Quartz Crystal Microbalance Platform for SARS-CoV-2 Immuno-Diagnostics
Abstract
:1. Introduction
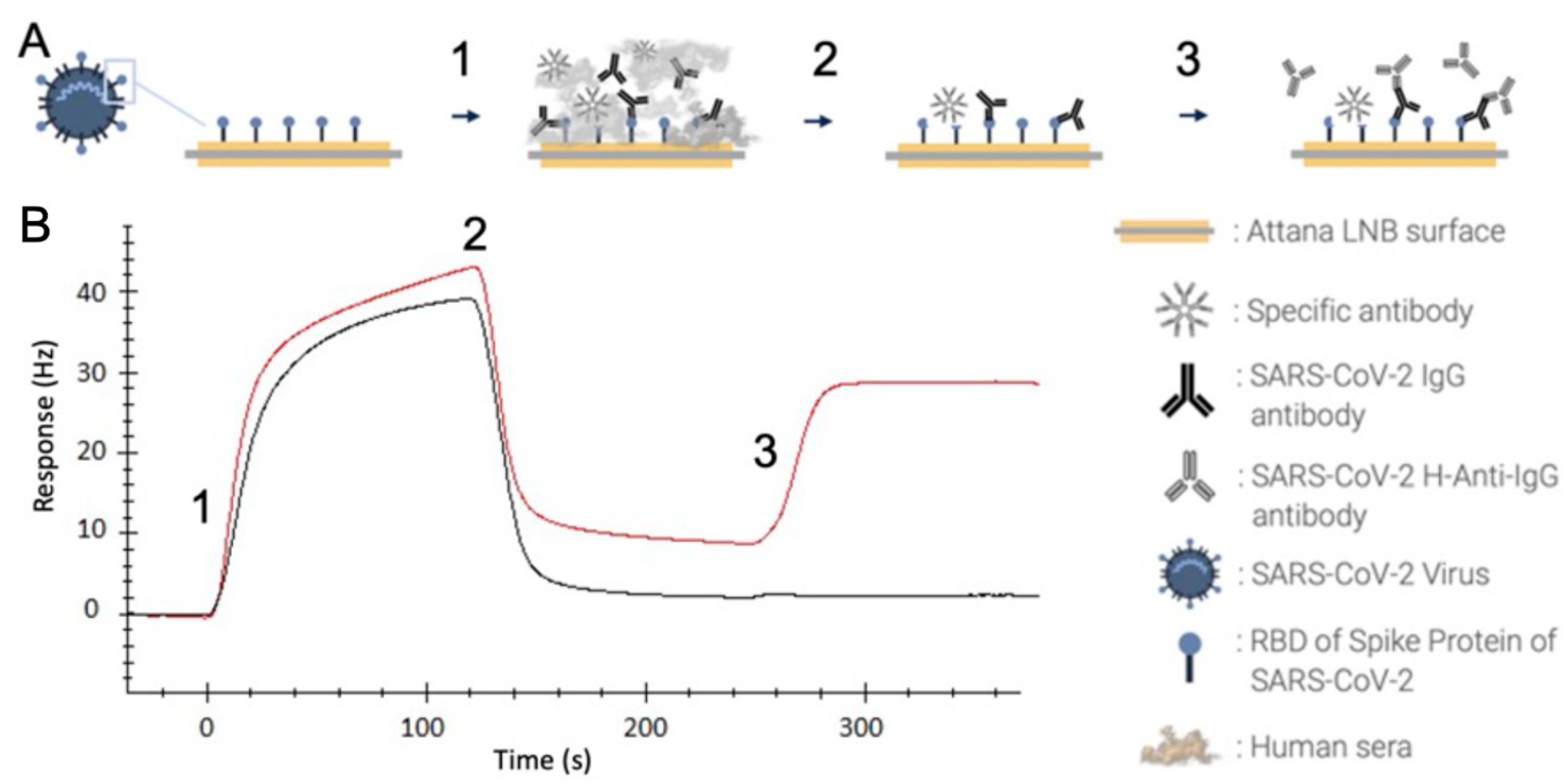
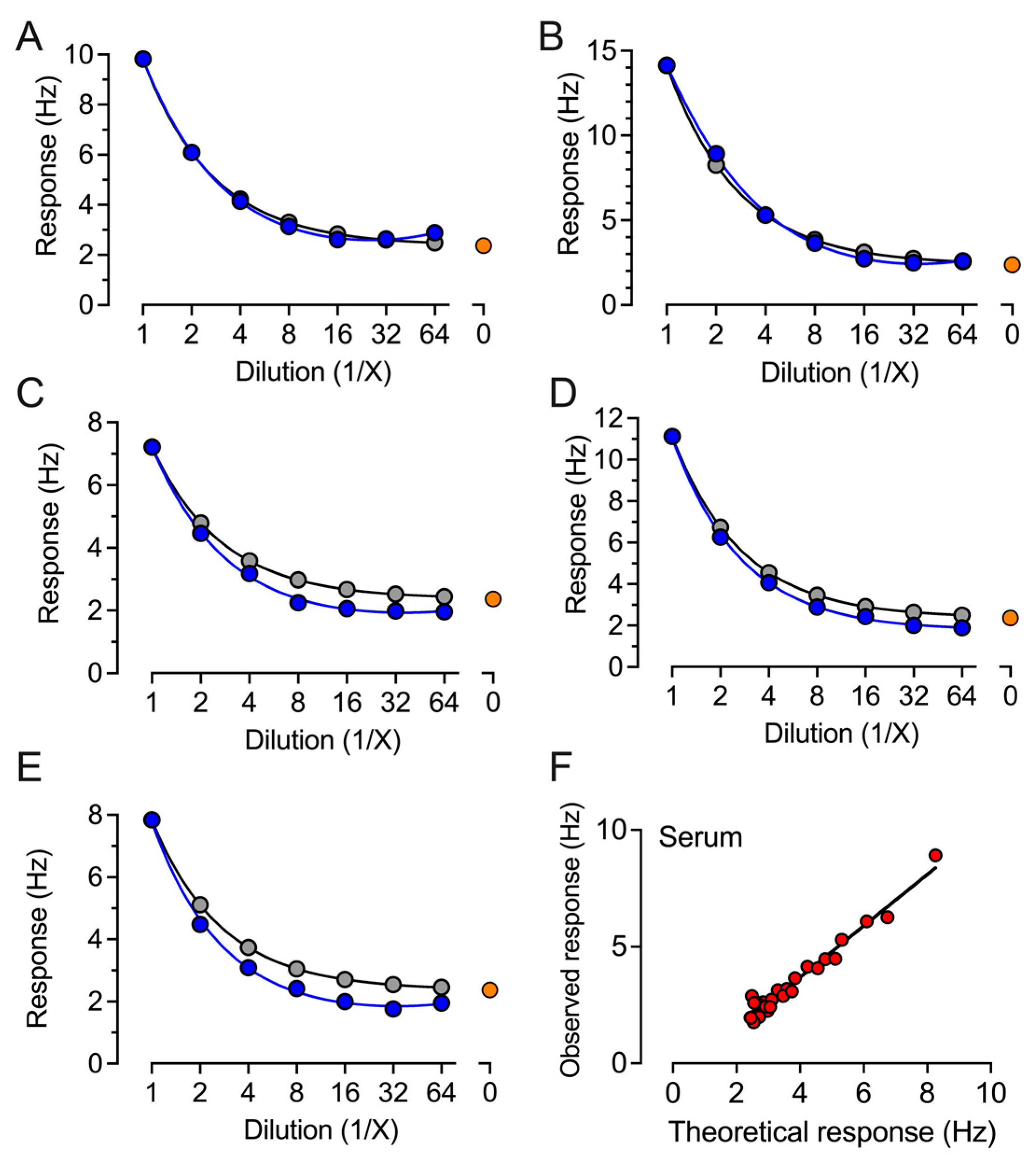
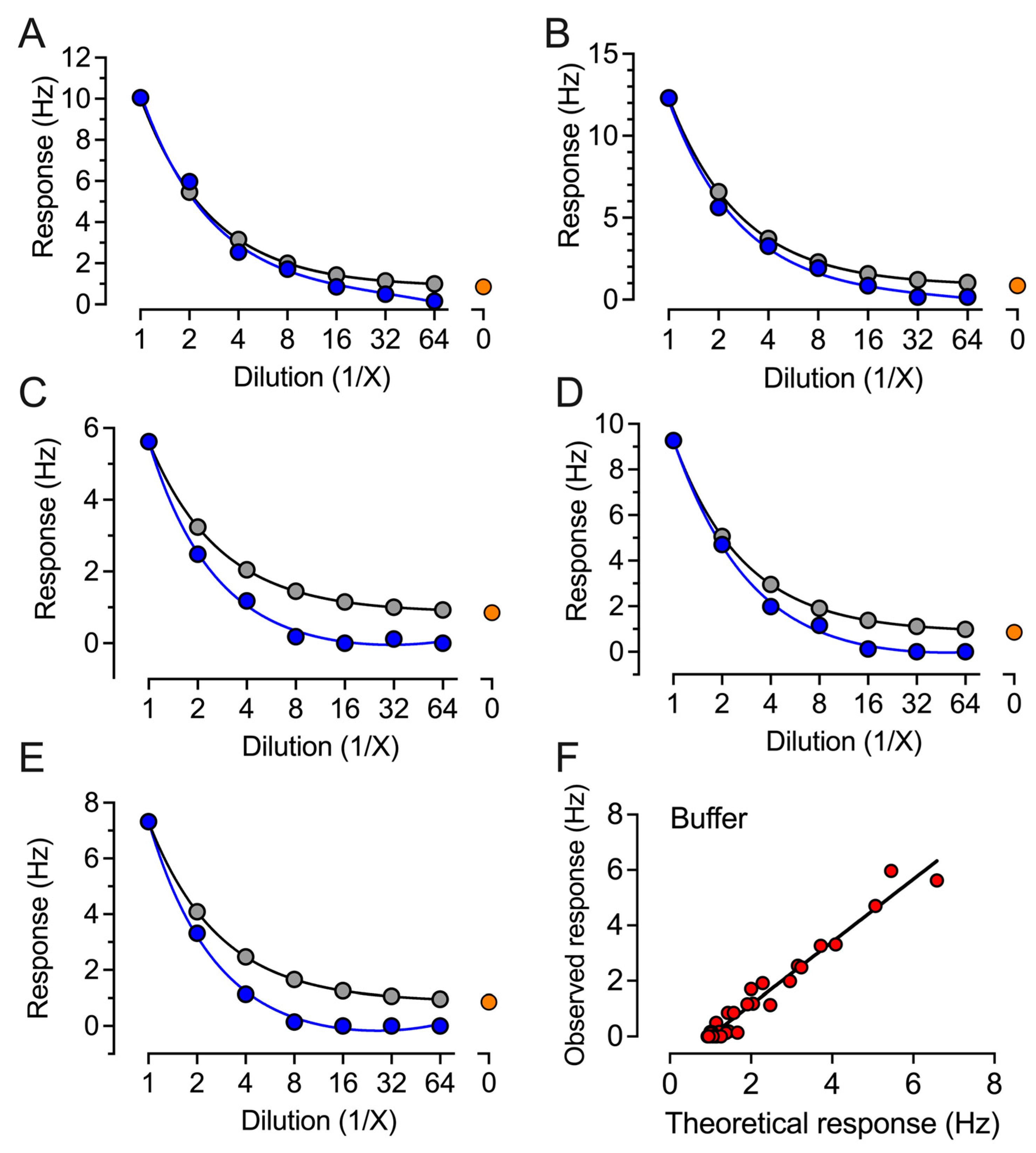
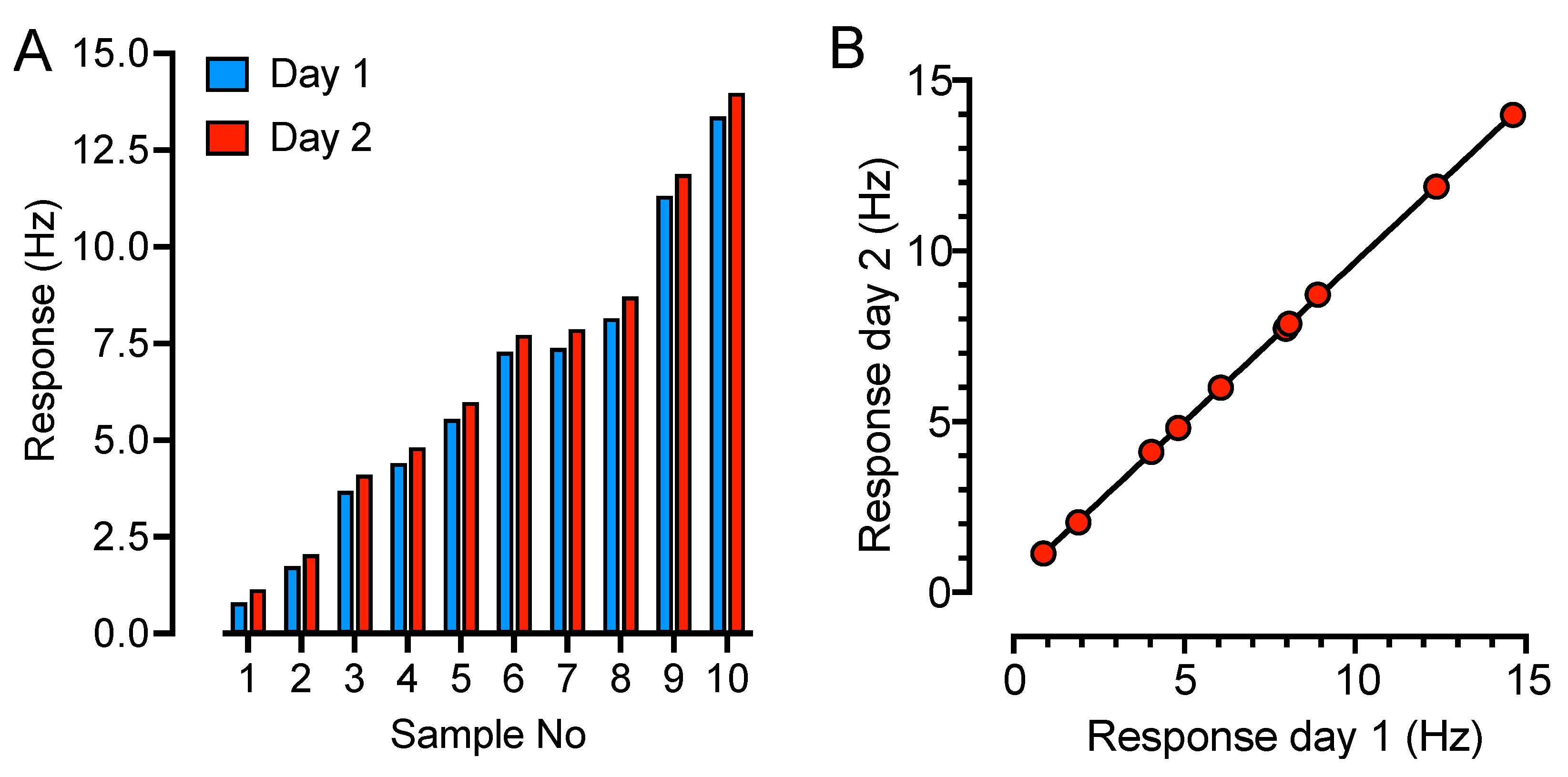
2. Results
3. Discussion
4. Materials and Methods
4.1. Blood Samples
4.2. QCM-Based SARS-CoV-2 IgG Immunoassay
4.3. ROCHE—Electrochemiluminescence Assay
4.4. YHLO Chemiluminescence Immunoassay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, R.; Babady, E.; Theel, E.S.; Storch, G.A.; Pinsky, B.A.; St George, K.; Smith, T.C.; Bertuzzi, S. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. mBio 2020, 11, e00722-20. [Google Scholar] [CrossRef]
- Caruana, G.; Croxatto, A.; Coste, A.T.; Opota, O.; Lamoth, F.; Jaton, K.; Greub, G. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol. Infect. 2020, 26, 1178–1182. [Google Scholar] [CrossRef]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. Profile of RT-PCR for SARS-CoV-2: A Preliminary Study From 56 COVID-19 Patients. Clin. Infect. Dis. 2020, 71, 2249–2251. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.; Rickard, H.; Stevenson, D.; Aranega-Bou, P.; Pitman, J.; Crook, A.; Davies, K.; Spencer, A.; Burton, C.; Easterbrook, L.; et al. Detection of SARS-CoV-2 within the healthcare environment: A multi-centre study conducted during the first wave of the COVID-19 outbreak in England. J. Hosp. Infect. 2021, 108, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Pujadas, E.; Chaudhry, F.; McBride, R.; Richter, F.; Zhao, S.; Wajnberg, A.; Nadkarni, G.; Glicksberg, B.S.; Houldsworth, J.; Cordon-Cardo, C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020, 8, e70. [Google Scholar] [CrossRef] [PubMed]
- Bhoyar, R.C.; Jain, A.; Sehgal, P.; Divakar, M.K.; Sharma, D.; Imran, M.; Jolly, B.; Ranjan, G.; Rophina, M.; Sharma, S.; et al. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS ONE 2021, 16, e0247115. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Worp, N.; Nieuwenhuijse, D.F.; Sikkema, R.S.; Haagmans, B.; Fouchier, R.A.M.; Koopmans, M. The next phase of SARS-CoV-2 surveillance: Real-time molecular epidemiology. Nat. Med. 2021, 27, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Stoesser, N.; Matthews, P.C.; Ayoubkhani, D.; Studley, R.; Bell, I.; Bell, J.I.; Newton, J.N.; Farrar, J.; Diamond, I.; et al. Antibody responses to SARS-CoV-2 vaccines in 45,965 adults from the general population of the United Kingdom. Nat. Microbiol. 2021, 6, 1140–1149. [Google Scholar] [CrossRef]
- Bryant, J.E.; Azman, A.S.; Ferrari, M.J.; Arnold, B.F.; Boni, M.F.; Boum, Y.; Hayford, K.; Luquero, F.J.; Mina, M.J.; Rodriguez-Barraquer, I.; et al. Serology for SARS-CoV-2: Apprehensions, opportunities, and the path forward. Sci. Immunol. 2020, 5, eabc6347. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay, Elisa. 3. Quantitation of specific antibodies by enzyme-labeled anti-immunoglobulin in antigen-coated tubes. J. Immunol. 1972, 109, 129–135. [Google Scholar] [CrossRef]
- Roche. Cobas® SARS-CoV-2 Test (for the COVID-19 Coronavirus). 2020. Available online: https://diagnostics.roche.com/us/en/products/params/cobas-sars-cov-2-test.html (accessed on 13 April 2022).
- Parai, D.; Dash, G.C.; Choudhary, H.R.; Peter, A.; Rout, U.K.; Nanda, R.R.; Kshatri, J.S.; Kanungo, S.; Pati, S.; Bhattacharya, D. Diagnostic accuracy comparison of three fully automated chemiluminescent immunoassay platforms for the detection of SARS-CoV-2 antibodies. J. Virol. Methods 2021, 292, 114121. [Google Scholar] [CrossRef]
- Alassi, A.; Benammar, M.; Brett, D. Quartz Crystal Microbalance Electronic Interfacing Systems: A Review. Sensors 2017, 17, 2799. [Google Scholar] [CrossRef]
- Pohanka, M. Quartz Crystal Microbalance (QCM) Sensing Materials in Biosensors Development. Int. J. Electrochem. Sci. 2021, 16, 211220. [Google Scholar] [CrossRef]
- Kurosawa, S.; Park, J.-W.; Aizawa, H.; Wakida, S.-I.; Tao, H.; Ishihara, K. Quartz crystal microbalance immunosensors for environmental monitoring. Biosens. Bioelectron. 2006, 22, 473–481. [Google Scholar] [CrossRef]
- Zhou, Q.a.; Zheng, C.; Zhu, L.; Wang, J. A review on rapid detection of modified quartz crystal microbalance sensors for food: Contamination, flavour and adulteration. TrAC Trends Anal. Chem. 2022, 157, 116805. [Google Scholar] [CrossRef]
- Migoń, D.; Wasilewski, T.; Suchy, D. Application of QCM in Peptide and Protein-Based Drug Product Development. Molecules 2020, 25, 3950. [Google Scholar] [CrossRef] [PubMed]
- Larsson, P.; Engqvist, H.; Biermann, J.; Werner Rönnerman, E.; Forssell-Aronsson, E.; Kovács, A.; Karlsson, P.; Helou, K.; Parris, T.Z. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci. Rep. 2020, 10, 5798. [Google Scholar] [CrossRef] [PubMed]
- Suriyanarayanan, S.; Lee, H.-H.; Liedberg, B.; Aastrup, T.; Nicholls, I.A. Protein-resistant hyperbranched polyethyleneimine brush surfaces. J. Colloid Interface Sci. 2013, 396, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Bragazzi, N.L.; Amicizia, D.; Panatto, D.; Tramalloni, D.; Valle, I.; Gasparini, R. Quartz-Crystal Microbalance (QCM) for Public Health: An Overview of Its Applications. Adv. Protein Chem. Struct. Biol. 2015, 101, 149–211. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.; Mujahid, A.; Schirhagl, R.; Bajwa, S.Z.; Latif, U.; Feroz, S. Gravimetric Viral Diagnostics: QCM Based Biosensors for Early Detection of Viruses. Chemosensors 2017, 5, 7. [Google Scholar] [CrossRef]
- Sauerbrey, G. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Clausen, T.M.; Pereira, M.A.; Oo, H.Z.; Resende, M.; Gustavson, T.; Mao, Y.; Sugiura, N.; Liew, J.; Fazli, L.; Theander, T.G.; et al. Real-time and label free determination of ligand binding-kinetics to primary cancer tissue specimens; a novel tool for the assessment of biomarker targeting. Sens. Bio-Sens. Res. 2016, 9, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Fenderico, N.; van Scherpenzeel, R.C.; Goldflam, M.; Proverbio, D.; Jordens, I.; Kralj, T.; Stryeck, S.; Bass, T.Z.; Hermans, G.; Ullman, C.; et al. Anti-LRP5/6 VHHs promote differentiation of Wnt-hypersensitive intestinal stem cells. Nat. Commun. 2019, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Elmlund, L.; Käck, C.; Aastrup, T.; Nicholls, I.A. Study of the Interaction of Trastuzumab and SKOV3 Epithelial Cancer Cells Using a Quartz Crystal Microbalance Sensor. Sensors 2015, 15, 5884–5894. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Benavides, D.A.; Sánchez-Fraga, R.; Mejía-Silva, I.; Alvarado-Orozco, J.M. Chapter 15—Microfluidic-based biosensor for SARS-CoV-2 antibodies. In Biomedical Innovations to Combat COVID-19; Rosales-Mendoza, S., Comas-Garcia, M., Gonzalez-Ortega, O., Eds.; Academic Press: Oxford, UK, 2022; pp. 253–270. [Google Scholar] [CrossRef]
- Bulut, A.; Temur, B.Z.; Kirimli, C.E.; Gok, O.; Balcioglu, B.K.; Ozturk, H.U.; Uyar, N.Y.; Kanlidere, Z.; Kocagoz, T.; Can, O. A Novel Peptide-Based Detection of SARS-CoV-2 Antibodies. Biomimetics 2023, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Plikusiene, I.; Maciulis, V.; Juciute, S.; Ramanavicius, A.; Ramanaviciene, A. Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum. Biosensors 2023, 13, 784. [Google Scholar] [CrossRef] [PubMed]
- YHLO. iFlash Immunoassay Analyzer SARS-CoV-2 IgG; YHLO Biotech Co., Ltd: Shenzhen, China, 2019. [Google Scholar]
- Röltgen, K.; Powell, A.E.; Wirz, O.F.; Stevens, B.A.; Hogan, C.A.; Najeeb, J.; Hunter, M.; Wang, H.; Sahoo, M.K.; Huang, C.; et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020, 5, eabe0240. [Google Scholar] [CrossRef]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef]
- Brien, J.D.; Sukupolvi-Petty, S.; Williams, K.L.; Lam, C.Y.; Schmid, M.A.; Johnson, S.; Harris, E.; Diamond, M.S. Protection by immunoglobulin dual-affinity retargeting antibodies against dengue virus. J. Virol. 2013, 87, 7747–7753. [Google Scholar] [CrossRef]
- Palladini, A.; Thrane, S.; Janitzek, C.M.; Pihl, J.; Clemmensen, S.B.; de Jongh, W.A.; Clausen, T.M.; Nicoletti, G.; Landuzzi, L.; Penichet, M.L.; et al. Virus-like particle display of HER2 induces potent anti-cancer responses. Oncoimmunology 2018, 7, e1408749. [Google Scholar] [CrossRef] [PubMed]

| QCM | Roche | YHLO | |||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| QCM | Positive | - | - | 47 | 1 | 45 | 3 |
| Negative | - | - | 12 | 59 | 7 | 64 | |
| Roche | Positive | 47 | 12 | - | - | 52 | 7 |
| Negative | 1 | 59 | - | - | 0 | 60 | |
| YHLO | Positive | 45 | 7 | 52 | 0 | - | - |
| Negative | 3 | 64 | 7 | 60 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nilsson, P.H.; Al-Majdoub, M.; Ibrahim, A.; Aseel, O.; Suriyanarayanan, S.; Andersson, L.; Fostock, S.; Aastrup, T.; Tjernberg, I.; Rydén, I.; et al. Quartz Crystal Microbalance Platform for SARS-CoV-2 Immuno-Diagnostics. Int. J. Mol. Sci. 2023, 24, 16705. https://doi.org/10.3390/ijms242316705
Nilsson PH, Al-Majdoub M, Ibrahim A, Aseel O, Suriyanarayanan S, Andersson L, Fostock S, Aastrup T, Tjernberg I, Rydén I, et al. Quartz Crystal Microbalance Platform for SARS-CoV-2 Immuno-Diagnostics. International Journal of Molecular Sciences. 2023; 24(23):16705. https://doi.org/10.3390/ijms242316705
Chicago/Turabian StyleNilsson, Per H., Mahmoud Al-Majdoub, Ahmed Ibrahim, Obaidullah Aseel, Subramanian Suriyanarayanan, Linnea Andersson, Samir Fostock, Teodor Aastrup, Ivar Tjernberg, Ingvar Rydén, and et al. 2023. "Quartz Crystal Microbalance Platform for SARS-CoV-2 Immuno-Diagnostics" International Journal of Molecular Sciences 24, no. 23: 16705. https://doi.org/10.3390/ijms242316705
APA StyleNilsson, P. H., Al-Majdoub, M., Ibrahim, A., Aseel, O., Suriyanarayanan, S., Andersson, L., Fostock, S., Aastrup, T., Tjernberg, I., Rydén, I., & Nicholls, I. A. (2023). Quartz Crystal Microbalance Platform for SARS-CoV-2 Immuno-Diagnostics. International Journal of Molecular Sciences, 24(23), 16705. https://doi.org/10.3390/ijms242316705









