia, I.; Garcia-Solano, J.; Radova, L.; Brchnelova, D.; Slaba, K.; Svoboda, M.; Halamkova, J.; et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis 2017, 6, 399. [Google Scholar] [CrossRef]
- Chen, Z.; Han, S.; Huang, W.; Wu, J.; Liu, Y.; Cai, S.; He, Y.; Wu, S.; Song, W. MicroRNA-215 suppresses cell proliferation, migration and invasion of colon cancer by repressing Yin-Yang 1. Biochem. Biophys. Res. Commun. 2016, 479, 482–488. [Google Scholar] [CrossRef]
- Lin, Y.; Jin, Y.; Xu, T.; Zhou, S.; Cui, M. MicroRNA-215 targets NOB1 and inhibits growth and invasion of epithelial ovarian cancer. Am. J. Transl. Res. 2017, 9, 466–477. [Google Scholar]
- Yao, J.; Zhang, P.; Li, J.; Xu, W. MicroRNA-215 acts as a tumor suppressor in breast cancer by targeting AKT serine/threonine kinase 1. Oncol. Lett. 2017, 14, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Rolver, M.G.; Holland, L.K.K.; Ponniah, M.; Prasad, N.S.; Yao, J.; Schnipper, J.; Kramer, S.; Elingaard-Larsen, L.; Pedraz-Cuesta, E.; Liu, B.; et al. Chronic acidosis rewires cancer cell metabolism through PPARalpha signaling. Int. J. Cancer 2023, 152, 1668–1684. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Wu, J.; Gao, Z.; Yu, S.; Cui, Y. Screening the key microRNAs and transcription factors in prostate cancer based on microRNA functional synergistic relationships. Medicine 2017, 96, e5679. [Google Scholar] [CrossRef]
- Lee, C.H.; Kuo, W.H.; Lin, C.C.; Oyang, Y.J.; Huang, H.C.; Juan, H.F. MicroRNA-regulated protein-protein interaction networks and their functions in breast cancer. Int. J. Mol. Sci. 2013, 14, 11560–11606. [Google Scholar] [CrossRef]
- Liu, W.; Song, H.; Xu, J.; Guo, Y.; Zhang, C.; Yao, Y.; Zhang, H.; Liu, Z.; Li, Y.C. Low shear stress inhibits endothelial mitophagy via caveolin-1/miR-7-5p/SQSTM1 signaling pathway. Atherosclerosis 2022, 356, 9–17. [Google Scholar] [CrossRef]
- Yang, Z.B.; Xiang, Y.; Zuo, M.L.; Mao, L.; Hu, G.H.; Song, G.L.; Sheikh, M.S.A.; Tan, L.M. lncRNA PINK1-AS Aggravates Cerebral Ischemia/Reperfusion Oxidative Stress Injury through Regulating ATF2 by Sponging miR-203. Oxidative Med. Cell. Longev. 2022, 2022, 1296816. [Google Scholar] [CrossRef] [PubMed]
- Asl, E.R.; Amini, M.; Najafi, S.; Mansoori, B.; Mokhtarzadeh, A.; Mohammadi, A.; Lotfinejad, P.; Bagheri, M.; Shirjang, S.; Lotfi, Z.; et al. Interplay between MAPK/ERK signaling pathway and MicroRNAs: A crucial mechanism regulating cancer cell metabolism and tumor progression. Life Sci. 2021, 278, 119499. [Google Scholar] [CrossRef]
- Babu, K.R.; Tay, Y. The Yin-Yang Regulation of Reactive Oxygen Species and MicroRNAs in Cancer. Int. J. Mol. Sci. 2019, 20, 5335. [Google Scholar] [CrossRef]
- Safa, A.; Abak, A.; Shoorei, H.; Taheri, M.; Ghafouri-Fard, S. MicroRNAs as regulators of ERK/MAPK pathway: A comprehensive review. Biomed. Pharmacother. 2020, 132, 110853. [Google Scholar] [CrossRef]
- Meng, Y.; Hu, X.; Li, S.; Zeng, X.; Qiu, L.; Wei, M.; Wang, Z.; Han, J. miR-203 inhibits cell proliferation and ERK pathway in prostate cancer by targeting IRS-1. BMC Cancer 2020, 20, 1028. [Google Scholar] [CrossRef]
- Rossi, M.; Tagliaferri, P.; Tassone, P. MicroRNAs in multiple myeloma and related bone disease. Ann. Transl. Med. 2015, 3, 334. [Google Scholar] [CrossRef]
- Sayed, D.; Abdellatif, M. AKT-ing via microRNA. Cell Cycle 2010, 9, 3213–3217. [Google Scholar] [CrossRef]
- Liu, X.; Li, G. MicroRNA-133b inhibits proliferation and invasion of ovarian cancer cells through Akt and Erk1/2 inactivation by targeting epidermal growth factor receptor. Int. J. Clin. Exp. Pathol. 2015, 8, 10605–10614. [Google Scholar]
- Adil, M.S.; Khulood, D.; Somanath, P.R. Targeting Akt-associated microRNAs for cancer therapeutics. Biochem. Pharmacol. 2021, 189, 114384. [Google Scholar] [CrossRef]
- Xu, M.; Mo, Y.Y. The Akt-associated microRNAs. Cell. Mol. Life Sci. 2012, 69, 3601–3612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, B.; Wang, H.Y.; Chang, A.; Zheng, X.F.S. Emerging Role of MicroRNAs in mTOR Signaling. Cell. Mol. Life Sci. 2017, 74, 2613–2625. [Google Scholar] [CrossRef] [PubMed]
- Sajjadi-Dokht, M.; Merza Mohamad, T.A.; Sulaiman Rahman, H.; Suliman Maashi, M.; Danshina, S.; Shomali, N.; Solali, S.; Marofi, F.; Zeinalzadeh, E.; Akbari, M.; et al. MicroRNAs and JAK/STAT3 signaling: A new promising therapeutic axis in blood cancers. Genes Dis. 2022, 9, 849–867. [Google Scholar] [CrossRef] [PubMed]
- Paroo, Z.; Ye, X.; Chen, S.; Liu, Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 2009, 139, 112–122. [Google Scholar] [CrossRef]
- Ikeda, Y.; Tanji, E.; Makino, N.; Kawata, S.; Furukawa, T. MicroRNAs associated with mitogen-activated protein kinase in human pancreatic cancer. Mol. Cancer Res. 2012, 10, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.H.; Zhang, C.; Shi, J.; Xu, M.H.; Liu, F.; Yuan, H.H.; Wang, J.Y.; Jiang, B.; Gao, F.H. Abnormal activation of the EGFR signaling pathway mediates the downregulation of miR-145 through the ERK1/2 in non-small cell lung cancer. Oncol. Rep. 2014, 31, 1940–1946. [Google Scholar] [CrossRef] [PubMed]
- Antoon, J.W.; Nitzchke, A.M.; Martin, E.C.; Rhodes, L.V.; Nam, S.; Wadsworth, S.; Salvo, V.A.; Elliott, S.; Collins-Burow, B.; Nephew, K.P.; et al. Inhibition of p38 mitogen-activated protein kinase alters microRNA expression and reverses epithelial-to-mesenchymal transition. Int. J. Oncol. 2013, 42, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- von Brandenstein, M.; Depping, R.; Schafer, E.; Dienes, H.P.; Fries, J.W. Protein kinase C alpha regulates nuclear pri-microRNA 15a release as part of endothelin signaling. Biochim. Biophys. Acta. 2011, 1813, 1793–1802. [Google Scholar] [CrossRef][Green Version]
- Sutcliffe, E.L.; Li, J.; Zafar, A.; Hardy, K.; Ghildyal, R.; McCuaig, R.; Norris, N.C.; Lim, P.S.; Milburn, P.J.; Casarotto, M.G.; et al. Chromatinized Protein Kinase C-theta: Can It Escape the Clutches of NF-kappaB? Front. Immunol. 2012, 3, 260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Lakshmanan, J.; Motameni, A.; Harbrecht, B.G. MicroRNA-203 suppresses proliferation in liver cancer associated with PIK3CA, p38 MAPK, c-Jun, and GSK3 signaling. Mol. Cell. Biochem. 2018, 441, 89–98. [Google Scholar] [CrossRef]
- Akamatsu, M.; Makino, N.; Ikeda, Y.; Matsuda, A.; Ito, M.; Kakizaki, Y.; Saito, Y.; Ishizawa, T.; Kobayashi, T.; Furukawa, T.; et al. Specific MAPK-Associated MicroRNAs in Serum Differentiate Pancreatic Cancer from Autoimmune Pancreatitis. PLoS ONE 2016, 11, e0158669. [Google Scholar] [CrossRef]
- Park, J.H.; Kho, C. MicroRNAs and Calcium Signaling in Heart Disease. Int. J. Mol. Sci. 2021, 22, 10582. [Google Scholar] [CrossRef]
- Diener, C.; Hart, M.; Alansary, D.; Poth, V.; Walch-Ruckheim, B.; Menegatti, J.; Grasser, F.; Fehlmann, T.; Rheinheimer, S.; Niemeyer, B.A.; et al. Modulation of intracellular calcium signaling by microRNA-34a-5p. Cell Death Dis. 2018, 9, 1008. [Google Scholar] [CrossRef] [PubMed]
- Sauvant, C.; Nowak, M.; Wirth, C.; Schneider, B.; Riemann, A.; Gekle, M.; Thews, O. Acidosis induces multi-drug resistance in rat prostate cancer cells (AT1) in vitro and in vivo by increasing the activity of the p-glycoprotein via activation of p38. Int. J. Cancer 2008, 123, 2532–2542. [Google Scholar] [CrossRef]
- Thews, O.; Dillenburg, W.; Fellner, M.; Buchholz, H.G.; Bausbacher, N.; Schreckenberger, M.; Rösch, F. Activation of P-glycoprotein (Pgp)-mediated drug efflux by extracellular acidosis: In vivo imaging with 68Ga-labelled PET tracer. Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 1935–1942. [Google Scholar] [CrossRef]
- Ullmann, P.; Nurmik, M.; Begaj, R.; Haan, S.; Letellier, E. Hypoxia- and MicroRNA-Induced Metabolic Reprogramming of Tumor-Initiating Cells. Cells 2019, 8, 528. [Google Scholar] [CrossRef]
- Qatrun Nada, D.; Masniza, M.L.; Abdullah, N.; Marlini, M.; Elias, M.H.; Pathmanathan, S.G.; Hayati, A.R.; Fadlul Azim, F.; Hamid, A.A.; Nur Fariha, M.M. Distinct microRNA expression pattern in breast cancer cells following anti-neoplastic treatment: A systematic review and functional analysis of microRNA target genes. Malays J. Pathol. 2022, 44, 367–385. [Google Scholar]
- Hu, J.; Sun, T.; Wang, H.; Chen, Z.; Wang, S.; Yuan, L.; Liu, T.; Li, H.R.; Wang, P.; Feng, Y.; et al. MiR-215 Is Induced Post-transcriptionally via HIF-Drosha Complex and Mediates Glioma-Initiating Cell Adaptation to Hypoxia by Targeting KDM1B. Cancer Cell 2016, 29, 49–60. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.C.; Ten Hoeve, J.J.; Grénman, R.; Wessels, L.F.; Kerkhoven, R.; Te Riele, H.; van den Brekel, M.W.; Verheij, M.; Begg, A.C. Pretreatment microRNA Expression Impacting on Epithelial-to-Mesenchymal Transition Predicts Intrinsic Radiosensitivity in Head and Neck Cancer Cell Lines and Patients. Clin. Cancer Res. 2015, 21, 5630–5638. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Sasayama, T.; Tanaka, K.; Nakamizo, S.; Nishihara, M.; Mizukawa, K.; Kohta, M.; Koyama, J.; Miyake, S.; Taniguchi, M.; et al. MicroRNA-183 upregulates HIF-1α by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J. Neurooncol. 2013, 111, 273–283. [Google Scholar] [CrossRef]
- Ullmann, P.; Nurmik, M.; Schmitz, M.; Rodriguez, F.; Weiler, J.; Qureshi-Baig, K.; Felten, P.; Nazarov, P.V.; Nicot, N.; Zuegel, N.; et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 2019, 450, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shen, Z.; Shen, Y.; Ma, M.; Jue, H.; Zhu, Y.; Guo, W. The regulatory role of MiR-203 in oxidative stress induced cell injury through the CBS/H(2)S pathway. Nitric Oxide 2022, 118, 31–38. [Google Scholar] [CrossRef]
- Fite, K.; Elkhadragy, L.; Gomez-Cambronero, J. A Repertoire of MicroRNAs Regulates Cancer Cell Starvation by Targeting Phospholipase D in a Feedback Loop That Operates Maximally in Cancer Cells. Mol. Cell. Biol. 2016, 36, 1078–1089. [Google Scholar] [CrossRef]
- Workman, P.; Aboagye, E.O.; Balkwill, F.; Balmain, A.; Bruder, G.; Chaplin, D.J.; Double, J.A.; Everitt, J.; Farningham, D.A.; Glennie, M.J.; et al. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer 2010, 102, 1555–1577. [Google Scholar] [CrossRef]
- Kalliomäki, T.; Hill, R.P. Effects of tumour acidification with glucose + MIBG on the spontaneous metastatic potential of two murine cell lines. Br. J. Cancer 2004, 90, 1842–1849. [Google Scholar] [CrossRef][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).