Transcriptome Dynamics of Brassica juncea Leaves in Response to Omnivorous Beet Armyworm (Spodoptera exigua, Hübner)
Abstract
:1. Introduction
2. Results
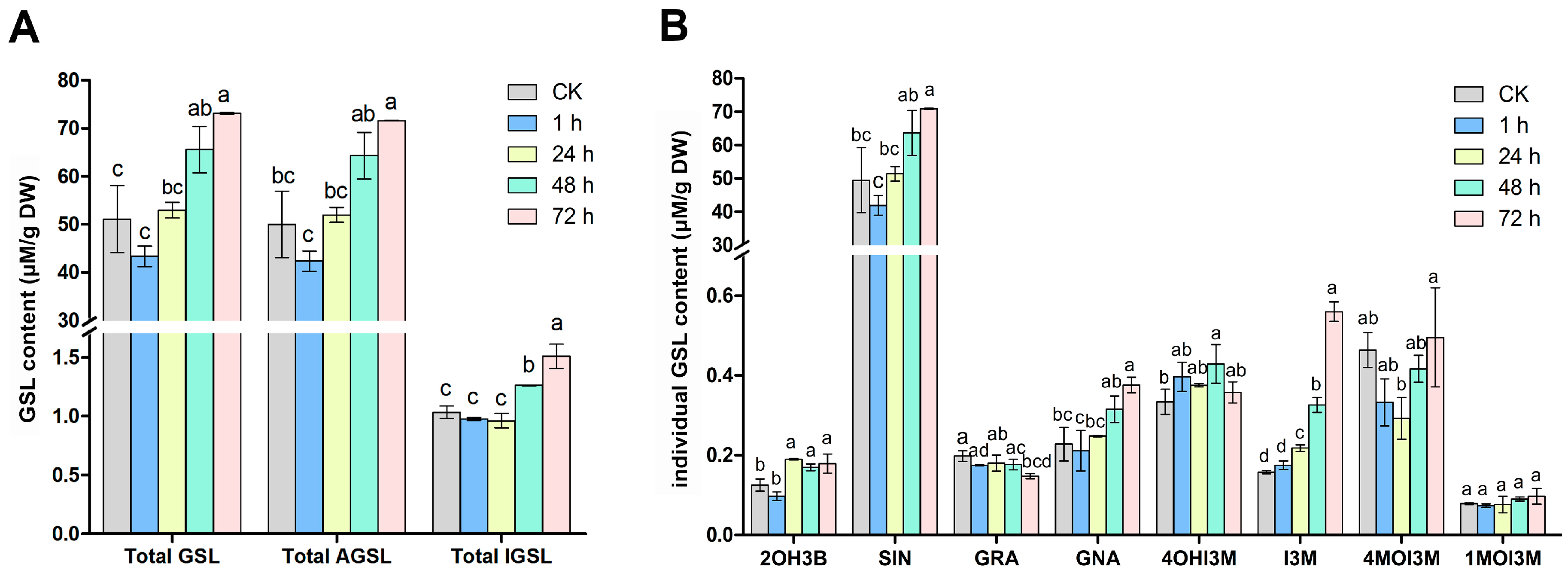
2.1. Changes in Glucosinolate Profiles in Mustard Leaves after Beet Armyworm Larvae Chewing
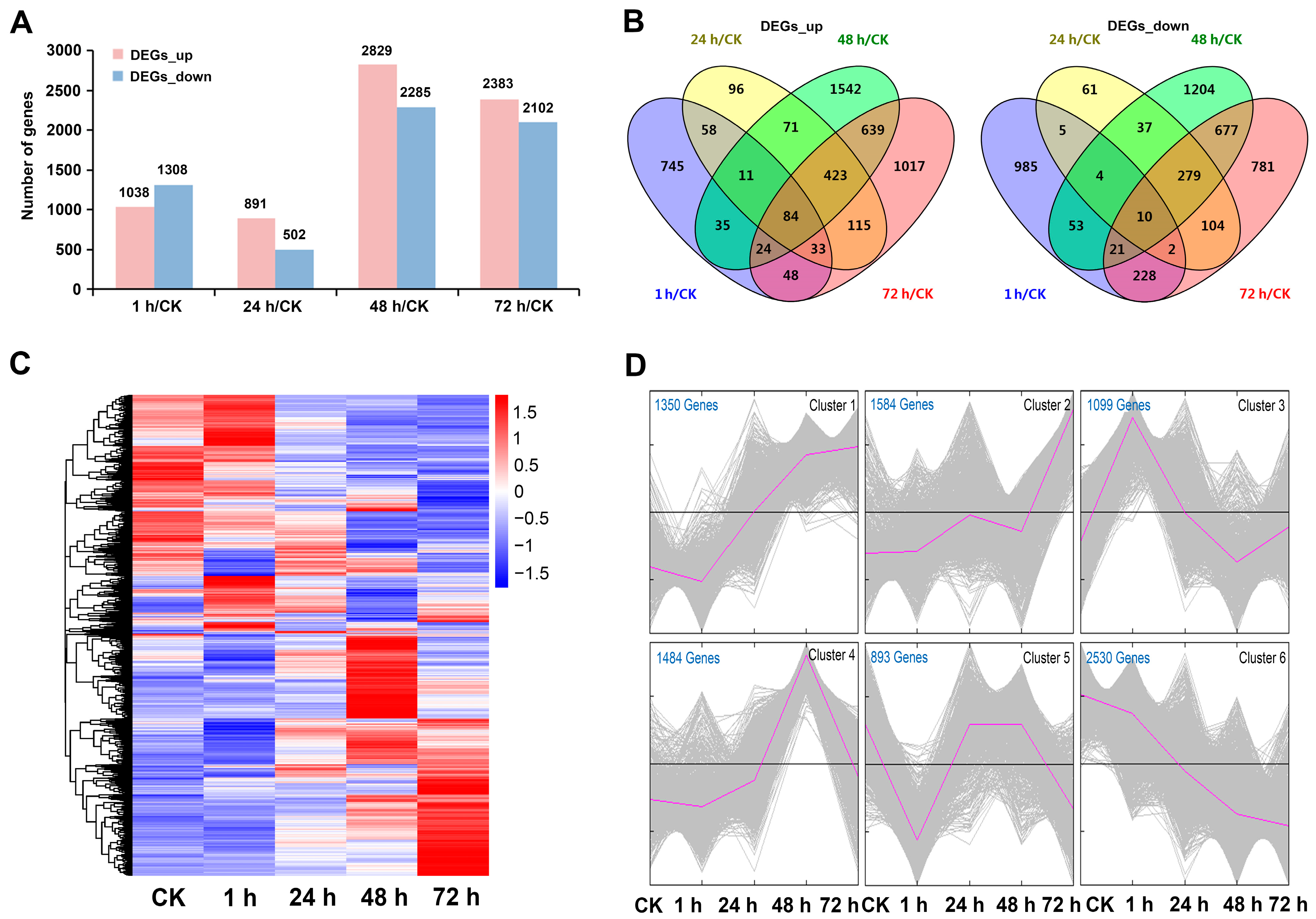
2.2. Transcriptome Sequencing Analysis
2.3. Dynamic Expression Pattern of DEGs
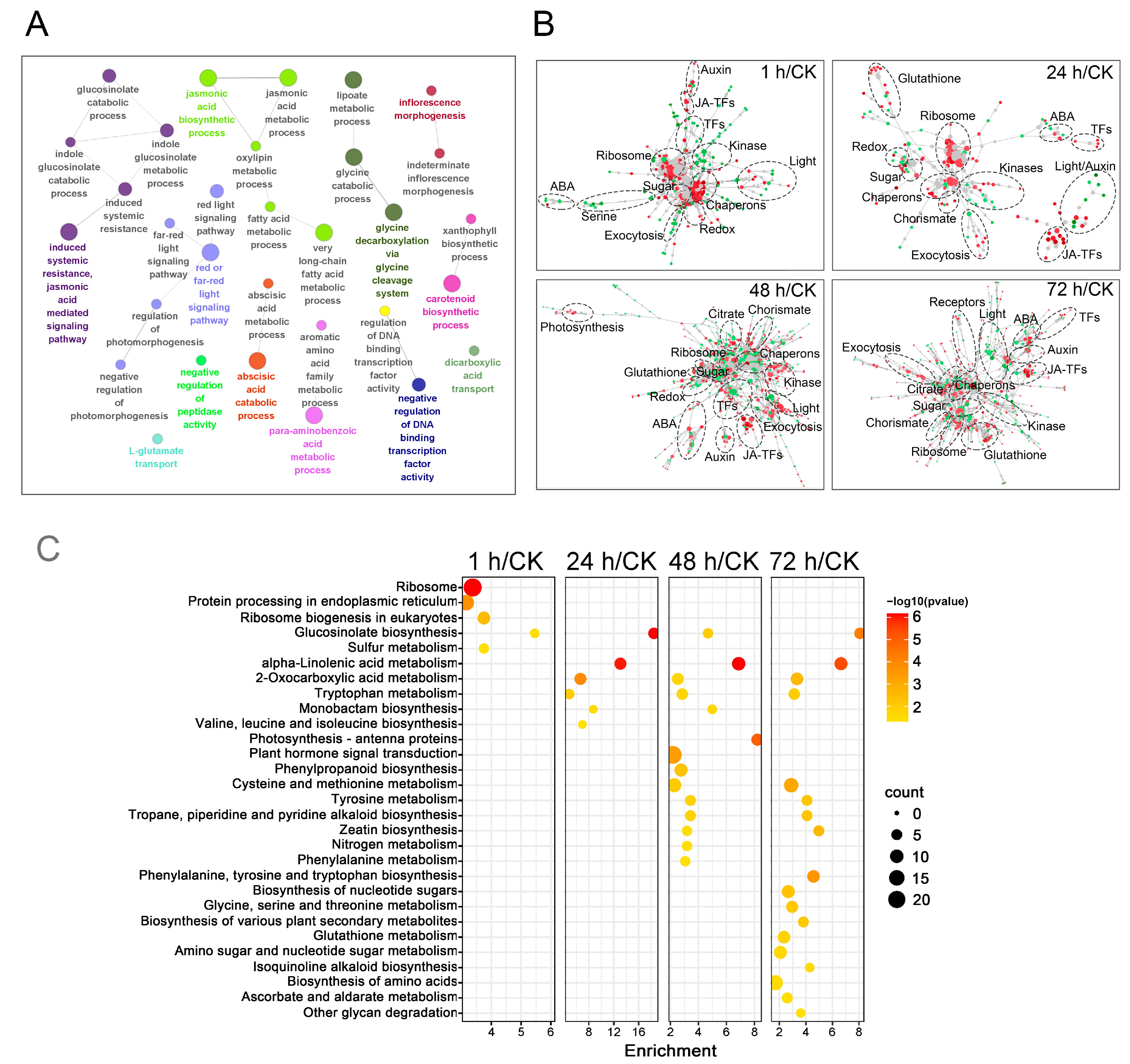
2.4. Functional Analysis of DEGs
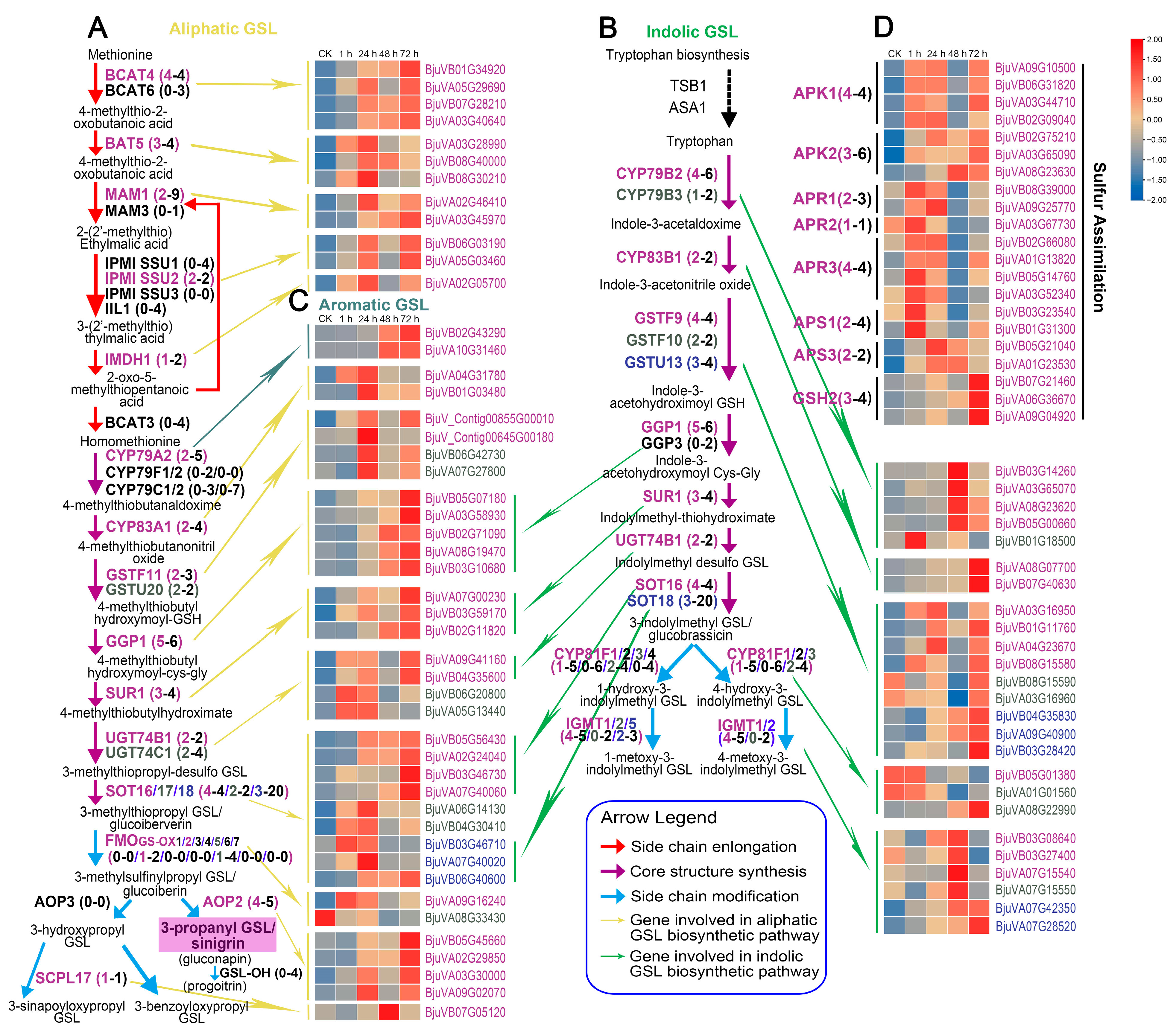
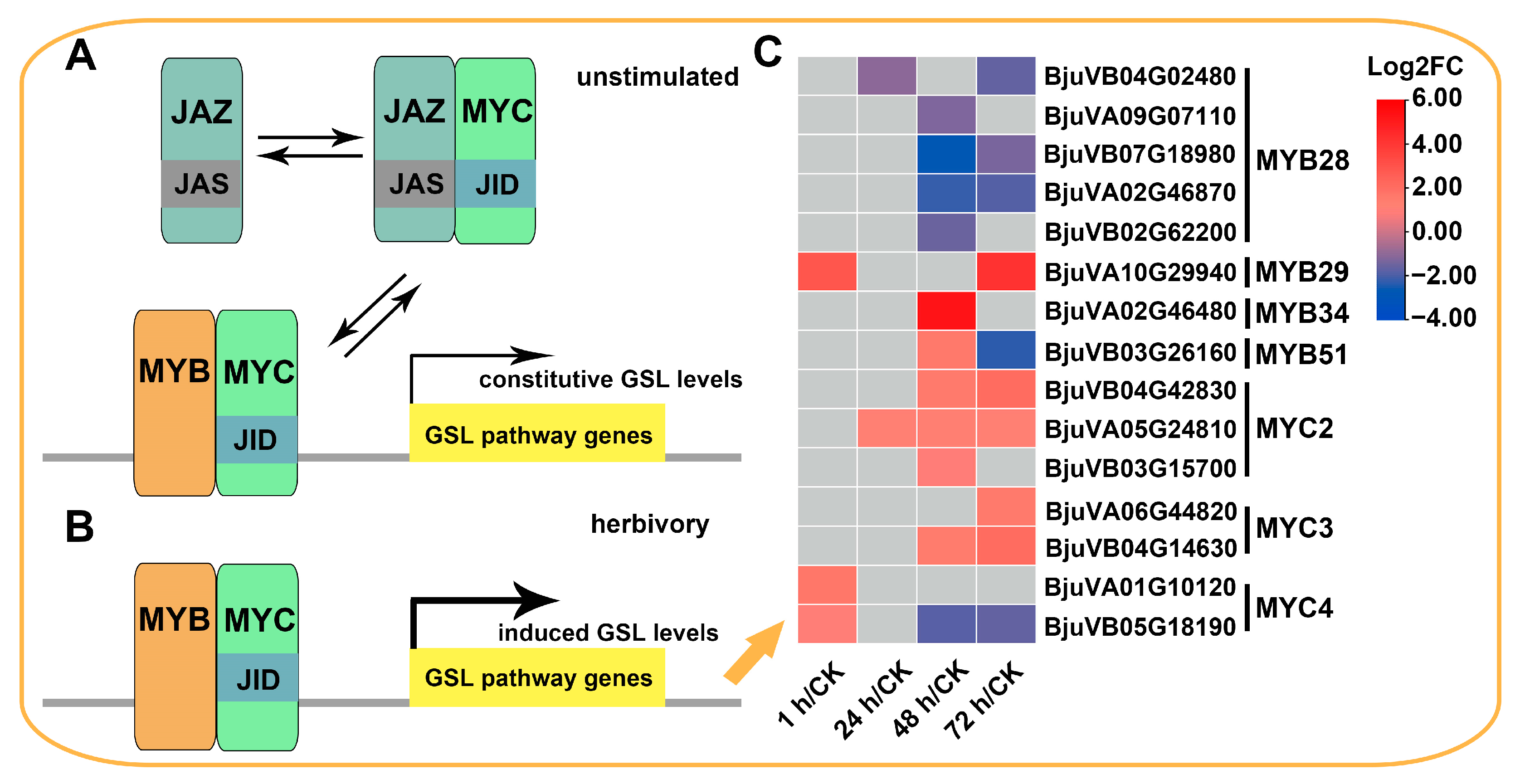
2.5. Expression Patterns of Genes Involved in Glucosinolate Biosynthesis
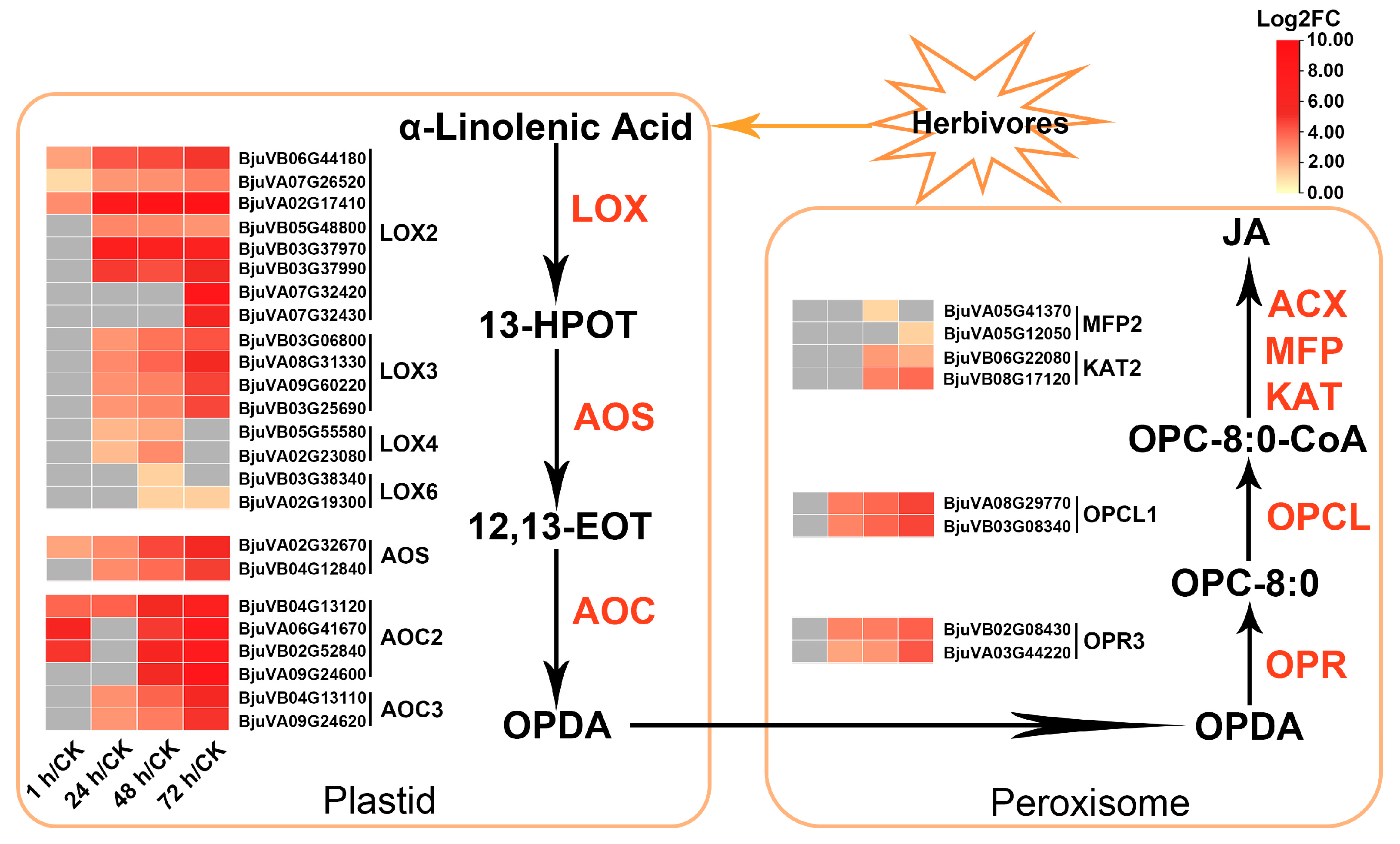
2.6. Multiple JA Biosynthesis-Related Genes in Mustard Were Upregulated after Insect Feeding
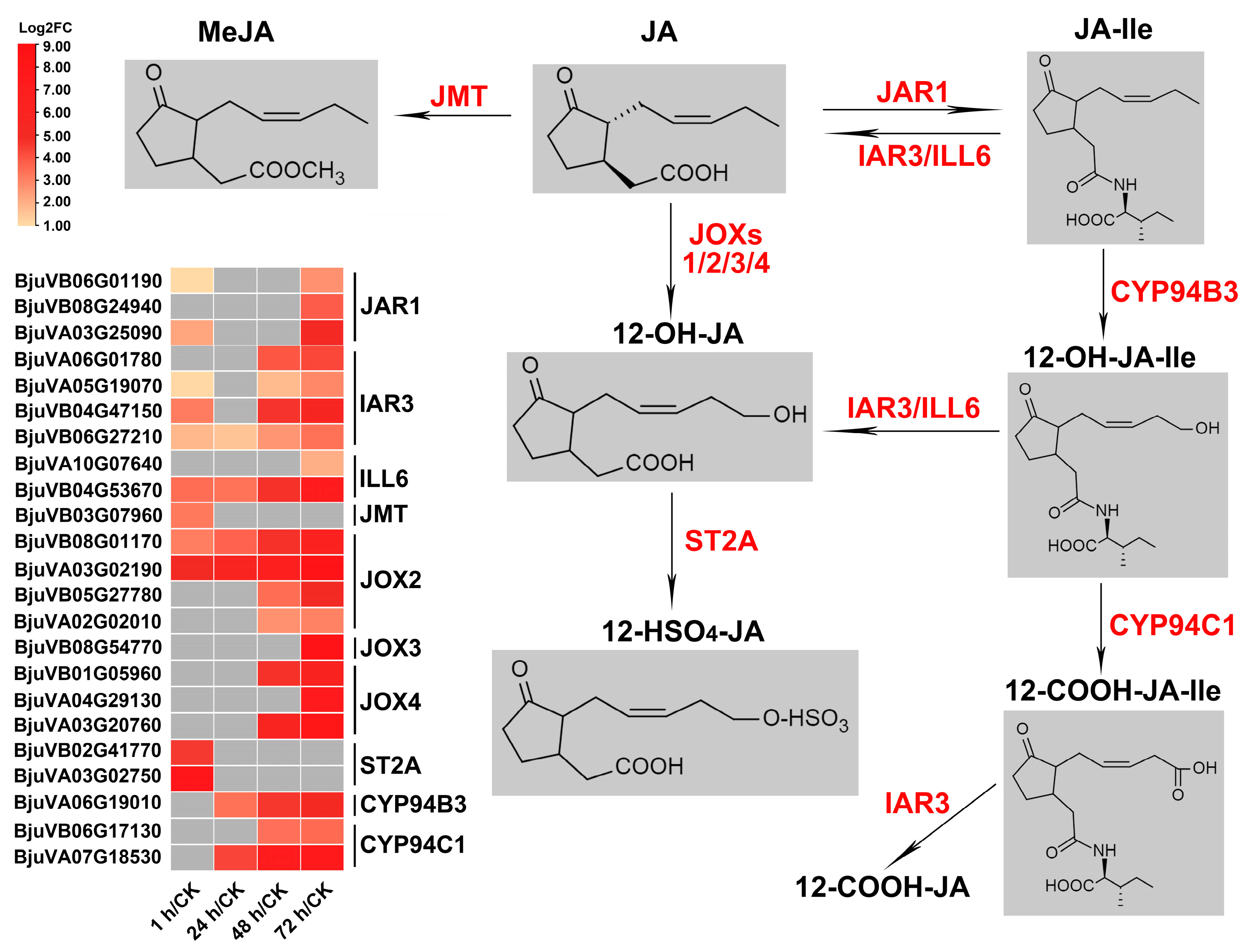
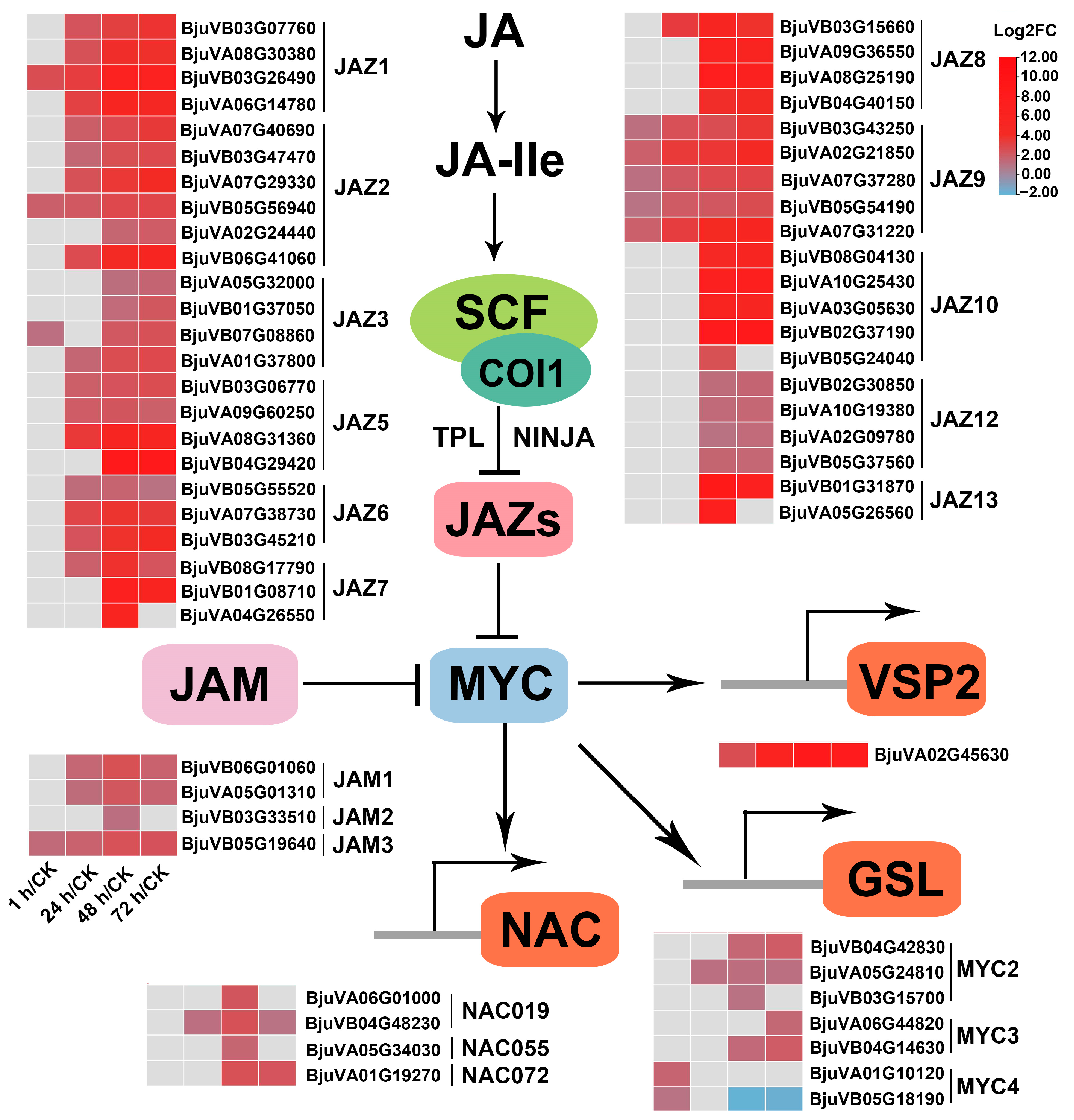
2.7. Genes Related to JA Metabolism and Repression of JA Signaling Were Significantly Induced after Insect Feeding
2.7.1. JA Metabolism
2.7.2. JA Signaling
3. Discussion
3.1. The Accumulation of GSLs in Mustard Leaves after Herbivory Depends on the Upregulation of a Large Number of BjuGSL Genes
3.2. The JA-Dependent Response Induced by Herbivory Is Fine-Tuned by Regulating the Homeostasis of JA Compounds and the Activation of JA Signaling Regulators
4. Materials and Methods
4.1. Plant and Insect Materials
4.2. Glucosinolate Analysis
4.3. Statistical Analysis
4.4. RNA Extraction, Library Construction, Sequencing and Assembly
4.5. Differential Expression Genes (DEGs)Analysis
4.6. Functional Analysis of DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rejeb, I.B.; Pastor, V.; Mauch-Mani, B. Plant responses to simultaneous biotic and abiotic stress: Molecular mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef]
- Nalam, V.; Louis, J.; Shah, J. Plant defense against aphids, the pest extraordinaire. Plant Sci. 2019, 279, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Wari, D.; Aboshi, T.; Shinya, T.; Galis, I. Integrated view of plant metabolic defense with particular focus on chewing herbivores. J. Integr. Plant Biol. 2022, 64, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, M.; Huang, J.; Li, L.; Huang, K.; Zhang, Y.; Li, Y.; Deng, Z.; Ni, X.; Li, X. Inductive and synergistic interactions between plant allelochemical flavone and Bt toxin Cry1Ac in Helicoverpa armigera. Insect Sci. 2021, 28, 1756–1765. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef]
- Schweiger, R.; Heise, A.M.; Persicke, M.; Muller, C. Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant Cell Environ. 2014, 37, 1574–1585. [Google Scholar] [CrossRef]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Li, C.; Xu, M.; Cai, X.; Han, Z.; Si, J.; Chen, D. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 2022, 23, 3945. [Google Scholar] [CrossRef]
- Truman, W.; Bennett, M.H.; Kubigsteltig, I.; Turnbull, C.; Grant, M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 2007, 104, 1075–1080. [Google Scholar] [CrossRef]
- Bouranis, D.L.; Malagoli, M.; Avice, J.C.; Bloem, E. Advances in plant sulfur research. Plants 2020, 9, 256. [Google Scholar] [CrossRef]
- Jessica, P.Y.; Hoang, V.T.; Jorrel, M.; Shawn, A.C.; Anna, K.B. Plant defense chemicals against insect pests. Agronomy 2020, 10, 1156. [Google Scholar]
- Li, R.; Jin, J.; Xu, J.; Wang, L.; Li, J.; Lou, Y.; Baldwin, I.T. Long non-coding RNAs associate with jasmonate-mediated plant defence against herbivores. Plant Cell Environ. 2021, 44, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Qiu, J.; Zhang, Y.; Li, M.; Liu, P. Jasmotes coordinate secondary with primary metabolism. Metabolites 2023, 13, 1008. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Blazevic, I.; Montaut, S.; Burcul, F.; Olsen, C.E.; Burow, M.; Rollin, P.; Agerbirk, N. Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 2020, 169, 112100. [Google Scholar] [CrossRef] [PubMed]
- Herr, I.; Buchler, M.W. Dietary constituents of broccoli and other cruciferous vegetables: Implications for prevention and therapy of cancer. Cancer Treat. Rev. 2010, 36, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Vartika, M.; Tom, O.G.T.; Rob, M.G.; Vinay, K.; Sankara Reddy, A.; Louise, E.M.V.; Nicole, M.D. Dealing with double trouble: Consequences of single and double herbivory in Brassica juncea. Chemoecology 2013, 23, 71–82. [Google Scholar]
- Kumar, P.; Augustine, R.; Singh, A.K.; Bisht, N.C. Feeding behaviour of generalist pests on Brassica juncea: Implication for manipulation of glucosinolate biosynthesis pathway for enhanced resistance. Plant Cell Environ. 2017, 40, 2109–2120. [Google Scholar] [CrossRef]
- Mitreiter, S.; Gigolashvili, T. Regulation of glucosinolate biosynthesis. J. Exp. Bot. 2021, 72, 70–91. [Google Scholar] [CrossRef]
- Pfalz, M.; Mikkelsen, M.D.; Bednarek, P.; Olsen, C.E.; Halkier, B.A.; Kroymann, J. Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant. Cell 2011, 23, 716–729. [Google Scholar] [CrossRef]
- Harun, S.; Abdullah-Zawawi, M.R.; Goh, H.H.; Mohamed-Hussein, Z.A. A comprehensive gene inventory for glucosinolate biosynthetic pathway in Arabidopsis thaliana. J. Agric. Food. Chem. 2020, 68, 7281–7297. [Google Scholar] [CrossRef]
- Huh, S.U. Functional analysis of hot pepper ethylene responsive factor 1A in plant defense. Plant Signal. Behav. 2022, 17, 2027137. [Google Scholar] [CrossRef]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant. 2014, 7, 814–828. [Google Scholar] [CrossRef]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant. Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Muller, C.; Flugge, U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant. J. 2007, 51, 247–261. [Google Scholar] [CrossRef]
- Sonderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Pislewska-Bednarek, M.; Sanchez-Vallet, A.; Molina, A.; Glawischnig, E.; Gigolashvili, T.; Bednarek, P. Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant. 2016, 9, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira, F.D.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wan, Z.; Nelson, M.N.; Chauhan, J.S.; Redden, R.; Burton, W.A.; Lin, P.; Salisbury, P.A.; Fu, T.; Cowling, W.A. Evidence from genome-wide simple sequence repeat markers for a polyphyletic origin and secondary centers of genetic diversity of Brassica juncea in China and India. J. Hered. 2013, 104, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Jixiang, C.; Weili, J.; Hongyan, H.; Xiaoyan, M.; Qian, L.; Xianpeng, S.; Xiangliang, R.; Yan, M. Joint toxicity of methoxyfenozide and lufenuron on larvae of Spodoptera exigua Hübner (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 2019, 22, 795–801. [Google Scholar]
- Tong, Y.; Gabriel-Neumann, E.; Ngwene, B.; Krumbein, A.; George, E.; Platz, S.; Rohn, S.; Schreiner, M. Topsoil drying combined with increased sulfur supply leads to enhanced aliphatic glucosinolates in Brassica juncea leaves and roots. Food Chem. 2014, 152, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, Y.; Xu, L.; Guo, E.; Zang, Y.; He, Y.; Zhu, Z. Transcriptome profiling reveals candidate key genes involved in sinigrin biosynthesis in Brassica nigra. Horticulturae 2021, 7, 173. [Google Scholar] [CrossRef]
- Mewis, I.; Appel, H.M.; Hom, A.; Raina, R.; Schultz, J.C. Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 2005, 138, 1149–1162. [Google Scholar] [CrossRef]
- Chen, C.Y.; Mao, Y.B. Research advances in plant-insect molecular interaction. F1000Research 2020, 9, F1000 Faculty Rev-198. [Google Scholar] [CrossRef]
- Divekar, P.A.; Narayana, S.; Divekar, B.A.; Kumar, R.; Gadratagi, B.G.; Ray, A.; Singh, A.K.; Rani, V.; Singh, V.; Singh, A.K.; et al. Plant secondary metabolites as defense tools against herbivores for sustainable crop protection. Int. J. Mol. Sci. 2022, 23, 2690. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Xie, J.; Lv, J.; Zhang, G.; Hu, L.; Luo, S.; Li, L.; Yu, J. The roles of Cruciferae glucosinolates in disease and pest resistance. Plants 2021, 10, 1097. [Google Scholar] [CrossRef]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef]
- Guo, R.; Shen, W.; Qian, H.; Zhang, M.; Liu, L.; Wang, Q. Jasmonic acid and glucose synergistically modulate the accumulation of glucosinolates in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 5707–5719. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Rekha, K.; Rajakumar, G.; Lee, T.J.; Kim, S.H.; Chung, I.M. Enhanced production of anthraquinones and phenolic compounds and biological activities in the cell suspension cultures of Polygonum multiflorum. Int. J. Mol. Sci. 2016, 17, 1912. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.; Halkier, B.A. How does a plant orchestrate defense in time and space? Using glucosinolates in Arabidopsis as case study. Curr. Opin. Plant Biol. 2017, 38, 142–147. [Google Scholar] [CrossRef]
- Ogran, A.; Faigenboim, A.; Barazani, O. Transcriptome responses to different herbivores reveal differences in defense strategies between populations of Eruca sativa. BMC Genomics 2019, 20, 843. [Google Scholar] [CrossRef] [PubMed]
- Gols, R.; van Dam, N.M.; Reichelt, M.; Gershenzon, J.; Raaijmakers, C.E.; Bullock, J.M.; Harvey, J.A. Seasonal and herbivore-induced dynamics of foliar glucosinolates in wild cabbage (Brassica oleracea). Chemoecology 2018, 28, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Pellissier, L.; Moreira, X.; Defossez, E.; Pfander, M.; Guyer, A.; van Dam, N.M.; Rasmann, S. Correlated induction of phytohormones and glucosinolates shapes insect herbivore resistance of cardamine species along elevational gradients. J. Chem. Ecol. 2019, 45, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Serena, S.; Tamara, S.; Pablo, V.; María, E.C. Antibiotic properties of the glucosinolates of Brassica oleracea var. acephala similarly affect generalist and specialist larvae of two lepidopteran pests. J. Pest. Sci. 2016, 89, 195–206. [Google Scholar]
- Santolamazza-Carbone, S.; Velasco, P.; Soengas, P.; Cartea, M.E. Bottom-up and top-down herbivore regulation mediated by glucosinolates in Brassica oleracea var. acephala. Oecologia 2014, 174, 893–907. [Google Scholar] [CrossRef]
- Bhandari, S.R.; Jo, J.S.; Lee, J.G. Comparison of glucosinolate profiles in different tissues of nine Brassica crops. Molecules 2015, 20, 15827–15841. [Google Scholar] [CrossRef]
- Huang, H.; Wang, J.; Mao, S.; Wu, Q.; Tian, Y.; Wang, F.; Wang, P.; Huang, K.; Wu, Q. Variation characteristics of glucosinolate contents in leaf mustard (Brassica juncea). Agronomy 2022, 12, 2287. [Google Scholar] [CrossRef]
- Augustine, R.; Mukhopadhyay, A.; Bisht, N.C. Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 2013, 11, 855–866. [Google Scholar] [CrossRef]
- Song, X.; Wei, Y.; Xiao, D.; Gong, K.; Sun, P.; Ren, Y.; Yuan, J.; Wu, T.; Yang, Q.; Li, X.; et al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021, 186, 388–406. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liang, J.; Cai, C.; Cai, X.; Wu, J.; Wang, X. Genome sequencing supports a multi-vertex model for Brassiceae species. Curr. Opin. Plant Biol. 2017, 36, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K.; Seol, Y.J.; Perumal, S.; Lee, J.; Waminal, N.E.; Jayakodi, M.; Lee, S.C.; Jin, S.; Choi, B.S.; Yu, Y.; et al. Re-exploration of U’s triangle Brassica species based on chloroplast genomes and 45S nrDNA sequences. Sci. Rep. 2018, 8, 7353. [Google Scholar] [CrossRef] [PubMed]
- Schuster, J.; Knill, T.; Reichelt, M.; Gershenzon, J.; Binder, S. Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant. Cell 2006, 18, 2664–2679. [Google Scholar] [CrossRef]
- Geu-Flores, F.; Moldrup, M.E.; Bottcher, C.; Olsen, C.E.; Scheel, D.; Halkier, B.A. Cytosolic gamma-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant. Cell 2011, 23, 2456–2469. [Google Scholar] [CrossRef]
- Hirschmann, F.; Papenbrock, J. The fusion of genomes leads to more options: A comparative investigation on the desulfo-glucosinolate sulfotransferases of Brassica napus and homologous proteins of Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 91, 10–19. [Google Scholar] [CrossRef]
- Burow, M.; Atwell, S.; Francisco, M.; Kerwin, R.E.; Halkier, B.A.; Kliebenstein, D.J. The Glucosinolate biosynthetic gene AOP2 mediates feed-back regulation of jasmonic acid signaling in Arabidopsis. Mol. Plant. 2015, 8, 1201–1212. [Google Scholar] [CrossRef]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [CrossRef]
- Koo, A.J.; Howe, G.A. The wound hormone jasmonate. Phytochemistry 2009, 70, 1571–1580. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, D.; Wang, Y.; Xie, D. Jasmonate action in plant defense against insects. J. Exp. Bot. 2019, 70, 3391–3400. [Google Scholar] [CrossRef]
- Schuman, M.C.; Baldwin, I.T. The layers of plant responses to insect herbivores. Annu. Rev. Entomol. 2016, 61, 373–394. [Google Scholar] [CrossRef]
- Bruinsma, M.; Posthumus, M.A.; Mumm, R.; Mueller, M.J.; van Loon, J.J.; Dicke, M. Jasmonic acid-induced volatiles of Brassica oleracea attract parasitoids: Effects of time and dose, and comparison with induction by herbivores. J. Exp. Bot. 2009, 60, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arif, Y.; Miszczuk, E.; Bajguz, A.; Hayat, S. Specific roles of lipoxygenases in development and responses to stress in plants. Plants 2022, 11, 979. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zhang, T.; Huangfu, J.; Li, R.; Lou, Y. Both allene oxide synthases genes are involved in the biosynthesis of herbivore-induced jasmonic acid and herbivore resistance in rice. Plants 2021, 10, 442. [Google Scholar] [CrossRef]
- Chen, H.; Wang, B.; Geng, S.; Arellano, C.; Chen, S.; Qu, R. Effects of overexpression of jasmonic acid biosynthesis genes on nicotine accumulation in tobacco. Plant Direct 2018, 2, e36. [Google Scholar] [CrossRef]
- Grover, S.; Varsani, S.; Kolomiets, M.V.; Louis, J. Maize defense elicitor, 12-oxo-phytodienoic acid, prolongs aphid salivation. Commun. Integr. Biol. 2020, 13, 63–66. [Google Scholar] [CrossRef]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Oliveira, F.D.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D.; et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7, 12570. [Google Scholar] [CrossRef]
- Koo, A.J.; Howe, G.A. Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 2012, 3, 19. [Google Scholar] [CrossRef]
- Heitz, T.; Smirnova, E.; Widemann, E.; Aubert, Y.; Pinot, F.; Menard, R. The rise and fall of jasmonate biological activities. Subcell. Biochem. 2016, 86, 405–426. [Google Scholar] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Suza, W.P.; Rowe, M.L.; Hamberg, M.; Staswick, P.E. A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta 2010, 231, 717–728. [Google Scholar] [CrossRef]
- Fukumoto, K.; Alamgir, K.; Yamashita, Y.; Mori, I.C.; Matsuura, H.; Galis, I. Response of rice to insect elicitors and the role of OsJAR1 in wound and herbivory-induced JA-Ile accumulation. J. Integr. Plant Biol. 2013, 55, 775–784. [Google Scholar] [CrossRef]
- Widemann, E.; Smirnova, E.; Aubert, Y.; Miesch, L.; Heitz, T. Dynamics of jasmonate metabolism upon flowering and across leaf stress responses in Arabidopsis thaliana. Plants 2016, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant. Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef]
- Sasaki-Sekimoto, Y.; Jikumaru, Y.; Obayashi, T.; Saito, H.; Masuda, S.; Kamiya, Y.; Ohta, H.; Shirasu, K. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 2013, 163, 291–304. [Google Scholar] [CrossRef]
- Fonseca, S.; Fernandez-Calvo, P.; Fernandez, G.M.; Diez-Diaz, M.; Gimenez-Ibanez, S.; Lopez-Vidriero, I.; Godoy, M.; Fernandez-Barbero, G.; Van Leene, J.; De Jaeger, G.; et al. bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS ONE 2014, 9, e86182. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef]
- Garg, R.; Singh, V.K.; Rajkumar, M.S.; Kumar, V.; Jain, M. Global transcriptome and coexpression network analyses reveal cultivar-specific molecular signatures associated with seed development and seed size/weight determination in chickpea. Plant. J. 2017, 91, 1088–1107. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.T.; Grimes, M.; Kutlu, B.; Bot, J.J.; Galas, D.J. RCytoscape: Tools for exploratory network analysis. BMC Bioinform. 2013, 14, 217. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic. Acids. Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic. Acids. Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
| 1 h/CK | 24 h/CK | 48 h/CK | 72 h/CK | |
|---|---|---|---|---|
| JAZs | 7 | 22 | 44 | 41 |
| JAMs | 1 | 3 | 4 | 3 |
| MYCs | 2 | 1 | 4 | 4 |
| NACs | 0 | 1 | 4 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, R.; Xu, L.; Hao, J.; Zhang, L.; Wang, S.; Zhu, Z.; Yu, Y. Transcriptome Dynamics of Brassica juncea Leaves in Response to Omnivorous Beet Armyworm (Spodoptera exigua, Hübner). Int. J. Mol. Sci. 2023, 24, 16690. https://doi.org/10.3390/ijms242316690
Xia R, Xu L, Hao J, Zhang L, Wang S, Zhu Z, Yu Y. Transcriptome Dynamics of Brassica juncea Leaves in Response to Omnivorous Beet Armyworm (Spodoptera exigua, Hübner). International Journal of Molecular Sciences. 2023; 24(23):16690. https://doi.org/10.3390/ijms242316690
Chicago/Turabian StyleXia, Rui, Liai Xu, Jiaojiao Hao, Lili Zhang, Shanyi Wang, Zhujun Zhu, and Youjian Yu. 2023. "Transcriptome Dynamics of Brassica juncea Leaves in Response to Omnivorous Beet Armyworm (Spodoptera exigua, Hübner)" International Journal of Molecular Sciences 24, no. 23: 16690. https://doi.org/10.3390/ijms242316690
APA StyleXia, R., Xu, L., Hao, J., Zhang, L., Wang, S., Zhu, Z., & Yu, Y. (2023). Transcriptome Dynamics of Brassica juncea Leaves in Response to Omnivorous Beet Armyworm (Spodoptera exigua, Hübner). International Journal of Molecular Sciences, 24(23), 16690. https://doi.org/10.3390/ijms242316690







