Enzyme Databases in the Era of Omics and Artificial Intelligence
Abstract
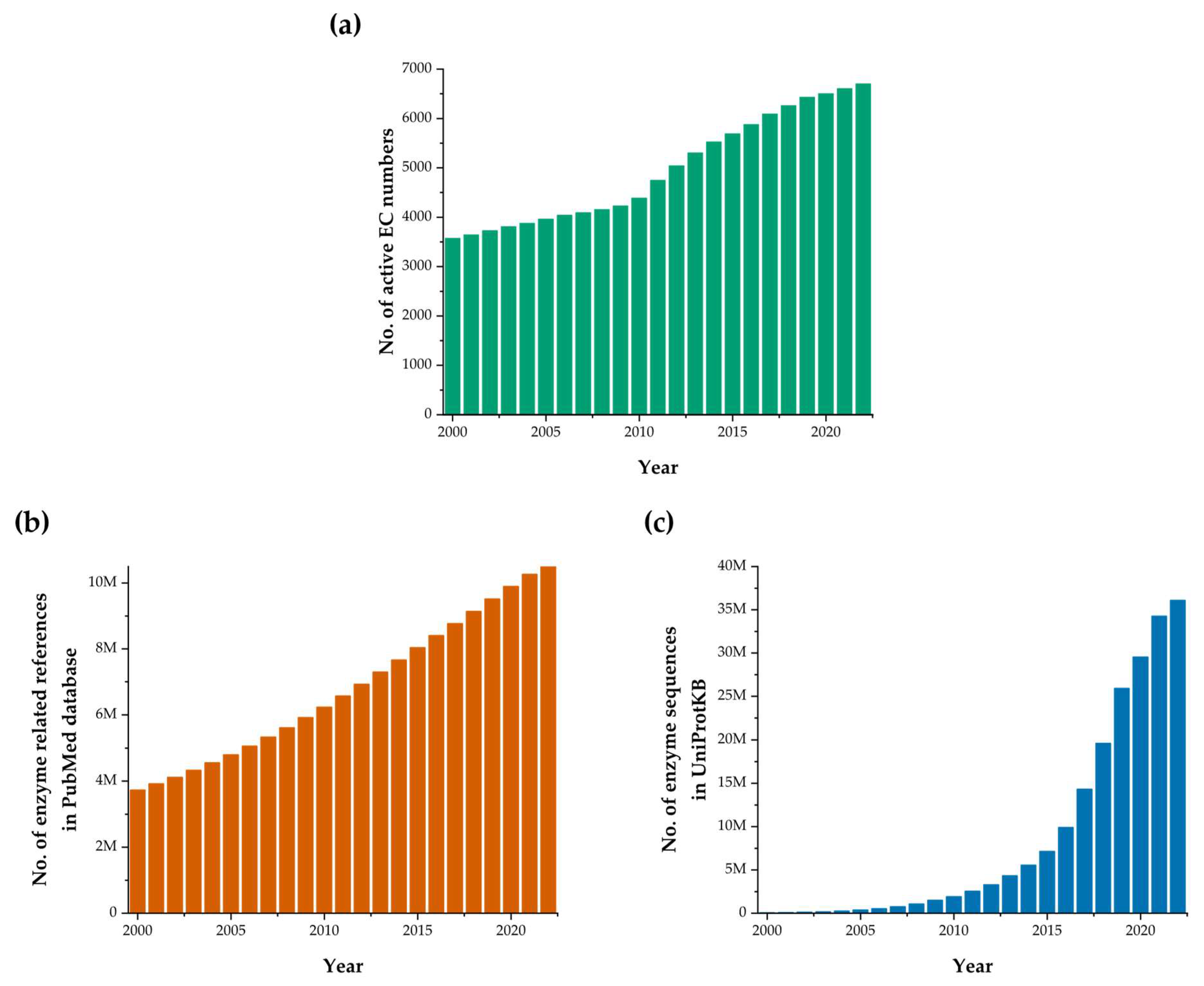
:1. Introduction
1.1. Enzyme Classification
1.2. Enzyme Databases
2. The Quest to Standardize Data Reporting
3. The Future of Enzyme Databases in the Light of Recent Artificial Intelligence Developments
4. Overview of General Enzyme Databases
4.1. Enzyme Nomenclature Databases: ExplorEnz, IntEnz, and ExPASy ENZYME
4.2. BRENDA
4.3. SABIO-RK
4.4. Reaction Mechanism Databases: M-CSA and EzCatDB
4.5. MetaCyc
4.6. KEGG
4.7. Reactome
4.8. GotEnzymes
4.9. TopEnzyme
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Strobel, S.A.; Cochrane, J.C. RNA catalysis: Ribozymes, ribosomes, and riboswitches. Curr. Opin. Chem. Biol. 2007, 11, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Geronikaki, A.; Eleutheriou, P.T. Enzymes and Enzyme Inhibitors—Applications in Medicine and Diagnosis. Int. J. Mol. Sci. 2023, 21, 5245. [Google Scholar] [CrossRef] [PubMed]
- Meghwanshi, G.K.; Kaur, N.; Verma, S.; Dabi, N.K.; Vashishtha, A.; Charan, P.D.; Purohit, P.; Bhandari, H.S.; Bhojak, N.; Kumar, R. Enzymes for pharmaceutical and therapeutic applications. Biotechnol. Appl. Biochem. 2020, 67, 586–601. [Google Scholar] [CrossRef] [PubMed]
- Mir Khan, U.; Selamoglu, Z. Use of Enzymes in Dairy Industry: A Review of Current Progress. Arch. Razi Inst. 2020, 75, 131–136. [Google Scholar]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef]
- McDonald, A.G.; Tipton, K.F. Enzyme nomenclature and classification: The state of the art. FEBS J. 2023, 290, 2214–2231. [Google Scholar] [CrossRef]
- Schomburg, D.; Schomburg, I. Enzyme Databases. In Data Mining Techniques for the Life Sciences; Carugo, O., Eisenhaber, F., Eds.; Humana Press: Totowa, NJ, USA, 2010; pp. 113–128. [Google Scholar]
- Alcántara, R.; Onwubiko, J.; Cao, H.; de Matos, P.; Cham, J.A.; Jacobsen, J.; Holliday, G.L.; Fischer, J.D.; Rahman, S.A.; Jassal, B.; et al. The EBI enzyme portal. Nucleic Acids Res. 2013, 41, D773–D780. [Google Scholar] [CrossRef]
- Ma, L.; Zou, D.; Liu, L.; Shireen, H.; Abbasi, A.A.; Bateman, A.; Xiao, J.; Zhao, W.; Bao, Y.; Zhang, Z. Database Commons: A Catalog of Worldwide Biological Databases. Genom. Proteom. Bioinform. 2022; in press. [Google Scholar] [CrossRef]
- McDonald, A.G.; Boyce, S.; Tipton, K.F. ExplorEnz: The primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009, 37, D593–D597. [Google Scholar] [CrossRef]
- Bairoch, A. The ENZYME database in 2000. Nucleic Acids Res. 2000, 28, 304–305. [Google Scholar] [CrossRef]
- Fleischmann, A.; Darsow, M.; Degtyarenko, K.; Fleischmann, W.; Boyce, S.; Axelsen, K.B.; Bairoch, A.; Schomburg, D.; Tipton, K.F.; Apweiler, R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004, 32, D434–D437. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef] [PubMed]
- Dudaš, D.; Wittig, U.; Rey, M.; Weidemann, A.; Müller, W. Improved insights into the SABIO-RK database via visualization. Database 2023, 2023, baad011. [Google Scholar] [CrossRef] [PubMed]
- Swainston, N.; Baici, A.; Bakker, B.M.; Cornish-Bowden, A.; Fitzpatrick, P.F.; Halling, P.; Leyh, T.S.; O’Donovan, C.; Raushel, F.M.; Reschel, U.; et al. STRENDA DB: Enabling the validation and sharing of enzyme kinetics data. FEBS J. 2018, 285, 2193–2204. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Ran, X.; Gollu, A.; Cheng, Z.; Zhou, X.; Chen, Y.; Yang, Z.J. IntEnzyDB: An Integrated Structure–Kinetics Enzymology Database. J. Chem. Inf. Model. 2022, 62, 5841–5848. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, X.; Peng, C.; Shi, Y.; Li, H.; Xu, Z.; Zhu, W. D3DistalMutation: A Database to Explore the Effect of Distal Mutations on Enzyme Activity. J. Chem. Inf. Model. 2021, 61, 2499–2508. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Anton, M.; Nielsen, J. GotEnzymes: An extensive database of enzyme parameter predictions. Nucleic Acids Res. 2023, 51, D583–D586. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- van der Weg, K.J.; Gohlke, H. TopEnzyme: A framework and database for structural coverage of the functional enzyme space. Bioinformatics 2023, 39, btad116. [Google Scholar] [CrossRef]
- Qi, G.; Hayward, S. Database of ligand-induced domain movements in enzymes. BMC Struct. Biol. 2009, 9, 13. [Google Scholar] [CrossRef]
- Fischer, J.D.; Holliday, G.L.; Thornton, J.M. The CoFactor database: Organic cofactors in enzyme catalysis. Bioinformatics 2010, 26, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Lingė, D.; Gedgaudas, M.; Merkys, A.; Petrauskas, V.; Vaitkus, A.; Grybauskas, A.; Paketurytė, V.; Zubrienė, A.; Zakšauskas, A.; Mickevičiūtė, A.; et al. PLBD: Protein–ligand binding database of thermodynamic and kinetic intrinsic parameters. Database 2023, 2023, baad040. [Google Scholar] [CrossRef] [PubMed]
- Sillitoe, I.; Furnham, N. FunTree: Advances in a resource for exploring and contextualising protein function evolution. Nucleic Acids Res. 2016, 44, D317–D323. [Google Scholar] [CrossRef] [PubMed]
- Hadadi, N.; Hafner, J.; Shajkofci, A.; Zisaki, A.; Hatzimanikatis, V. ATLAS of Biochemistry: A Repository of All Possible Biochemical Reactions for Synthetic Biology and Metabolic Engineering Studies. ACS Synth. Biol. 2016, 5, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Stelzer, M.; Schomburg, D. BKM-react, an integrated biochemical reaction database. BMC Biochem. 2011, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Cheng, X.; Tian, Y.; Ding, S.; Zhang, D.; Cai, P.; Hu, Q. EnzyMine: A comprehensive database for enzyme function annotation with enzymatic reaction chemical feature. Database 2020, baaa065. [Google Scholar] [CrossRef]
- Bansal, P.; Morgat, A.; Axelsen, K.B.; Muthukrishnan, V.; Coudert, E.; Aimo, L.; Hyka-Nouspikel, N.; Gasteiger, E.; Kerhornou, A.; Neto, T.B.; et al. Rhea, the reaction knowledgebase in 2022. Nucleic Acids Res. 2022, 50, D693–D700. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.G.; Tipton, K.F.; Boyce, S. Tracing metabolic pathways from enzyme data. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.J.M.; Holliday, G.L.; Furnham, N.; Tyzack, J.D.; Ferris, K.; Thornton, J.M. Mechanism and Catalytic Site Atlas (M-CSA): A database of enzyme reaction mechanisms and active sites. Nucleic Acids Res. 2018, 46, D618–D623. [Google Scholar] [CrossRef] [PubMed]
- Nagano, N.; Nakayama, N.; Ikeda, K.; Fukuie, M.; Yokota, K.; Doi, T.; Kato, T.; Tomii, K. EzCatDB: The enzyme reaction database, 2015 update. Nucleic Acids Res. 2015, 43, D453–D458. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Wishart, D.S.; Li, C.; Marcu, A.; Badran, H.; Pon, A.; Budinski, Z.; Patron, J.; Lipton, D.; Cao, X.; Oler, E.; et al. PathBank: A comprehensive pathway database for model organisms. Nucleic Acids Res. 2020, 48, D470–D478. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2023; in press. [Google Scholar] [CrossRef]
- Wittig, U.; Rey, M.; Kania, R.; Bittkowski, M.; Shi, L.; Golebiewski, M.; Weidemann, A.; Müller, W.; Rojas, I. Challenges for an enzymatic reaction kinetics database. FEBS J. 2014, 281, 572–582. [Google Scholar] [CrossRef]
- Halling, P.; Fitzpatrick, P.F.; Raushel, F.M.; Rohwer, J.; Schnell, S.; Wittig, U.; Wohlgemuth, R.; Kettner, C. An empirical analysis of enzyme function reporting for experimental reproducibility: Missing/incomplete information in published papers. Biophys. Chem. 2018, 242, 22–27. [Google Scholar] [CrossRef]
- Wittig, U.; Kania, R.; Bittkowski, M.; Wetsch, E.; Shi, L.; Jong, L.; Golebiewski, M.; Rey, M.; Weidemann, A.; Rojas, I.; et al. Data extraction for the reaction kinetics database SABIO-RK. Perspect. Sci. 2014, 1, 33–40. [Google Scholar] [CrossRef]
- Gardossi, L.; Poulsen, P.B.; Ballesteros, A.; Hult, K.; Svedas, V.K.; Vasić-Racki, D.; Carrea, G.; Magnusson, A.; Schmid, A.; Wohlgemuth, R.; et al. Guidelines for reporting of biocatalytic reactions. Trends Biotechnol. 2010, 28, 171–180. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Range, J.; Halupczok, C.; Lohmann, J.; Swainston, N.; Kettner, C.; Bergmann, F.T.; Weidemann, A.; Wittig, U.; Schnell, S.; Pleiss, J. EnzymeML—A data exchange format for biocatalysis and enzymology. FEBS J. 2022, 289, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, S.; Dienhart, H.; Range, J.; Malzacher, S.; Spöring, J.D.; Rother, D.; Pinto, M.F.; Martins, P.; Lagerman, C.E.; Bommarius, A.S.; et al. EnzymeML: Seamless data flow and modeling of enzymatic data. Nat. Methods 2023, 20, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.Y.; Kim, H.U.; Lee, S.Y. Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers. Proc. Natl. Acad. Sci. USA 2019, 116, 13996–14001. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, S.; Umarov, R.; Xie, B.; Fan, M.; Li, L.; Gao, X. DEEPre: Sequence-based enzyme EC number prediction by deep learning. Bioinformatics 2018, 34, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Dalkiran, A.; Rifaioglu, A.S.; Martin, M.J.; Cetin-Atalay, R.; Atalay, V.; Doğan, T. ECPred: A tool for the prediction of the enzymatic functions of protein sequences based on the EC nomenclature. BMC Bioinform. 2018, 19, 334. [Google Scholar] [CrossRef]
- Zou, Z.; Tian, S.; Gao, X.; Li, Y. mlDEEPre: Multi-functional enzyme function prediction with hierarchical multi-label deep learning. Front. Genet. 2019, 9, 714. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mulnaes, D.; Porta, N.; Clemens, R.; Apanasenko, I.; Reiners, J.; Gremer, L.; Neudecker, P.; Smits, S.H.J.; Gohlke, H. TopModel: Template-Based Protein Structure Prediction at Low Sequence Identity Using Top-Down Consensus and Deep Neural Networks. J. Chem. Theory Comput. 2020, 16, 1953–1967. [Google Scholar] [CrossRef]
- Torng, W.; Altman, R.B. High precision protein functional site detection using 3D convolutional neural networks. Bioinformatics 2019, 35, 1503–1512. [Google Scholar] [CrossRef]
- Song, J.; Li, F.; Takemoto, K.; Haffari, G.; Akutsu, T.; Chou, K.-C.; Webb, G.I. PREvaIL, an integrative approach for inferring catalytic residues using sequence, structural, and network features in a machine-learning framework. J. Theor. Biol. 2018, 443, 125–137. [Google Scholar] [CrossRef]
- Kroll, A.; Rousset, Y.; Hu, X.-P.; Liebrand, N.A.; Lercher, M.J. Turnover number predictions for kinetically uncharacterized enzymes using machine and deep learning. Nat. Commun. 2023, 14, 4139. [Google Scholar] [CrossRef] [PubMed]
- Kroll, A.; Engqvist, M.K.M.; Heckmann, D.; Lercher, M.J. Deep learning allows genome-scale prediction of Michaelis constants from structural features. PLoS Biol. 2021, 19, e3001402. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yuan, L.; Lu, H.; Li, G.; Chen, Y.; Engqvist, M.K.M.; Kerkhoven, E.J.; Nielsen, J. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nat. Catal. 2022, 5, 662–672. [Google Scholar] [CrossRef]
- Yeh, A.H.-W.; Norn, C.; Kipnis, Y.; Tischer, D.; Pellock, S.J.; Evans, D.; Ma, P.; Lee, G.R.; Zhang, J.Z.; Anishchenko, I.; et al. De novo design of luciferases using deep learning. Nature 2023, 614, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lisanza, S.; Juergens, D.; Tischer, D.; Watson, J.L.; Castro, K.M.; Ragotte, R.; Saragovi, A.; Milles, L.F.; Baek, M.; et al. Scaffolding protein functional sites using deep learning. Science 2022, 377, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Thapa, S.; Adhikari, S. ChatGPT, Bard, and Large Language Models for Biomedical Research: Opportunities and Pitfalls. Ann. Biomed. Eng. 2023, 51, 2647–2651. [Google Scholar] [CrossRef]
- Qian, C.; Tang, H.; Yang, Z.-J.; Liang, H.; Liu, Y. Can Large Language Models Empower Molecular Property Prediction? arXiv 2023, arXiv:2307.07443. [Google Scholar]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, I.; Hofmann, O.; Baensch, C.; Chang, A.; Schomburg, D. Enzyme data and metabolic information: BRENDA, a resource for research in biology, biochemistry, and medicine. Gene Funct. Dis. 2000, 1, 109–118. [Google Scholar] [CrossRef]
- Jeske, L.; Placzek, S.; Schomburg, I.; Chang, A.; Schomburg, D. BRENDA in 2019: A European ELIXIR core data resource. Nucleic Acids Res. 2019, 47, D542–D549. [Google Scholar] [CrossRef]
- Gremse, M.; Chang, A.; Schomburg, I.; Grote, A.; Scheer, M.; Ebeling, C.; Schomburg, D. The BRENDA Tissue Ontology (BTO): The first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011, 39, D507–D513. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Consortium, T.G.O.; Aleksander, S.A.; Balhoff, J.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Scheer, M.; Grote, A.; Chang, A.; Schomburg, I.; Munaretto, C.; Rother, M.; Söhngen, C.; Stelzer, M.; Thiele, J.; Schomburg, D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011, 39, D670–D676. [Google Scholar] [CrossRef] [PubMed]
- Barthelmes, J.; Ebeling, C.; Chang, A.; Schomburg, I.; Schomburg, D. BRENDA, AMENDA and FRENDA: The enzyme information system in 2007. Nucleic Acids Res. 2007, 35, D511–D514. [Google Scholar] [CrossRef]
- Chang, A.; Scheer, M.; Grote, A.; Schomburg, I.; Schomburg, D. BRENDA, AMENDA and FRENDA the enzyme information system: New content and tools in 2009. Nucleic Acids Res. 2009, 37, D588–D592. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, I.; Chang, A.; Placzek, S.; Söhngen, C.; Rother, M.; Lang, M.; Munaretto, C.; Ulas, S.; Stelzer, M.; Grote, A.; et al. BRENDA in 2013: Integrated reactions, kinetic data, enzyme function data, improved disease classification: New options and contents in BRENDA. Nucleic Acids Res. 2013, 41, D764–D772. [Google Scholar] [CrossRef]
- Chang, A.; Schomburg, I.; Placzek, S.; Jeske, L.; Ulbrich, M.; Xiao, M.; Sensen, C.W.; Schomburg, D. BRENDA in 2015: Exciting developments in its 25th year of existence. Nucleic Acids Res. 2015, 43, D439–D446. [Google Scholar] [CrossRef]
- Placzek, S.; Schomburg, I.; Chang, A.; Jeske, L.; Ulbrich, M.; Tillack, J.; Schomburg, D. BRENDA in 2017: New perspectives and new tools in BRENDA. Nucleic Acids Res. 2017, 45, D380–D388. [Google Scholar] [CrossRef] [PubMed]
- Quester, S.; Schomburg, D. EnzymeDetector: An integrated enzyme function prediction tool and database. BMC Bioinform. 2011, 12, 376. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Heijne, G.; von Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed]
- Wittig, U.; Rey, M.; Weidemann, A.; Kania, R.; Müller, W. SABIO-RK: An updated resource for manually curated biochemical reaction kinetics. Nucleic Acids Res. 2018, 46, D656–D660. [Google Scholar] [CrossRef]
- Krebs, O.; Golebiewski, M.; Kania, R.; Mir, S.; Saric, J.; Weidemann, A.; Wittig, U.; Rojas, I. SABIO-RK: A data warehouse for biochemical reactions and their kinetics. J. Integr. Bioinform. 2007, 4, 22–30. [Google Scholar] [CrossRef]
- Wittig, U.; Golebiewski, M.; Kania, R.; Krebs, O.; Mir, S.; Weidemann, A.; Anstein, S.; Saric, J.; Rojas, I. SABIO-RK: Integration and curation of reaction kinetics data. In Proceedings of the Lecture Notes in Computer Science (including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 4075 LNBI, Hinxton, UK, 20–22 July 2006; pp. 94–103. [Google Scholar]
- Furnham, N.; Holliday, G.L.; de Beer, T.A.P.; Jacobsen, J.O.B.; Pearson, W.R.; Thornton, J.M. The Catalytic Site Atlas 2.0: Cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 2014, 42, D485–D489. [Google Scholar] [CrossRef]
- Holliday, G.L.; Bartlett, G.J.; Almonacid, D.E.; O’Boyle, N.M.; Murray-Rust, P.; Thornton, J.M.; Mitchell, J.B.O. MACiE: A database of enzyme reaction mechanisms. Bioinformatics 2005, 21, 4315–4316. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Riley, M.; Paley, S.M.; Pellegrini-Toole, A. The MetaCyc Database. Nucleic Acids Res. 2002, 30, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Midford, P.E.; Billington, R.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Ong, W.K.; Subhraveti, P.; Caspi, R.; Fulcher, C.; et al. Pathway Tools version 23.0 update: Software for pathway/genome informatics and systems biology. Brief. Bioinform. 2021, 22, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Paley, S.; Karp, P.D. The BioCyc Metabolic Network Explorer. BMC Bioinform. 2021, 22, 208. [Google Scholar] [CrossRef]
- Krummenacker, M.; Latendresse, M.; Karp, P.D. Metabolic route computation in organism communities. Microbiome 2019, 7, 89. [Google Scholar] [CrossRef]
- Paley, S.; Parker, K.; Spaulding, A.; Tomb, J.-F.; O’Maille, P.; Karp, P.D. The Omics Dashboard for interactive exploration of gene-expression data. Nucleic Acids Res. 2017, 45, 12113–12124. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Enzyme Annotation and Metabolic Reconstruction Using KEGG. In Protein Function Prediction: Methods and Protocols; Kihara, D., Ed.; Springer: New York, NY, USA, 2017; pp. 135–145. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Altman, T.; Travers, M.; Kothari, A.; Caspi, R.; Karp, P.D. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinform. 2013, 14, 112. [Google Scholar] [CrossRef]
- Vastrik, I.; D’Eustachio, P.; Schmidt, E.; Gopinath, G.; Croft, D.; de Bono, B.; Gillespie, M.; Jassal, B.; Lewis, S.; Matthews, L.; et al. Reactome: A knowledge base of biologic pathways and processes. Genome Biol. 2007, 8, R39. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef]
- Rothfels, K.; Milacic, M.; Matthews, L.; Haw, R.; Sevilla, C.; Gillespie, M.; Stephan, R.; Gong, C.; Ragueneau, E.; May, B.; et al. Using the Reactome Database. Curr. Protoc. 2023, 3, e722. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Sauveplane, V.; Olry, A.; Ginglinger, J.-F.; Provart, N.J.; Werck-Reichhart, D. An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 2008, 8, 47. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, X.; Shi, T.; Gu, Z.; Yang, Z.; Liu, M.; Xu, Y.; Yang, Y.; Ren, L.; Song, X.; et al. P450Rdb: A manually curated database of reactions catalyzed by cytochrome P450 enzymes. J. Adv. Res. 2023; online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Q.; Liu, Y.; Liao, X.; Chu, H.; Chang, H.; Cao, Y.; Li, Z.; Zhang, T.; Cheng, J.; et al. PCPD: Plant cytochrome P450 database and web-based tools for structural construction and ligand docking. Synth. Syst. Biotechnol. 2021, 6, 102–109. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Corcoran, C.C.; Grady, C.R.; Pisitkun, T.; Parulekar, J.; Knepper, M.A. From 20th century metabolic wall charts to 21st century systems biology: Database of mammalian metabolic enzymes. Am. J. Physiol. Renal Physiol. 2016, 312, F533–F542. [Google Scholar] [CrossRef]
- Sajed, T.; Marcu, A.; Ramirez, M.; Pon, A.; Guo, A.C.; Knox, C.; Wilson, M.; Grant, J.R.; Djoumbou, Y.; Wishart, D.S. ECMDB 2.0: A richer resource for understanding the biochemistry of E. coli. Nucleic Acids Res. 2016, 44, D495–D501. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Le Boulch, M.; Déhais, P.; Combes, S.; Pascal, G. The MACADAM database: A MetAboliC pAthways DAtabase for Microbial taxonomic groups for mining potential metabolic capacities of archaeal and bacterial taxonomic groups. Database 2019, 2019, baz049. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Brewer, L.K.; Jones, J.W.; Nguyen, A.T.; Marcu, A.; Wishart, D.S.; Oglesby-Sherrouse, A.G.; Kane, M.A.; Wilks, A. PAMDB: A comprehensive Pseudomonas aeruginosa metabolome database. Nucleic Acids Res. 2018, 46, D575–D580. [Google Scholar] [CrossRef]
- Hawkins, C.; Ginzburg, D.; Zhao, K.; Dwyer, W.; Xue, B.; Xu, A.; Rice, S.; Cole, B.; Paley, S.; Karp, P.; et al. Plant Metabolic Network 15: A resource of genome-wide metabolism databases for 126 plants and algae. J. Integr. Plant Biol. 2021, 63, 1888–1905. [Google Scholar] [CrossRef] [PubMed]
- Shameer, S.; Logan-Klumpler, F.J.; Vinson, F.; Cottret, L.; Merlet, B.; Achcar, F.; Boshart, M.; Berriman, M.; Breitling, R.; Bringaud, F.; et al. TrypanoCyc: A community-led biochemical pathways database for Trypanosoma brucei. Nucleic Acids Res. 2015, 43, D637–D644. [Google Scholar] [CrossRef]
- Ramirez-Gaona, M.; Marcu, A.; Pon, A.; Guo, A.C.; Sajed, T.; Wishart, N.A.; Karu, N.; Djoumbou Feunang, Y.; Arndt, D.; Wishart, D.S. YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 2017, 45, D440–D445. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Egorova, K.S.; Smirnova, N.S.; Toukach, P.V. CSDB_GT, a curated glycosyltransferase database with close-to-full coverage on three most studied nonanimal species. Glycobiology 2021, 31, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Ausland, C.; Zheng, J.; Yi, H.; Yang, B.; Li, T.; Feng, X.; Zheng, B.; Yin, Y. dbCAN-PUL: A database of experimentally characterized CAZyme gene clusters and their substrates. Nucleic Acids Res. 2021, 49, D523–D528. [Google Scholar] [CrossRef]
- Zheng, J.; Hu, B.; Zhang, X.; Ge, Q.; Yan, Y.; Akresi, J.; Piyush, V.; Huang, L.; Yin, Y. dbCAN-seq update: CAZyme gene clusters and substrates in microbiomes. Nucleic Acids Res. 2023, 51, D557–D563. [Google Scholar] [CrossRef]
- d’Acierno, A.; Scafuri, B.; Facchiano, A.; Marabotti, A. The evolution of a Web resource: The Galactosemia Proteins Database 2.0. Hum. Mutat. 2018, 39, 52–60. [Google Scholar] [CrossRef]
- Srivastava, J.; Sunthar, P.; Balaji, P.V. Monosaccharide biosynthesis pathways database. Glycobiology 2021, 31, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, A.; Taujale, R.; McGinn, N.; Yin, Y. PlantCAZyme: A database for plant carbohydrate-active enzymes. Database 2014, 2014, bau079. [Google Scholar] [CrossRef] [PubMed]
- Adler, B.A.; Trinidad, M.I.; Bellieny-Rabelo, D.; Zhang, E.; Karp, H.M.; Skopintsev, P.; Thornton, B.W.; Weissman, R.F.; Yoon, P.H.; Chen, L.; et al. CasPEDIA Database: A functional classification system for class 2 CRISPR-Cas enzymes. Nucleic Acids Res. 2023; in press. gkad890. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chen, S.; Chen, A.; He, B.; Zhou, Y.; Chai, G.; Guo, F.; Huang, J. CasPDB: An integrated and annotated database for Cas proteins from bacteria and archaea. Database 2019, 2019, baz093. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Salvatierra, A.; Boccaletto, P.; Bujnicki, J.M. DNAmoreDB, a database of DNAzymes. Nucleic Acids Res. 2021, 49, D76–D81. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Jiang, H.; Liu, X.; Chen, Y.; Wong, J.; Wang, Q.; Huang, W.; Shi, T.; Zhang, J. HEMD: An Integrated Tool of Human Epigenetic Enzymes and Chemical Modulators for Therapeutics. PLoS ONE 2012, 7, e39917. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.K.; Petrucci, L.H.; Lambert, A.R.; Baxter, S.K.; Jarjour, J.; Stoddard, B.L. LAHEDES: The LAGLIDADG homing endonuclease database and engineering server. Nucleic Acids Res. 2012, 40, W110–W116. [Google Scholar] [CrossRef]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE: A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2023, 51, D629–D630. [Google Scholar] [CrossRef]
- Milanowska, K.; Krwawicz, J.; Papaj, G.; Kosiński, J.; Poleszak, K.; Lesiak, J.; Osińska, E.; Rother, K.; Bujnicki, J.M. REPAIRtoire—A database of DNA repair pathways. Nucleic Acids Res. 2011, 39, D788–D792. [Google Scholar] [CrossRef]
- Deng, J.; Shi, Y.; Peng, X.; He, Y.; Chen, X.; Li, M.; Lin, X.; Liao, W.; Huang, Y.; Jiang, T.; et al. Ribocentre: A database of ribozymes. Nucleic Acids Res. 2023, 51, D262–D268. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.; Tang, Q.; Liu, Y.; Qin, H.; Liu, S.; Wu, M.; Feng, P.; Chen, W. RNAME: A comprehensive database of RNA modification enzymes. Comput. Struct. Biotechnol. J. 2022, 20, 6244–6249. [Google Scholar] [CrossRef] [PubMed]
- Podlevsky, J.D.; Bley, C.J.; Omana, R.V.; Qi, X.; Chen, J.J.-L. The Telomerase Database. Nucleic Acids Res. 2008, 36, D339–D343. [Google Scholar] [CrossRef]
- Wild, A.R.; Hogg, P.W.; Flibotte, S.; Nasseri, G.G.; Hollman, R.B.; Abazari, D.; Haas, K.; Bamji, S.X. Exploring the expression patterns of palmitoylating and de-palmitoylating enzymes in the mouse brain using the curated RNA-seq database BrainPalmSeq. eLife 2022, 11, e75804. [Google Scholar] [CrossRef] [PubMed]
- Wild, A.R.; Hogg, P.W.; Flibotte, S.; Kochhar, S.; Hollman, R.B.; Haas, K.; Bamji, S.X. CellPalmSeq: A curated RNAseq database of palmitoylating and de-palmitoylating enzyme expression in human cell types and laboratory cell lines. Front. Physiol. 2023, 14, 1110550. [Google Scholar] [CrossRef]
- Grinshpon, R.D.; Williford, A.; Titus-McQuillan, J.; Clay Clark, A. The CaspBase: A curated database for evolutionary biochemical studies of caspase functional divergence and ancestral sequence inference. Protein Sci. 2018, 27, 1857–1870. [Google Scholar] [CrossRef]
- Damle, N.P.; Köhn, M. The human DEPhOsphorylation Database DEPOD: 2019 update. Database 2019, 2019, baz133. [Google Scholar] [CrossRef]
- Xue, Z.; Chen, J.-X.; Zhao, Y.; Medvar, B.; Knepper, M.A. Data integration in physiology using Bayes’ rule and minimum Bayes’ factors: Deubiquitylating enzymes in the renal collecting duct. Physiol. Genom. 2016, 49, 151–159. [Google Scholar] [CrossRef]
- Huang, H.; Arighi, C.N.; Ross, K.E.; Ren, J.; Li, G.; Chen, S.-C.; Wang, Q.; Cowart, J.; Vijay-Shanker, K.; Wu, C.H. iPTMnet: An integrated resource for protein post-translational modification network discovery. Nucleic Acids Res. 2018, 46, D542–D550. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, Y.; Lin, S.; Guo, Y.; Deng, W.; Zhang, Y.; Guo, A.; Xue, Y. iUUCD 2.0: An update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res. 2018, 46, D447–D453. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Garavelli, J.S. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics 2004, 4, 1527–1533. [Google Scholar] [CrossRef]
- Scietti, L.; Campioni, M.; Forneris, F. SiMPLOD, a Structure-Integrated Database of Collagen Lysyl Hydroxylase (LH/PLOD) Enzyme Variants. J. Bone Miner. Res. 2019, 34, 1376–1382. [Google Scholar] [CrossRef]
- Li, Z.; Chen, S.; Jhong, J.-H.; Pang, Y.; Huang, K.-Y.; Li, S.; Lee, T.-Y. UbiNet 2.0: A verified, classified, annotated and updated database of E3 ubiquitin ligase–substrate interactions. Database 2021, 2021, baab010. [Google Scholar] [CrossRef]
- Jorge, P.; Alves, D.; Pereira, M.O. Catalysing the way towards antimicrobial effectiveness: A systematic analysis and a new online resource for antimicrobial–enzyme combinations against Pseudomonas aeruginosa and Staphylococcus aureus. Int. J. Antimicrob. Agents 2019, 53, 598–605. [Google Scholar] [CrossRef]
- Vivek, K.; Diene, S.M.; Adrien, E.; Justine, D.; Olivier, C.; Laurent, T.; Jean-Marc, R.; Didier, R.; Pierre, P. An Integrative Database of β-Lactamase Enzymes: Sequences, Structures, Functions, and Phylogenetic Trees. Antimicrob. Agents Chemother. 2019, 63, e02319-18. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Li, F.; Yin, J.; Lu, M.; Mou, M.; Li, Z.; Zeng, Z.; Tan, Y.; Wang, S.; Chu, X.; Dai, H.; et al. DrugMAP: Molecular atlas and pharma-information of all drugs. Nucleic Acids Res. 2023, 51, D1288–D1299. [Google Scholar] [CrossRef]
- Yin, J.; Li, F.; Zhou, Y.; Mou, M.; Lu, Y.; Chen, K.; Xue, J.; Luo, Y.; Fu, J.; He, X.; et al. INTEDE: Interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021, 49, D1233–D1243. [Google Scholar] [CrossRef]
- Zhou, J.; Ouyang, J.; Gao, Z.; Qin, H.; Jun, W.; Shi, T. MagMD: Database summarizing the metabolic action of gut microbiota to drugs. Comput. Struct. Biotechnol. J. 2022, 20, 6427–6430. [Google Scholar] [CrossRef]
- Gao, J.; Ellis, L.B.M.; Wackett, L.P. The University of Minnesota Biocatalysis/Biodegradation Database: Improving public access. Nucleic Acids Res. 2010, 38, D488–D491. [Google Scholar] [CrossRef]
- Rojas-Vargas, J.; Castelán-Sánchez, H.G.; Pardo-López, L. HADEG: A curated hydrocarbon aerobic degradation enzymes and genes database. Comput. Biol. Chem. 2023, 107, 107966. [Google Scholar] [CrossRef]
- Arora, P.K.; Kumar, M.; Chauhan, A.; Raghava, G.P.S.; Jain, R.K. OxDBase: A database of oxygenases involved in biodegradation. BMC Res. Notes 2009, 2, 67. [Google Scholar] [CrossRef]
- Buchholz, P.C.F.; Feuerriegel, G.; Zhang, H.; Perez-Garcia, P.; Nover, L.-L.; Chow, J.; Streit, W.R.; Pleiss, J. Plastics degradation by hydrolytic enzymes: The plastics-active enzymes database—PAZy. Proteins 2022, 90, 1443–1456. [Google Scholar] [CrossRef]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. PlasticDB: A database of microorganisms and proteins linked to plastic biodegradation. Database 2022, 2022, baac008. [Google Scholar] [CrossRef]
- Gan, Z.; Zhang, H. PMBD: A Comprehensive Plastics Microbial Biodegradation Database. Database 2019, 2019, baz119. [Google Scholar] [CrossRef]
- Chakraborty, J.; Jana, T.; Saha, S.; Dutta, T.K. Ring-Hydroxylating Oxygenase database: A database of bacterial aromatic ring-hydroxylating oxygenases in the management of bioremediation and biocatalysis of aromatic compounds. Environ. Microbiol. Rep. 2014, 6, 519–523. [Google Scholar] [CrossRef]
- Deckers, M.; Van Braeckel, J.; Vanneste, K.; Deforce, D.; Fraiture, M.-A.; Roosens N h., c. Food Enzyme Database (FEDA): A web application gathering information about food enzyme preparations available on the European market. Database 2021, 2021, baab060. [Google Scholar] [CrossRef]
- Mariano, D.; Pantuza, N.; Santos, L.H.; Rocha, R.E.O.; de Lima, L.H.F.; Bleicher, L.; de Melo-Minardi, R.C. Glutantβase: A database for improving the rational design of glucose-tolerant β-glucosidases. BMC Mol. Cell Biol. 2020, 21, 50. [Google Scholar] [CrossRef]
- Wu, H.; Huang, J.; Lu, H.; Li, G.; Huang, Q. GMEnzy: A Genetically Modified Enzybiotic Database. PLoS ONE 2014, 9, e103687. [Google Scholar] [CrossRef]
- Sunny, J.S.; Nisha, K.; Natarajan, A.; Saleena, L.M. IND-enzymes: A repository for hydrolytic enzymes derived from thermophilic and psychrophilic bacterial species with potential industrial usage. Extremophiles 2021, 25, 319–325. [Google Scholar] [CrossRef]
- Sharma, V.K.; Kumar, N.; Prakash, T.; Taylor, T.D. MetaBioME: A database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 2010, 38, D468–D472. [Google Scholar] [CrossRef]
- Wang, C.Y.; Chang, P.M.; Ary, M.L.; Allen, B.D.; Chica, R.A.; Mayo, S.L.; Olafson, B.D. ProtaBank: A repository for protein design and engineering data. Protein Sci. 2018, 27, 1113–1124. [Google Scholar] [CrossRef]
- Finnigan, W.; Lubberink, M.; Hepworth, L.J.; Citoler, J.; Mattey, A.P.; Ford, G.J.; Sangster, J.; Cosgrove, S.C.; da Costa, B.Z.; Heath, R.S.; et al. RetroBioCat Database: A Platform for Collaborative Curation and Automated Meta-Analysis of Biocatalysis Data. ACS Catal. 2023, 13, 11771–11780. [Google Scholar] [CrossRef]
- Duigou, T.; du Lac, M.; Carbonell, P.; Faulon, J.-L. RetroRules: A database of reaction rules for engineering biology. Nucleic Acids Res. 2019, 47, D1229–D1235. [Google Scholar] [CrossRef]
- Percudani, R.; Peracchi, A. The B6 database: A tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinform. 2009, 10, 273. [Google Scholar] [CrossRef]
- Buchholz, P.C.F.; Vogel, C.; Reusch, W.; Pohl, M.; Rother, D.; Spieß, A.C.; Pleiss, J. BioCatNet: A Database System for the Integration of Enzyme Sequences and Biocatalytic Experiments. ChemBioChem 2016, 17, 2093–2098. [Google Scholar] [CrossRef]
- Tao, X.B.; LaFrance, S.; Xing, Y.; Nava, A.A.; Martin, H.G.; Keasling, J.D.; Backman, T.W.H. ClusterCAD 2.0: An updated computational platform for chimeric type I polyketide synthase and nonribosomal peptide synthetase design. Nucleic Acids Res. 2023, 51, D532–D538. [Google Scholar] [CrossRef]
- Bretaudeau, A.; Coste, F.; Humily, F.; Garczarek, L.; Le Corguillé, G.; Six, C.; Ratin, M.; Collin, O.; Schluchter, W.M.; Partensky, F. CyanoLyase: A database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res. 2013, 41, D396–D401. [Google Scholar] [CrossRef]
- Lenfant, N.; Hotelier, T.; Velluet, E.; Bourne, Y.; Marchot, P.; Chatonnet, A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: Tools to explore diversity of functions. Nucleic Acids Res. 2013, 41, D423–D429. [Google Scholar] [CrossRef]
- Amata, E.; Marrazzo, A.; Dichiara, M.; Modica, M.N.; Salerno, L.; Prezzavento, O.; Nastasi, G.; Rescifina, A.; Romeo, G.; Pittalà, V. Heme Oxygenase Database (HemeOxDB) and QSAR Analysis of Isoform 1 Inhibitors. ChemMedChem 2017, 12, 1873–1881. [Google Scholar] [CrossRef]
- Yu, J.-L.; Wu, S.; Zhou, C.; Dai, Q.-Q.; Schofield, C.J.; Li, G.-B. MeDBA: The Metalloenzyme Data Bank and Analysis platform. Nucleic Acids Res. 2023, 51, D593–D602. [Google Scholar] [CrossRef]
- Kishore, S.; Khosla, C. Genomic mining and diversity of assembly line polyketide synthases. Open Biol. 2023, 13, 230096. [Google Scholar] [CrossRef]
- Gunera, J.; Kindinger, F.; Li, S.-M.; Kolb, P. PrenDB, a Substrate Prediction Database to Enable Biocatalytic Use of Prenyltransferases. J. Biol. Chem. 2017, 292, 4003–4021. [Google Scholar] [CrossRef]
- Velez Rueda, A.J.; Palopoli, N.; Zacarías, M.; Sommese, L.M.; Parisi, G. ProtMiscuity: A database of promiscuous proteins. Database 2019, 2019, baz103. [Google Scholar] [CrossRef]
- Oberg, N.; Precord, T.W.; Mitchell, D.A.; Gerlt, J.A. RadicalSAM.org: A Resource to Interpret Sequence-Function Space and Discover New Radical SAM Enzyme Chemistry. ACS Bio Med Chem Au 2022, 2, 22–35. [Google Scholar] [CrossRef]
- Akhter, S.; Kaur, H.; Agrawal, P.; Raghava, G.P.S. RareLSD: A manually curated database of lysosomal enzymes associated with rare diseases. Database 2019, 2019, baz112. [Google Scholar] [CrossRef]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef]
- Stam, M.; Lelièvre, P.; Hoebeke, M.; Corre, E.; Barbeyron, T.; Michel, G. SulfAtlas, the sulfatase database: State of the art and new developments. Nucleic Acids Res. 2023, 51, D647–D653. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, R.; Zeng, T.; Zhang, X.; Wu, R. Developing TeroENZ and TeroMAP modules for the terpenome research platform TeroKit. Database 2023, 2023, baad020. [Google Scholar] [CrossRef]
- Caswell, B.T.; de Carvalho, C.C.; Nguyen, H.; Roy, M.; Nguyen, T.; Cantu, D.C. Thioesterase enzyme families: Functions, structures, and mechanisms. Protein Sci. 2022, 31, 652–676. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, K.; Iñigo, S.; Kreft, L.; Pollier, J.; De Bo, C.; Botzki, A.; Coppens, F.; Bak, S.; Goossens, A. The TriForC database: A comprehensive up-to-date resource of plant triterpene biosynthesis. Nucleic Acids Res. 2018, 46, D586–D594. [Google Scholar] [CrossRef]
| Database Type | Database | Scope of Database | Data Source | Curation |
|---|---|---|---|---|
| Enzyme nomenclature | ExplorEnz | IUBMB classification [12] | IUBMB enzyme list | Manual |
| ExPASy ENZYME | IUBMB classification with references to UniProt entries [13] | IUBMB enzyme list | Manual | |
| IntEnz | IUBMB classification with references to UniProt and ChEBI entries [14] | IUBMB enzyme list | Manual | |
| Kinetics | BRENDA | Function and kinetic parameters, enzyme–ligand interactions, organism-related information, isolation methods [15] | Experimental (implementation of some prediction tools) | Manual and automated * |
| SABIO-RK | Kinetic parameters with experimental conditions [16] | Experimental | Manual (option of data submission by experimenters) | |
| STRENDA-DB | Standardized kinetic data [17] | Experimental | Submission of data by experimenters | |
| IntEnzyDB | A comparison of kinetic parameters between wildtype and mutant enzymes [18] | Integration from multiple databases | Automated | |
| D3DistalMutation | Effects of mutations on enzyme activity [19] | Integration from multiple databases | Automated (mostly) | |
| GotEnzymes | Kinetic parameters predicted with a computer algorithm [20] | Predicted | Automated | |
| Structure | UniProt | Protein sequence and functional information [3] | Experimental and predicted | Manual and automated |
| PDB ** | Experimentally verified protein structures [21] | Experimental | Manual | |
| AlphaFold DB ** | Protein structures predicted with a computer algorithm [22] | Predicted | Automated | |
| TopEnzyme | Enzyme structures predicted with a computer algorithm [23] | Predicted | Automated | |
| Ligand-induced domain movements in enzymes | Data on movements of enzyme domains upon ligand binding [24] | Experimental | Manual and automated | |
| CoFactor | Data on organic enzyme cofactors [25] | Experimental | Manual and automated | |
| Natural Ligand DataBase | Structural data on enzyme–ligand interactions [26] | Experimental and predicted | Automated | |
| Phylogeny | FunTree | Sequence, structural, and phylogenetic data on enzymes and other proteins fun [27] | Integration from multiple databases | Automated |
| Reactions (general) | ATLAS of Biochemistry | A database of all theoretical biochemical reactions [28] | Experimental and predicted | Automated |
| BKMS-react | List of biochemical reactions from BRENDA, KEGG, MetaCyc, and SABIO-RK [29] | Integration from multiple databases | Automated | |
| EnzyMine | Mining of enzymatic reactions linked to sequence and structural annotations [30] | Integration from multiple databases | Manual | |
| Rhea | A resource of biochemical reactions [31] | IUBMB enzyme list | Manual | |
| Reaction explorer | Biochemical reactions derived from IUBMB enzyme list [32] | IUBMB enzyme list | Manual | |
| Reaction mechanism | M-CSA | Information on position and role of catalytic residues and annotated step-by-step reaction mechanisms [33] | Experimental | Manual |
| EzCatDB | A hierarchical classification of catalytic mechanisms [34] | Experimental | Manual | |
| Metabolic pathways | KEGG | Information about metabolic pathways, reactions, metabolites, enzymes, and genes [35] | Experimental | Manual |
| MetaCyc | Information about metabolic pathways, reactions, metabolites, enzymes, and genes [36] | Experimental | Manual | |
| PathBank | A metabolic pathway resource for model organisms [37] | Experimental | Manual | |
| Reactome | Information about biological pathways in human and model organisms [38] | Experimental | Manual and automated | |
| Secondary information resource | Enzyme Portal | Integration of publicly available enzyme information [10] | Integration from multiple databases | Automated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prešern, U.; Goličnik, M. Enzyme Databases in the Era of Omics and Artificial Intelligence. Int. J. Mol. Sci. 2023, 24, 16918. https://doi.org/10.3390/ijms242316918
Prešern U, Goličnik M. Enzyme Databases in the Era of Omics and Artificial Intelligence. International Journal of Molecular Sciences. 2023; 24(23):16918. https://doi.org/10.3390/ijms242316918
Chicago/Turabian StylePrešern, Uroš, and Marko Goličnik. 2023. "Enzyme Databases in the Era of Omics and Artificial Intelligence" International Journal of Molecular Sciences 24, no. 23: 16918. https://doi.org/10.3390/ijms242316918
APA StylePrešern, U., & Goličnik, M. (2023). Enzyme Databases in the Era of Omics and Artificial Intelligence. International Journal of Molecular Sciences, 24(23), 16918. https://doi.org/10.3390/ijms242316918







