Identification of Flavanone 3-Hydroxylase Gene Family in Strawberry and Expression Analysis of Fruit at Different Coloring Stages
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of F3H Gene Family in Strawberry
2.2. Phylogenetic Tree, Secondary Structure and Subcellular Localization of Strawberry F3H Family
2.3. Analysis of Gene Structure, Motif, Domain and Cis-Acting Elements
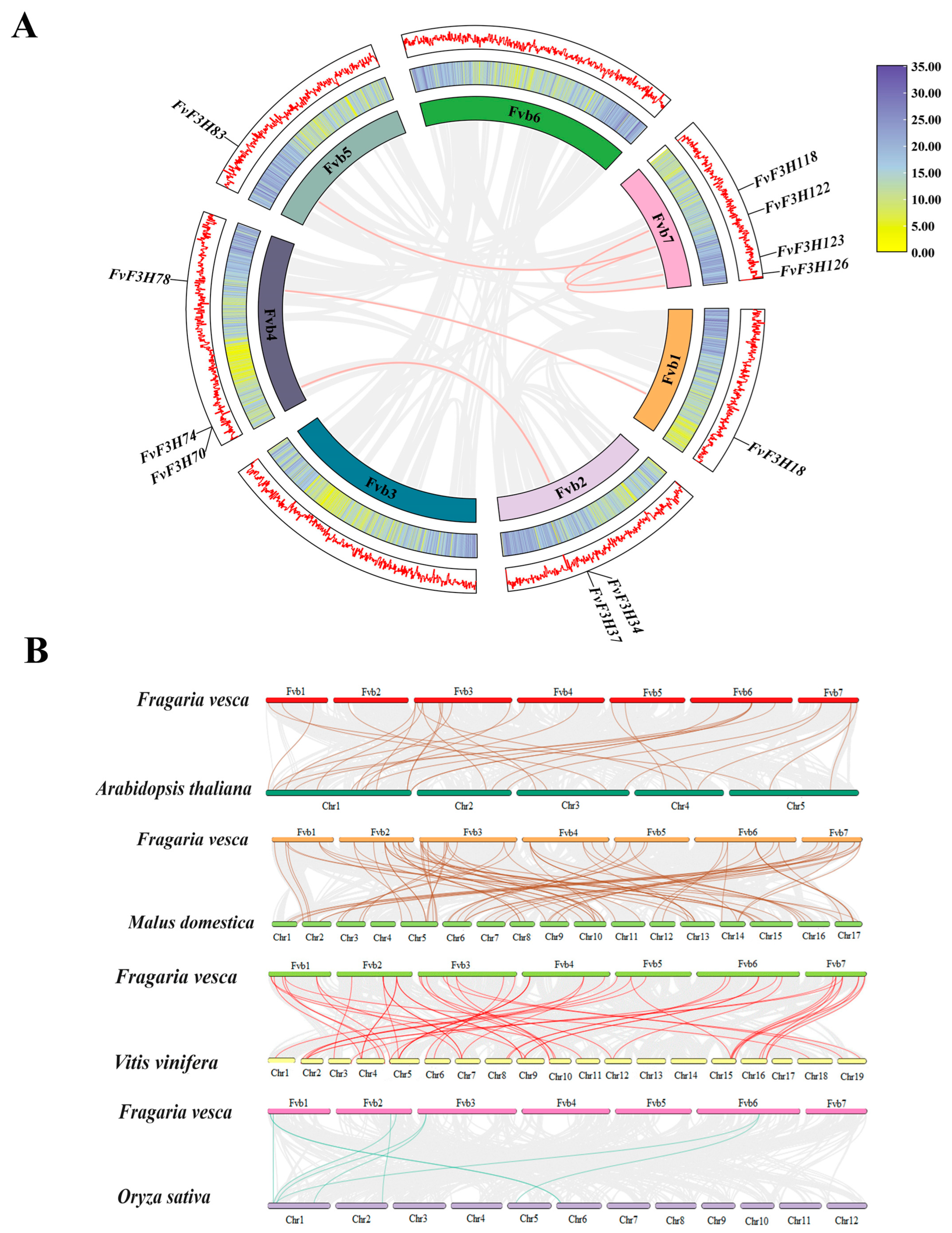
2.4. Chromosome Localization and Collinearity Analysis
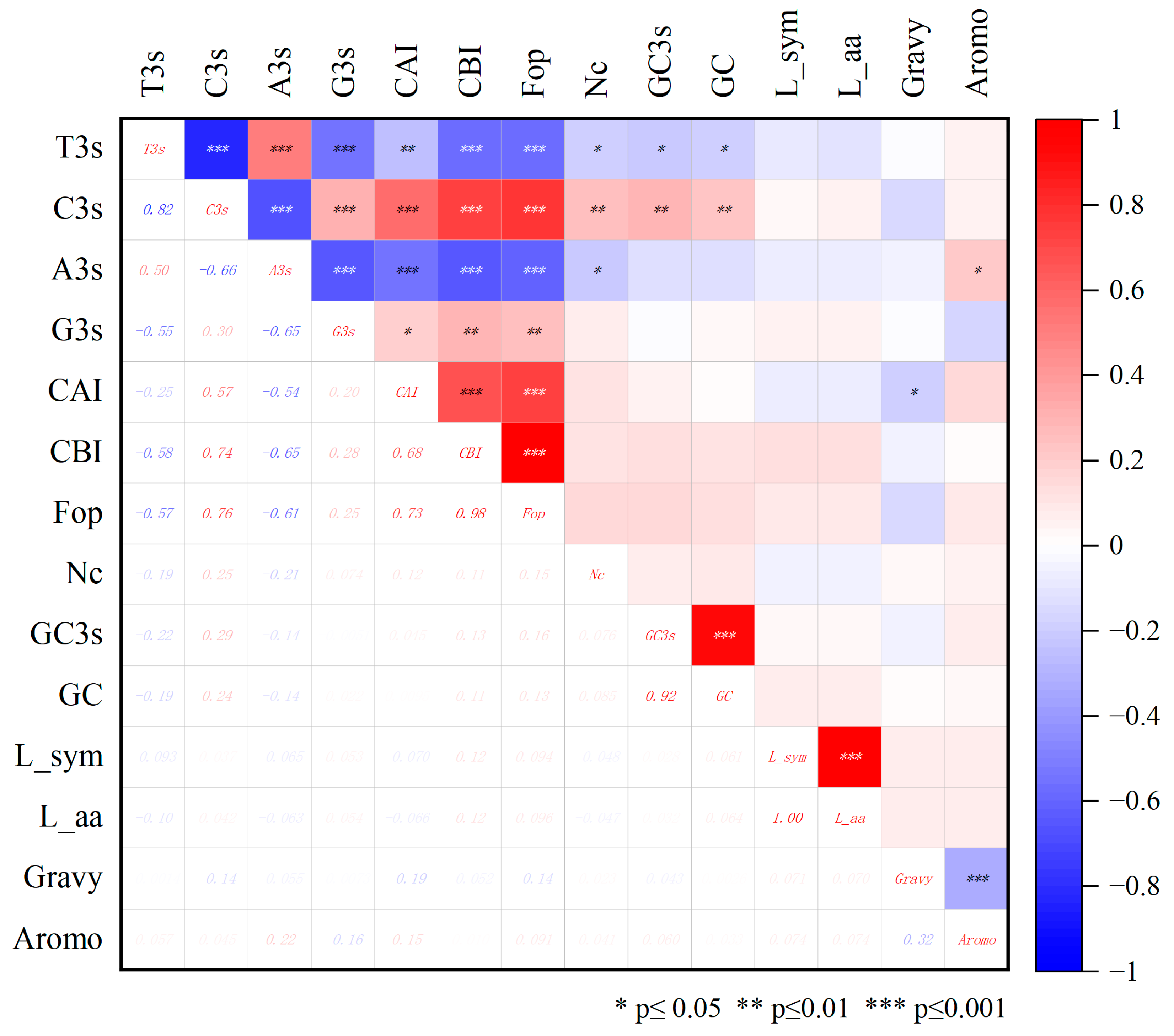
2.5. Codon Bias
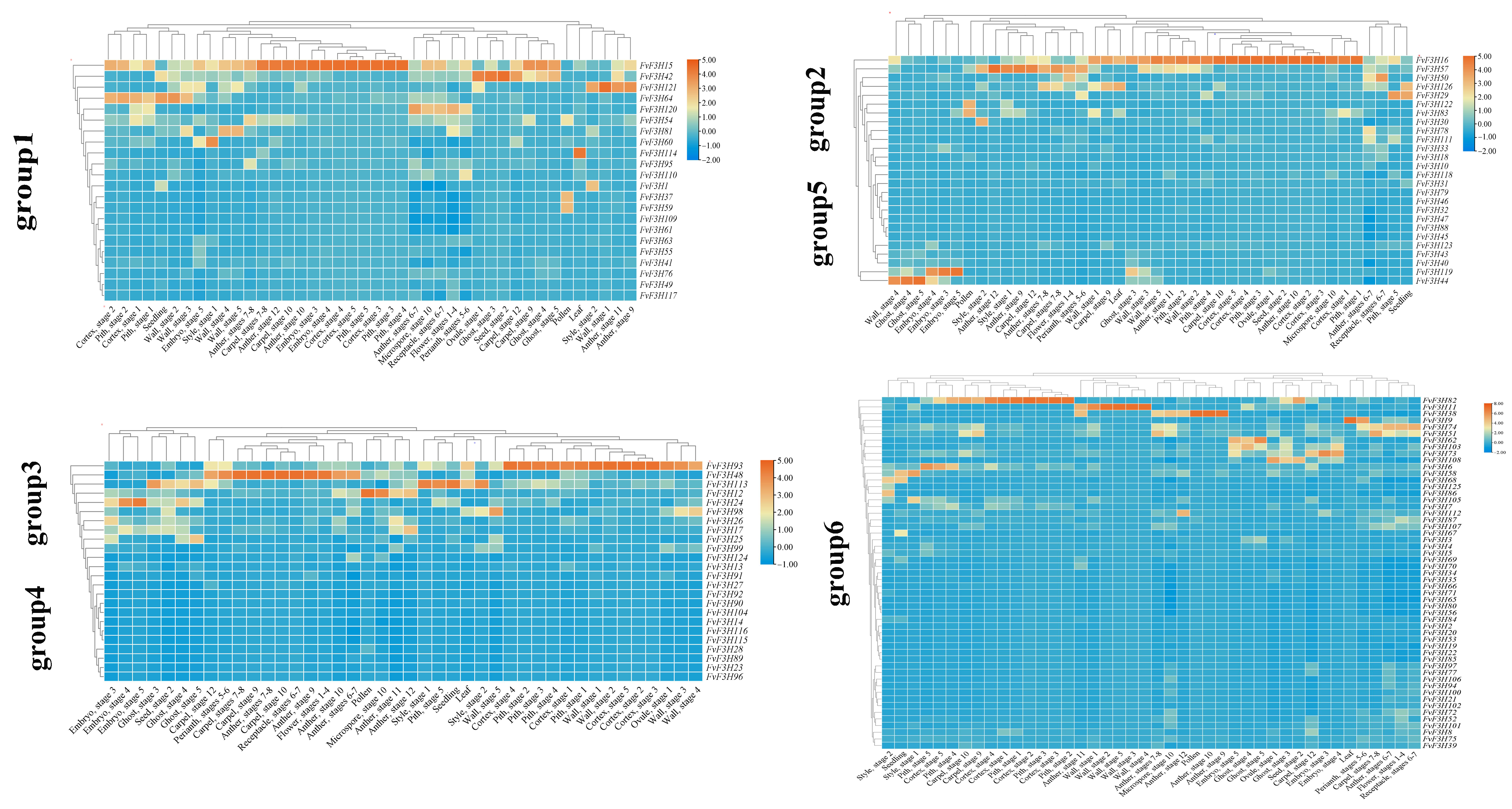
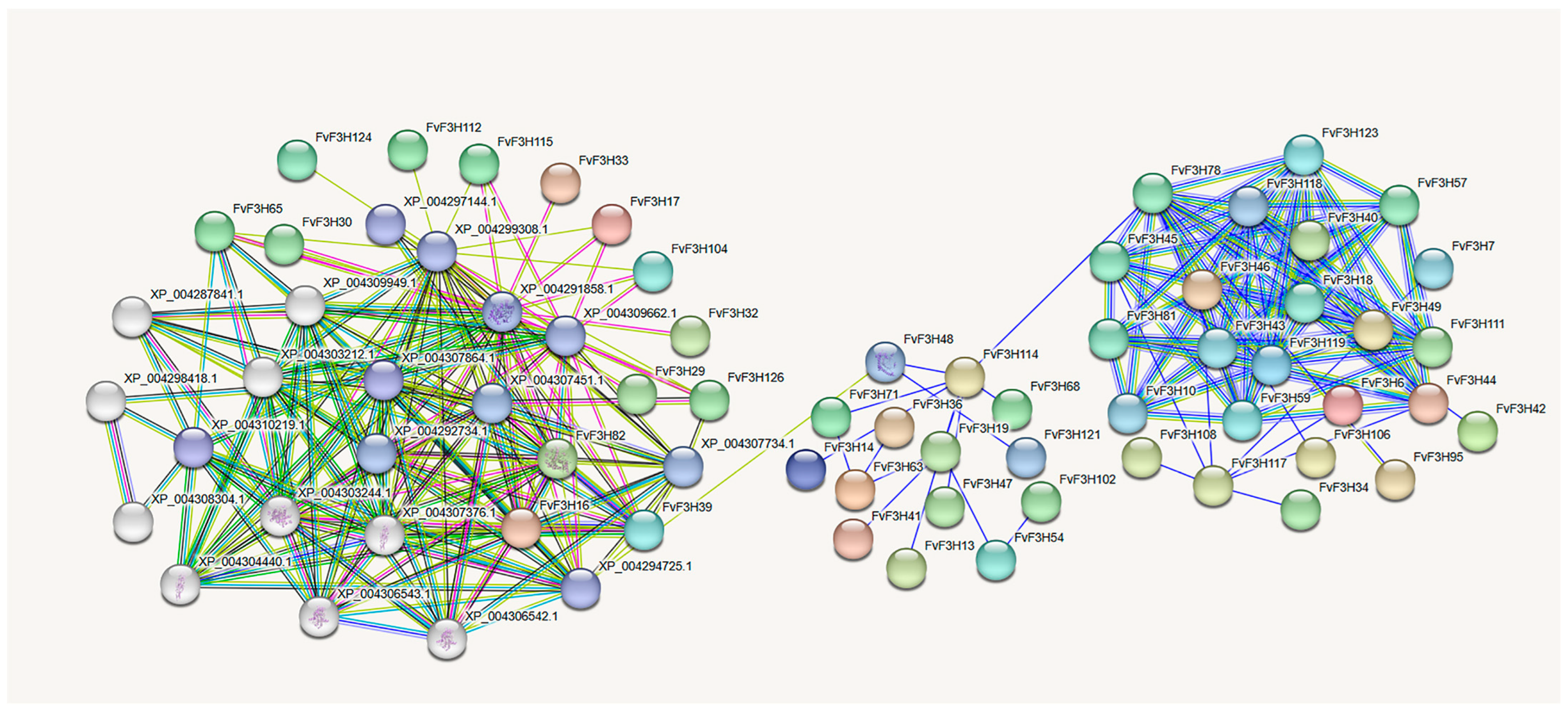
2.6. Tissue-Specific Expression Analysis and Protein Interaction Prediction
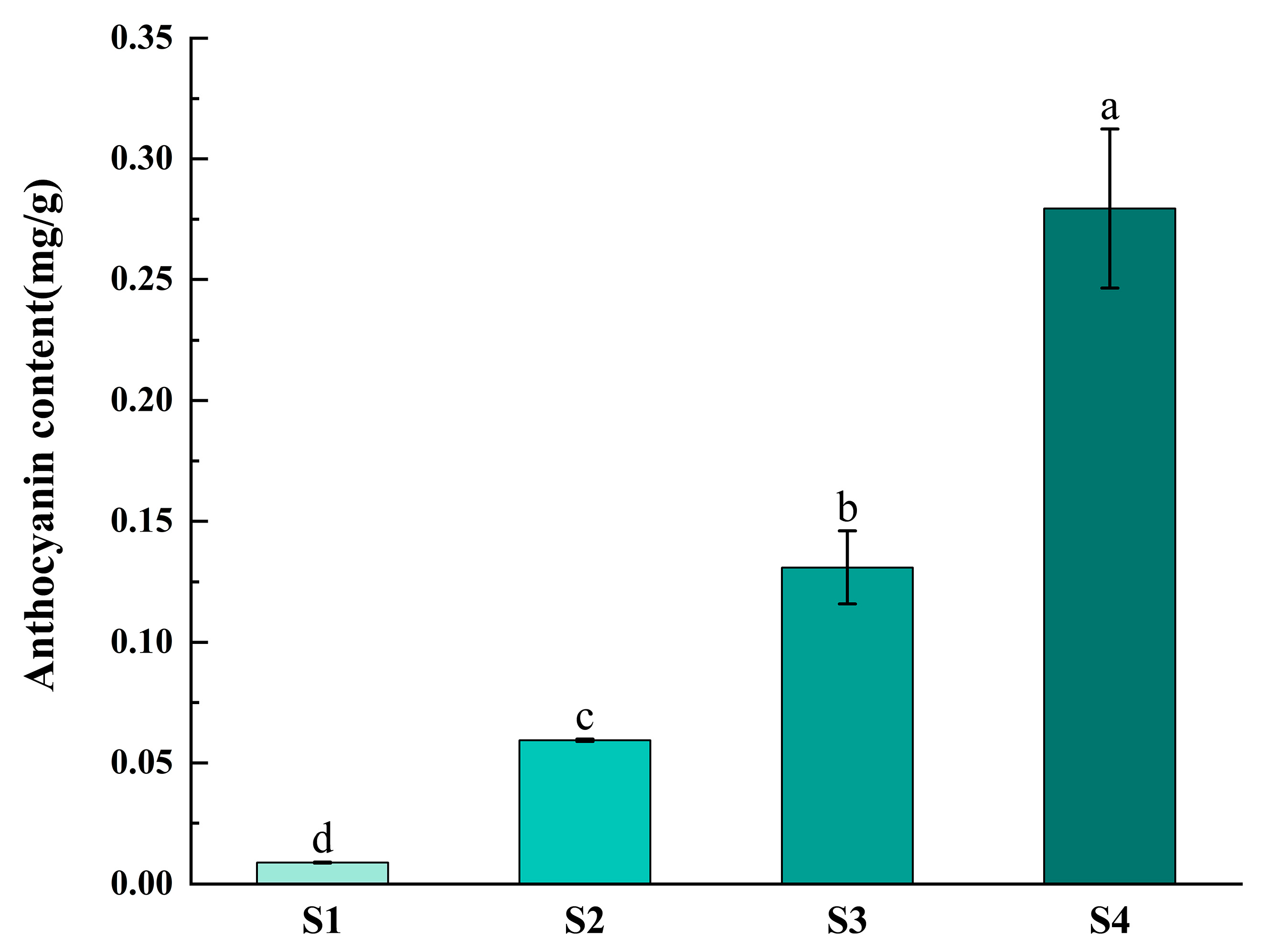
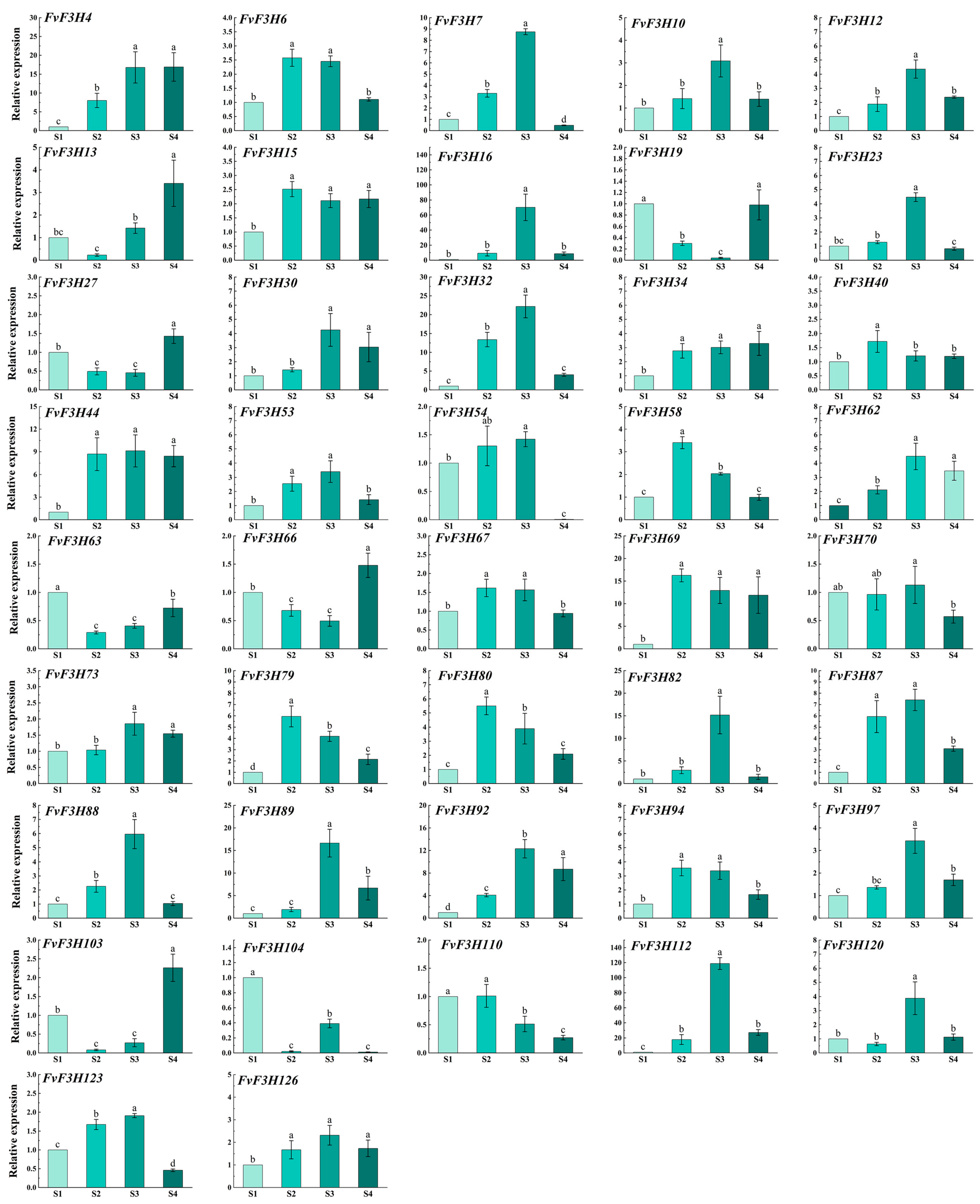
2.7. Determination of Anthocyanin Content and Expression Analysis of FvF3H Gene Family in Strawberry at Different Coloring Stages
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of F3H Gene Family in Strawberry
4.3. Evolutionary Tree Construction, Secondary Structure and Subcellular Localization
4.4. Analysis of Gene Structure, Motif and Cis-Acting Elements
4.5. Chromosome Localization and Collinearity Analysis
4.6. Codon Bias
4.7. Tissue-Specific Expression Analysis and Protein Interaction Prediction
4.8. Determination of Anthocyanin Content in Strawberry Peel at Different Developmental Stages
4.9. qRT-PCR Analysis
4.10. Statistical Analysis of Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shashank, K.; Sanjay, G. Dietary phytochemicals and their role in cancer chemoprevention. J. Cancer Metastasis Treat. 2021, 7. [Google Scholar] [CrossRef]
- Béatrice, D.; Alexandre, P.; Johann, P.; Christophe, R. Deciphering the genetic architecture of fruit color in strawberry. J. Exp. Bot. 2023, 74, 6306–6320. [Google Scholar]
- Yan, H.; Pang, F.; Wang, J.; Cai, W.; Xia, J.; Zhao, M. Identification of anthocyanin compositions and expression analysis of key related genes in Fragaria × ananassa. Acta Hortic. Sin. 2023, 50, 791–801. [Google Scholar]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, X.; Zhang, J.; Yang, B.; Yu, Y.; Liu, T.; Nie, B.; Song, B. Functional analysis of an anthocyanin synthase gene StANS in potato. Sci. Hortic. 2020, 272, 109569. [Google Scholar] [CrossRef]
- Zhou, P.; Li, J.; Jiang, H.; Jin, Q.; Wang, Y.; Xu, Y. Analysis of bZIP gene family in lotus (Nelumbo) and functional study of NnbZIP36 in regulating anthocyanin synthesis. BMC Plant Biol. 2023, 23, 429. [Google Scholar] [CrossRef]
- Britsch, L.; Ruhnau-Brich, B.; Forkmann, G. Molecular cloning, sequence analysis, and in vitro expression of flavanone 3 betahy-droxylase from Petunia hybrida. J. Biol. Chem. 1992, 267, 5380–5387. [Google Scholar] [CrossRef]
- Zuker, A.; Tzfira, T.; Ben-Meir, H.; Ovadis, M.; Shklarman, E.; Itzhaki, H.; Forkmann, G.; Martens, S.; Neta-Sharir, I.; Weiss, D.; et al. Modification of flower color and fragrance by antisense suppression of the flavanone 3-hydroxylase gene. Mol. Breed. 2002, 9, 33–34. [Google Scholar] [CrossRef]
- Wang, H.; Yan, H.; Xu, Q. Molecular cloning and expression analysis of f3h gene in Ribes rubrum and R. albrum during fruit mature. Acta Hortic. Sin. 2016, 43, 2003–2011. [Google Scholar]
- Kobayashi, S.; Ishimaru, M.; Ding, C.; Yakushiji, H.; Goto, N. Comparison of UDP-glucose: Flavornoid-3-o-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant. Sci. 2001, 160, 543–550. [Google Scholar] [CrossRef]
- Zhang, D.; Cui, L.; Wang, J.; Wu, S. Cloning f3h Gene from “Red Ballet” Fruit and Its Construction of Plant Express Vector. J. Nanjing For. Univ. 2005, 48, 65–68. [Google Scholar]
- Shen, G.; Pang, Y.Z.; Wu, W.; Deng, Z.; Zhao, L.; Cao, Y.; Sun, X.; Tang, K. Cloning and characterization of a flavanone-3-hydroxylase gene from Ginkgo biloba. Biosci. Rep. 2006, 26, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, W.; Jiang, S. Research progress of anthocyanin biosynthesis and molecular regulation. Chin. J. Hortic. 2008, 35, 909–916. [Google Scholar]
- Cheng, S.; Song, Q.; Yu, T.; Liu, X.; Wang, L.; Mao, D.; Zhang, W.; Xu, F. Characterization and expression analysis of four members genes of flavanone 3-hydroxylase families from Chamaemelum nobile. Not. Bot. Horti Agrobot. Cluj. Napoca 2020, 48, 102–115. [Google Scholar] [CrossRef]
- Li, M.; Hong, Y.; Dai, S.; Shen, R.; Zhou, Y. Effects of different light qualities on anthocyanin biosynthesis and coloration in chrysanthemum. In Proceedings of the 2016 China Ornamental Horticulture Symposium, Changsha, China, 19 July 2016. [Google Scholar]
- An, X.; Tan, T.; Zhang, X.; Guo, X.; Zhu, Y.; Song, Z.; Wang, D. Effects of light intensity on endogenous hormones and key enzyme activities of anthocyanin synthesis in blueberry leaves. Horticulturae 2023, 9, 618. [Google Scholar] [CrossRef]
- Zhang, G.; Li, P. Regulating mechanism of low night temperature on anthocyanin metabolism in ‘Fuji’ apple peel. In Proceedings of the Abstracts of the First World Apple Congress, Yangling/Yanan, China, 10 October 2016. [Google Scholar]
- Fan, M.; Jin, L.; Huang, S.; Xie, K.; Liu, Q.; Qu, D. Effect of drought on gene expressions of key enzymes in carotenoid and flavonoid biosynthesis in potato. Acta Hortic. 2008, 4, 535–542. [Google Scholar]
- Enrique, G.A.; Ana, G.; Antonia, G.; Ana, G.V.; Javier, G.M.F.; Beatriz, R.S. Elicitation with Bacillus QV15 reveals a pivotal role of F3H on flavonoid metabolism improving adaptation to biotic stress in blackberry. PLoS ONE 2020, 15, e0232626. [Google Scholar]
- Saniewski, M.; Szablińska-Piernik, J.; Marasek-Ciołakowska, A.; Mitrus, J.; Góraj-Koniarska, J.; Lahuta, L.B.; Wiczkowski, W.; Miyamoto, K.; Ueda, J.; Horbowicz, M. Accumulation of anthocyanins in detached leaves of Kalanchoë blossfeldiana: Relevance to the effect of methyl jasmonate on this process. Int. J. Mol. Sci. 2022, 24, 626. [Google Scholar] [CrossRef]
- Xue, C.; Han, J.; Peng, J.; Yang, Y.; Xiao, H.; Huang, S.; Dong, X.; Cao, F. Cloning and expression analysis of HmF3H gene related to flower color of hydrangea macrophylla. J. Mol. Plant Breed. 2021, 12, 59–64. [Google Scholar]
- Zhou, X.; Yin, H.; Zhu, Y.; Li, J. Cloning and expression analysis of flavanone 3-hydroxylase gene CnF3H from Camellia nitidissima. Mol. Plant Breed. 2015, 13, 2051–2057. [Google Scholar]
- Wang, R.; Zhang, J.; Li, Y.; Xie, Y. Cloning and expression analysis of flavanone 3-hydroxylase gene from Syringa oblata Lindl. J. Mol. Plant Breed. 2018, 16, 3863–3869. [Google Scholar]
- Song, X.; Huang, K.; Wang, R. Cloning and expression analysis of flavanone 3-hydroxylase gene in Bletilla striata. Mol. Plant Breed. 2019, 17, 4586–4591. [Google Scholar]
- Qin, S.; Zhao, X.; Cui, B.; Cheng, J.; Liu, S.; Liu, H. Functional characterization of chalcone isomerase gene HvCHI revealing its role in anthocyanin accumulation in Hosta ventricosa. Braz. J. Bot. 2022, 45, 635–643. [Google Scholar]
- Chao, N.; Wang, R.; Hou, C.; Yu, T.; Miao, K.; Cao, F.; Fang, R.; Liu, L. Functional characterization of two chalcone isomerase (CHI) revealing their responsibility for anthocyanins accumulation in mulberry. Plant Physiol. Biochem. 2021, 161, 65–73. [Google Scholar] [CrossRef]
- Kumari, G.; Nigam, V.K.; Pandey, D.M. The molecular docking and molecular dynamics study of flavonol synthase and flavonoid 3 0 -monooxygenase enzymes involved for the enrichment of kaempferol. Biomol. Struct. Dyn. 2023, 41, 2478–2491. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Kang, X.; Wang, Y.; Huang, S.; Guo, Y.; Wang, R.; Chao, N.; Liu, L. Functional characterization of flavanone 3-hydroxylase (F3H) and its role in anthocyanin and flavonoid biosynthesis in mulberry. Molecules 2022, 27, 3341. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red bartlett pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Liu, K.; Zhao, Z.; Gao, A.; Chen, Y.; Huang, J.; Dang, Z.; Luo, R. Cloning and expression analysis of F3H gene from mango (Mangifera indica). Mol. Plant Breed. 2017, 15, 2106–2111. [Google Scholar]
- Yang, H.; Wang, Y.; Huang, H. Construction of DNA library and expression of F3H gene in “red Yang” monkey. Yi Chuan 2009, 31, 1265–1272. [Google Scholar]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for in-teractive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Zheng, C.; Wu, X.; Wang, W. Protein subcellular location: The gap between prediction and experimentation. Plant Mol. Biol. Rep. 2015, 34, 52–61. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607−D613. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Feng, Y.; Yang, S.; Qiao, H.; Wu, A.; Yang, J.; Ma, Z. Identification of Flavanone 3-Hydroxylase Gene Family in Strawberry and Expression Analysis of Fruit at Different Coloring Stages. Int. J. Mol. Sci. 2023, 24, 16807. https://doi.org/10.3390/ijms242316807
Zhang Y, Feng Y, Yang S, Qiao H, Wu A, Yang J, Ma Z. Identification of Flavanone 3-Hydroxylase Gene Family in Strawberry and Expression Analysis of Fruit at Different Coloring Stages. International Journal of Molecular Sciences. 2023; 24(23):16807. https://doi.org/10.3390/ijms242316807
Chicago/Turabian StyleZhang, Yanqi, Yongqing Feng, Shangwen Yang, Huilan Qiao, Aiyuan Wu, Jinghua Yang, and Zonghuan Ma. 2023. "Identification of Flavanone 3-Hydroxylase Gene Family in Strawberry and Expression Analysis of Fruit at Different Coloring Stages" International Journal of Molecular Sciences 24, no. 23: 16807. https://doi.org/10.3390/ijms242316807
APA StyleZhang, Y., Feng, Y., Yang, S., Qiao, H., Wu, A., Yang, J., & Ma, Z. (2023). Identification of Flavanone 3-Hydroxylase Gene Family in Strawberry and Expression Analysis of Fruit at Different Coloring Stages. International Journal of Molecular Sciences, 24(23), 16807. https://doi.org/10.3390/ijms242316807




