Bioengineering of the Marine Diatom Phaeodactylum tricornutum with Cannabis Genes Enables the Production of the Cannabinoid Precursor, Olivetolic Acid
Abstract
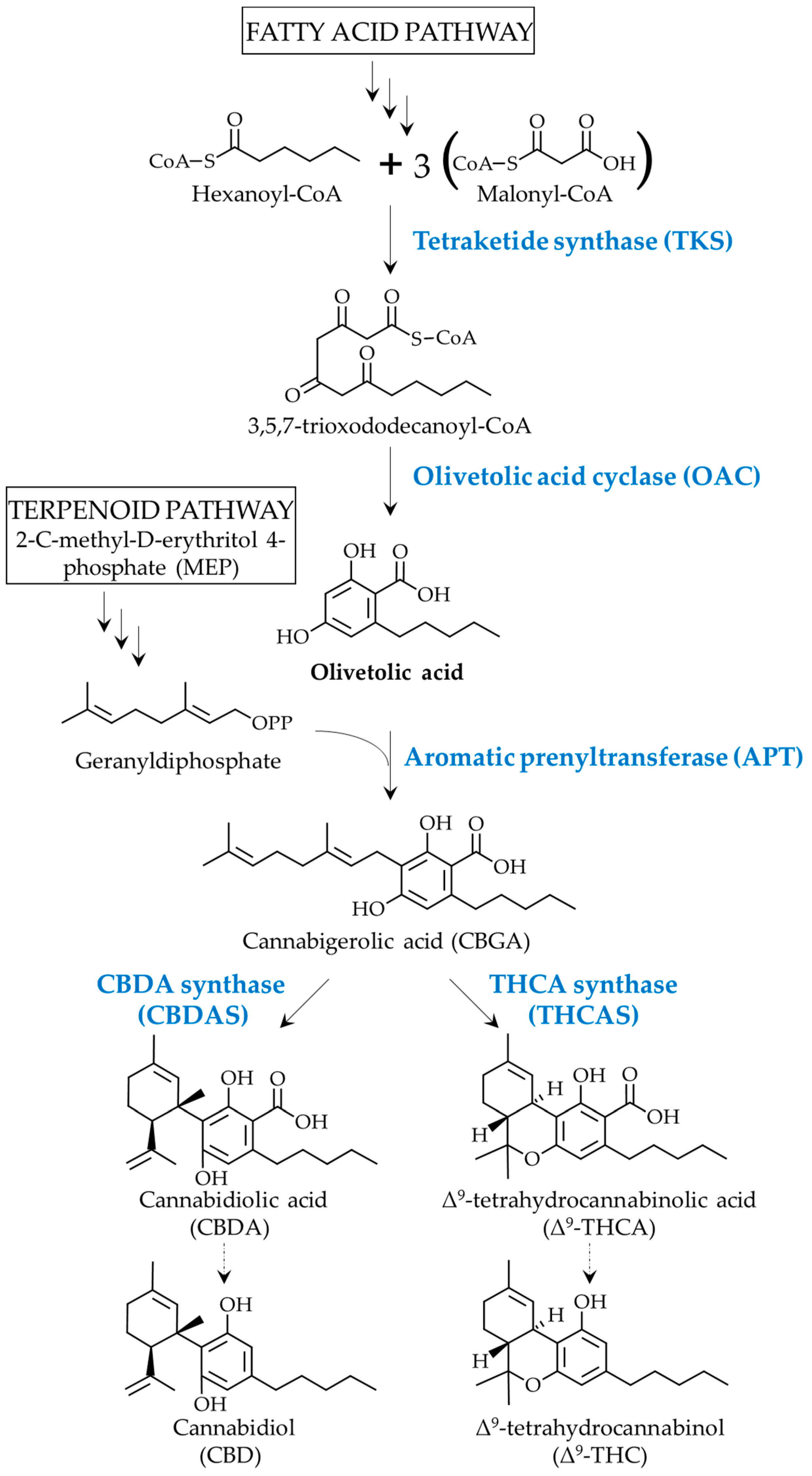
1. Introduction
2. Results
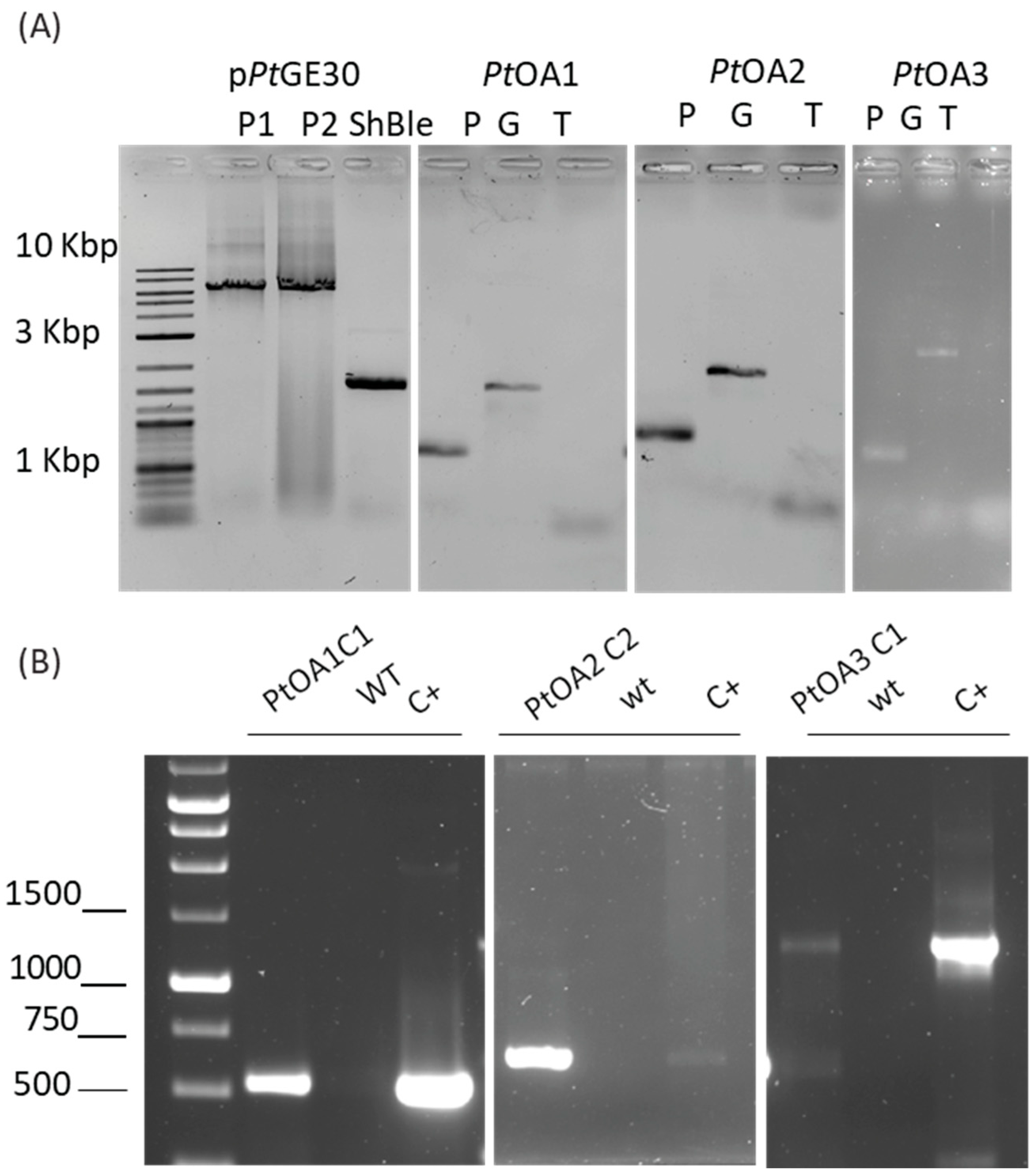
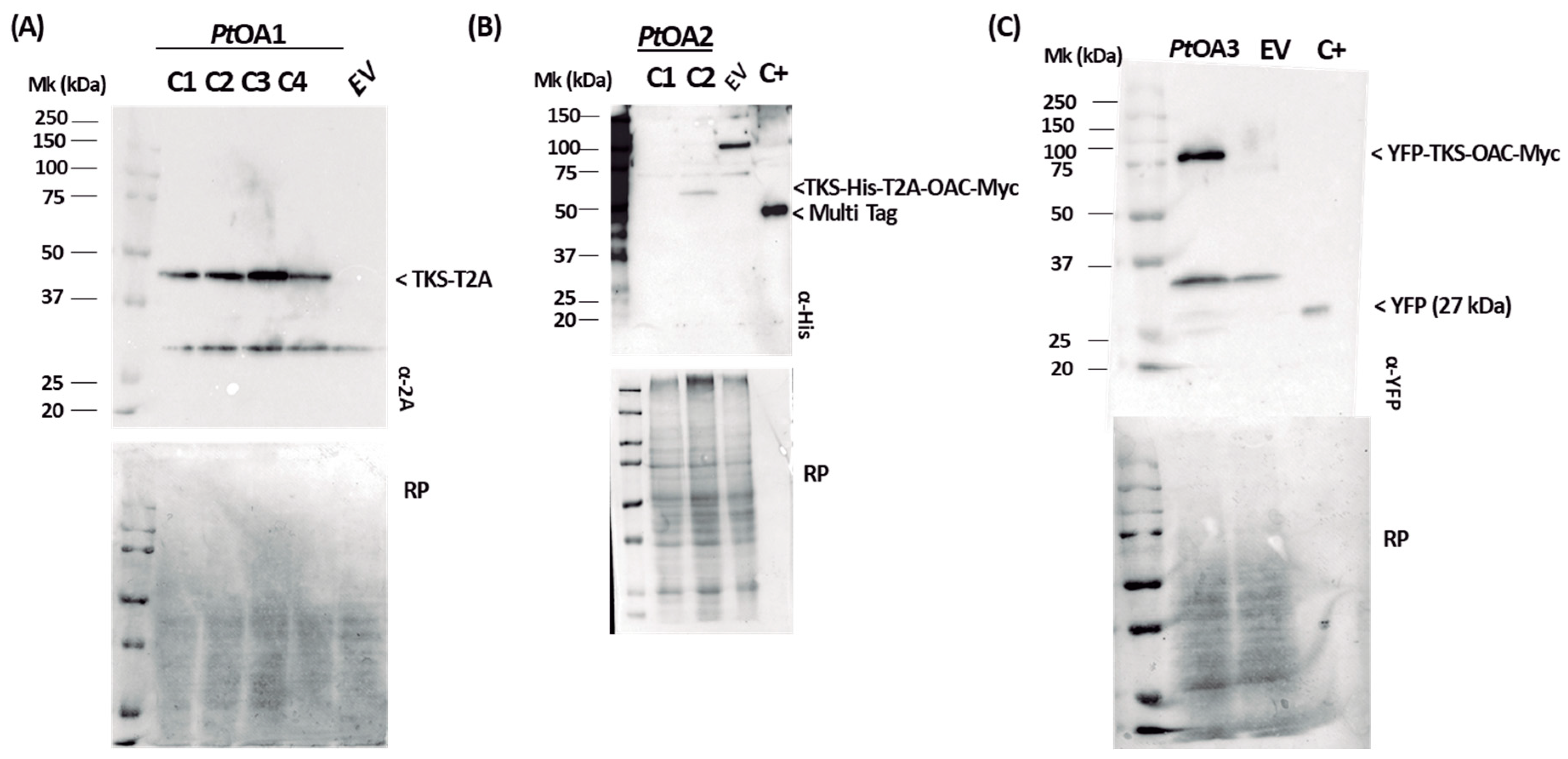
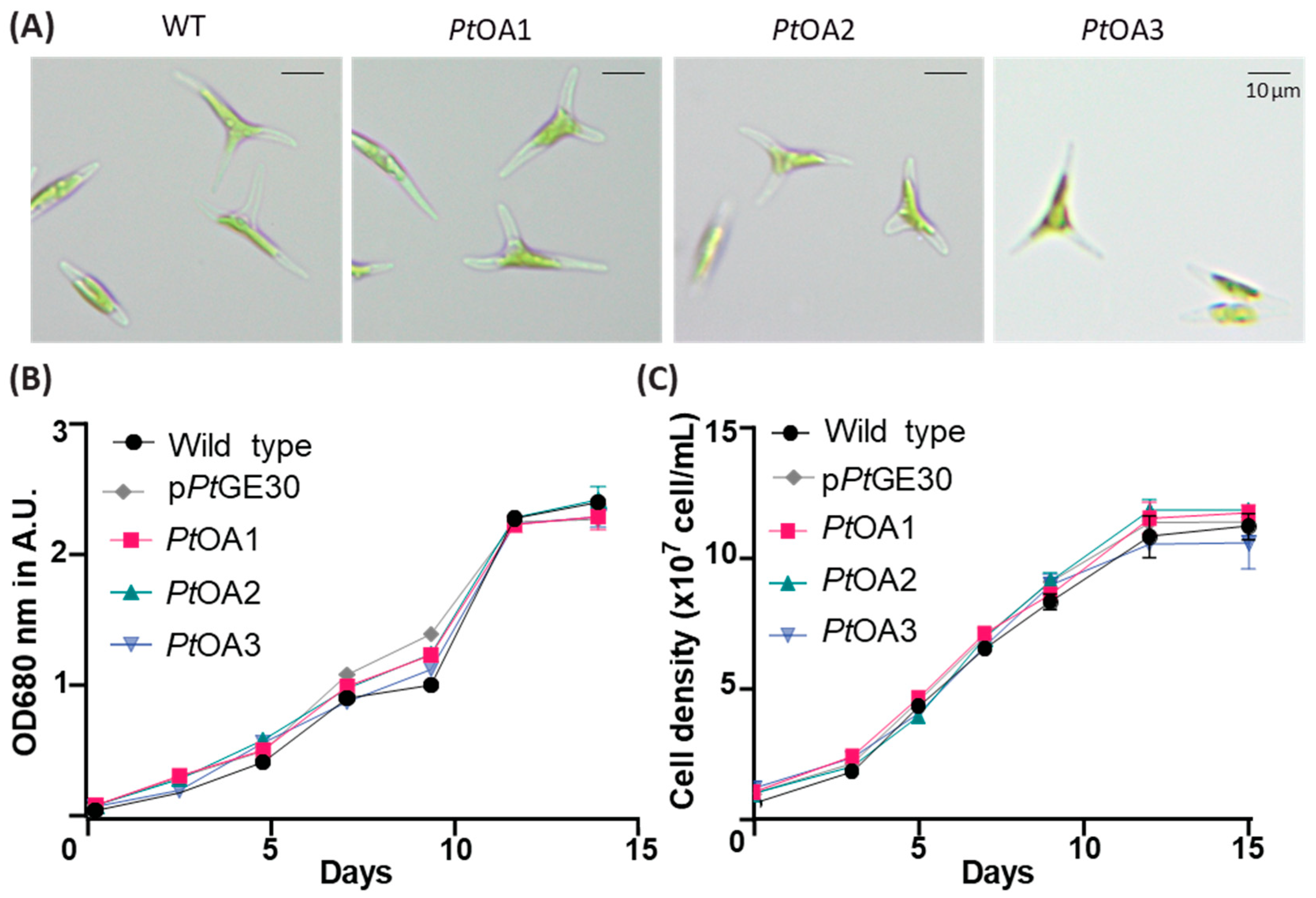
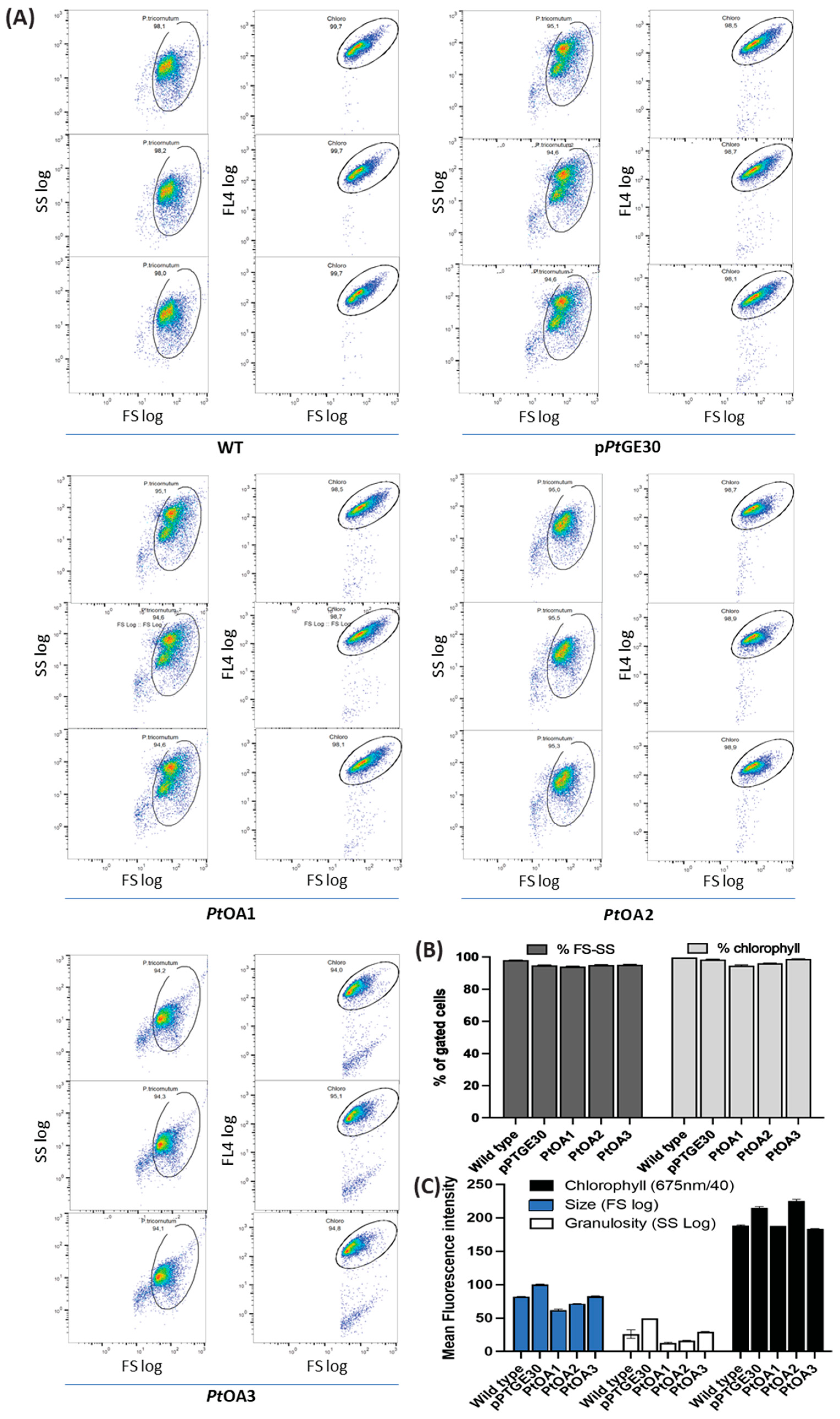
2.1. Heterologous Protein Expression and Localization in the Diatom P. tricornutum
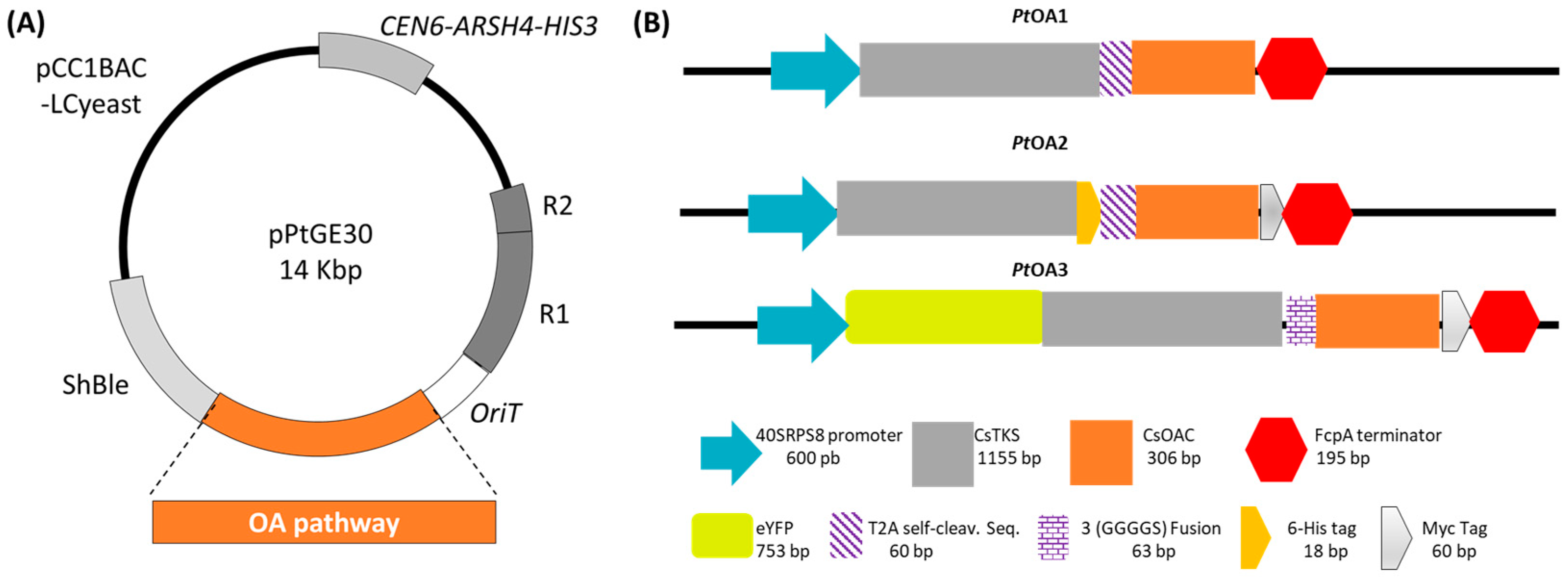
2.1.1. Transformation Validation and Transconjugant Characterization
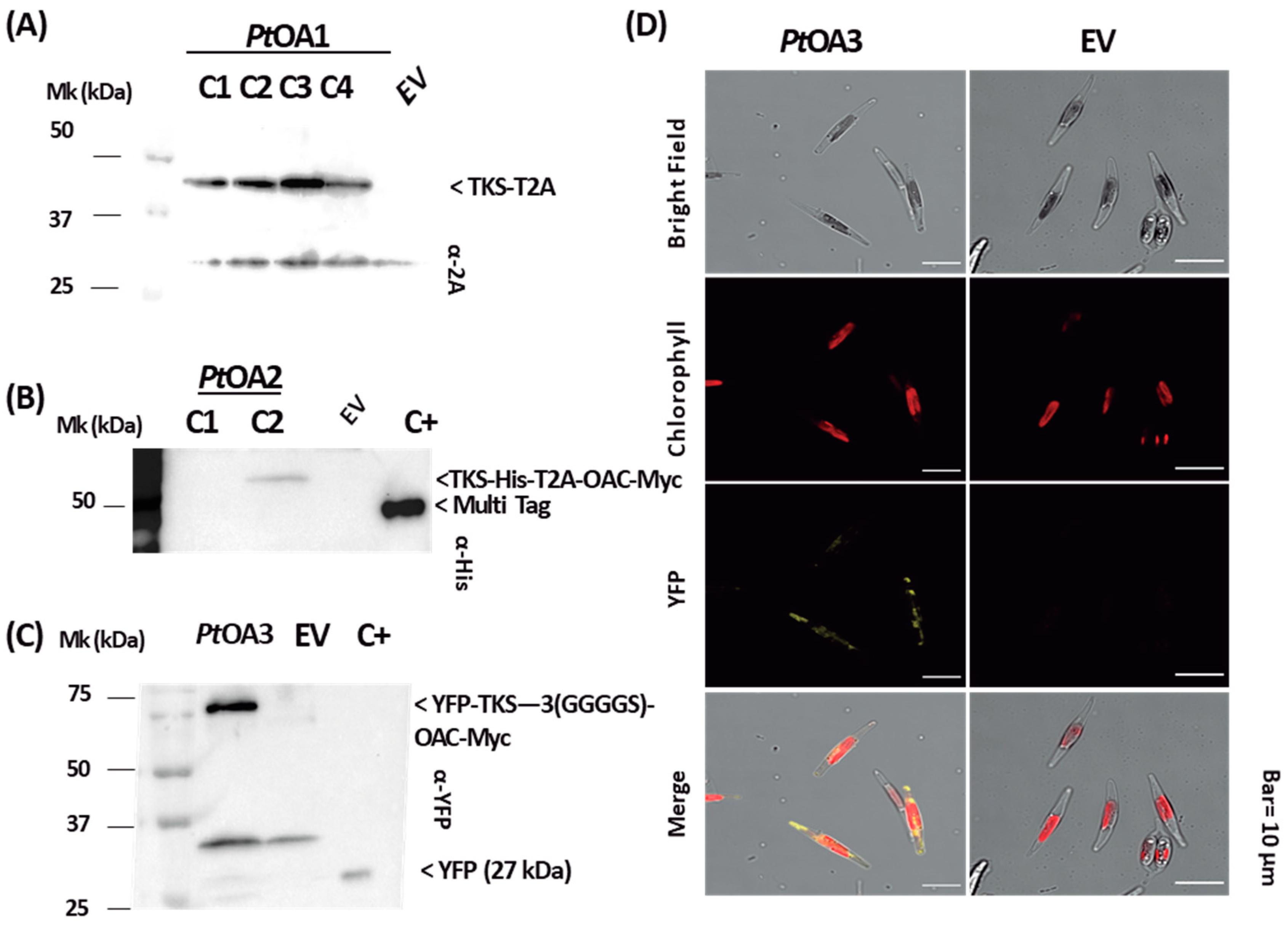
2.1.2. Heterologous Protein Detection and Localization
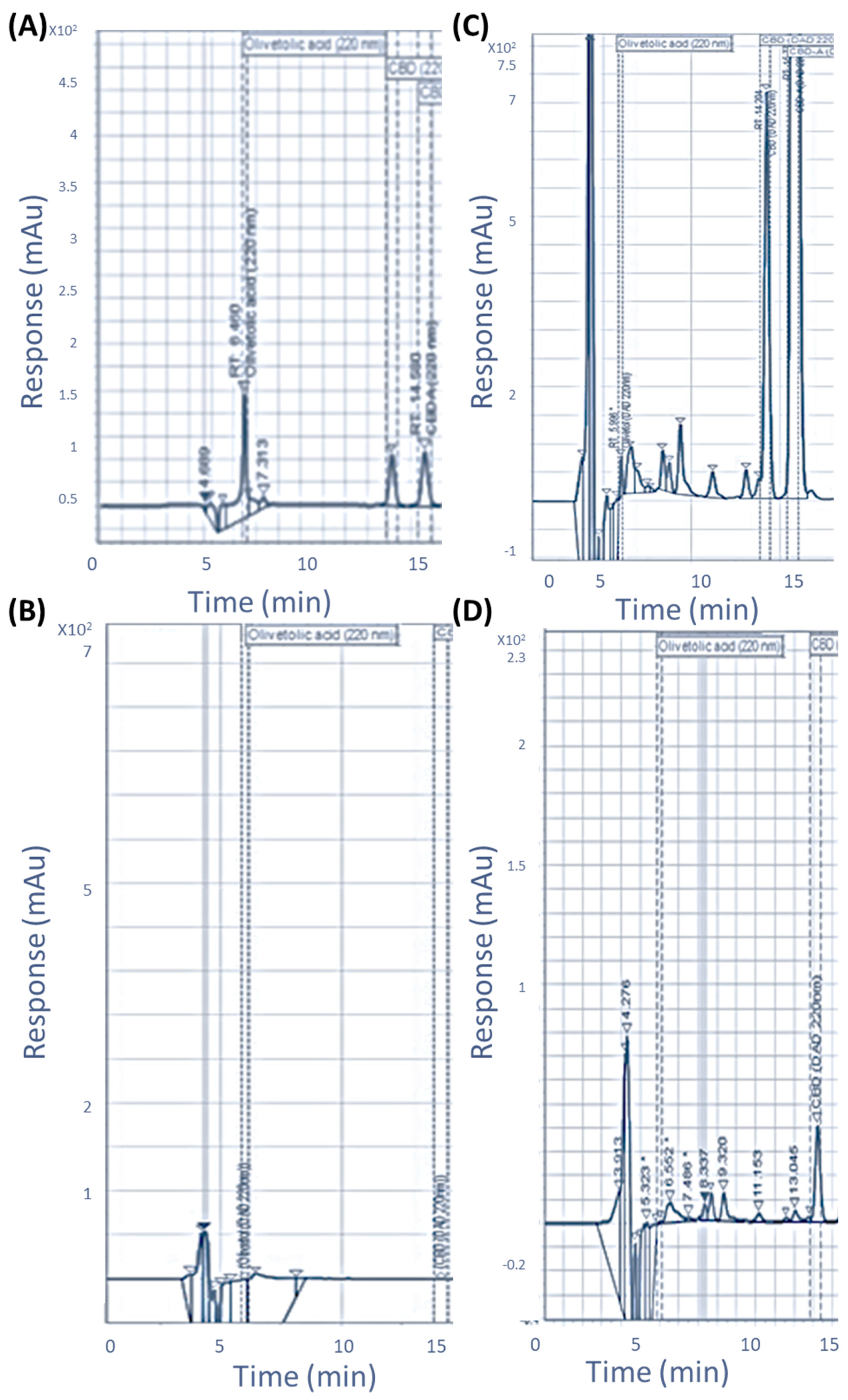
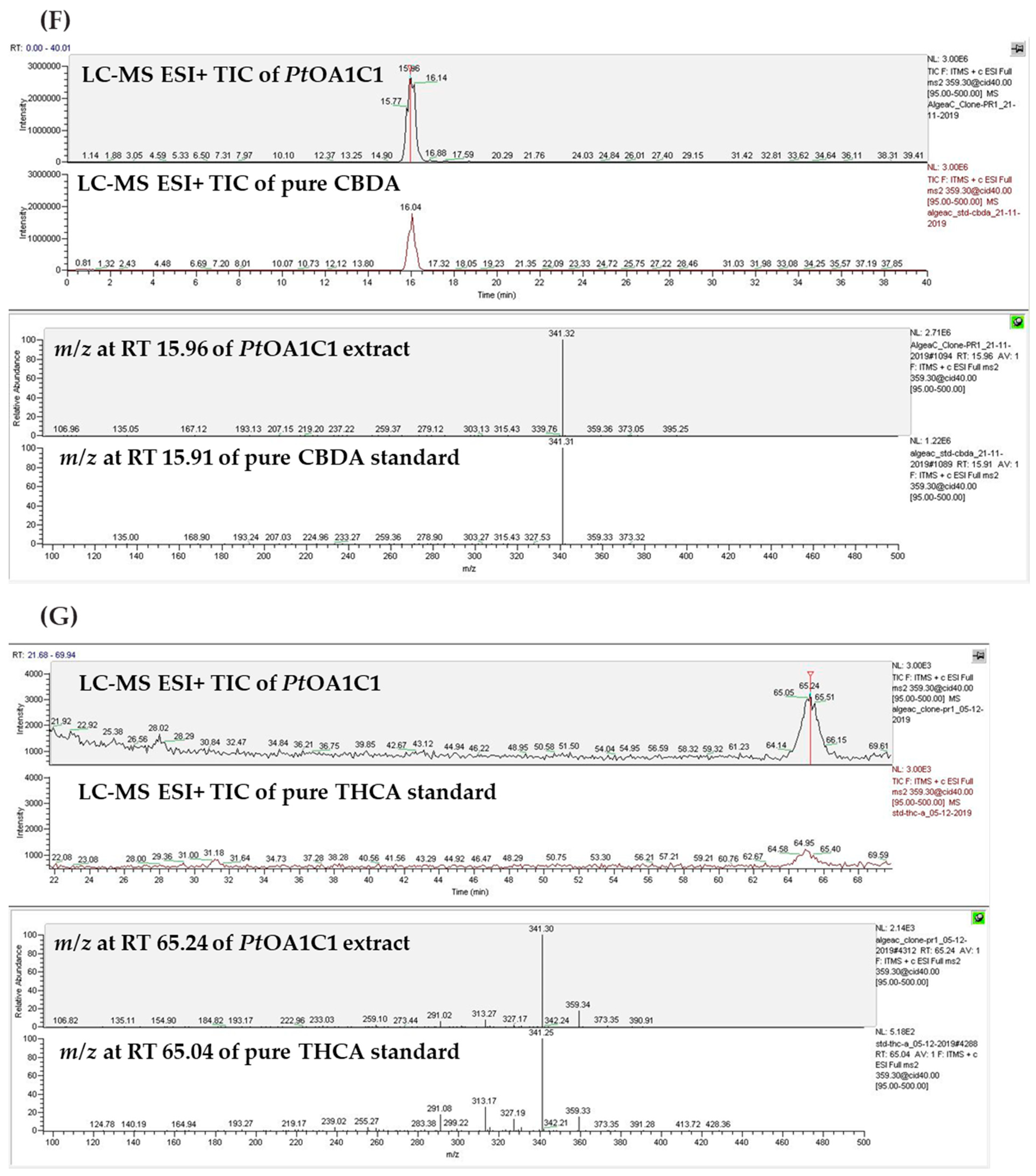
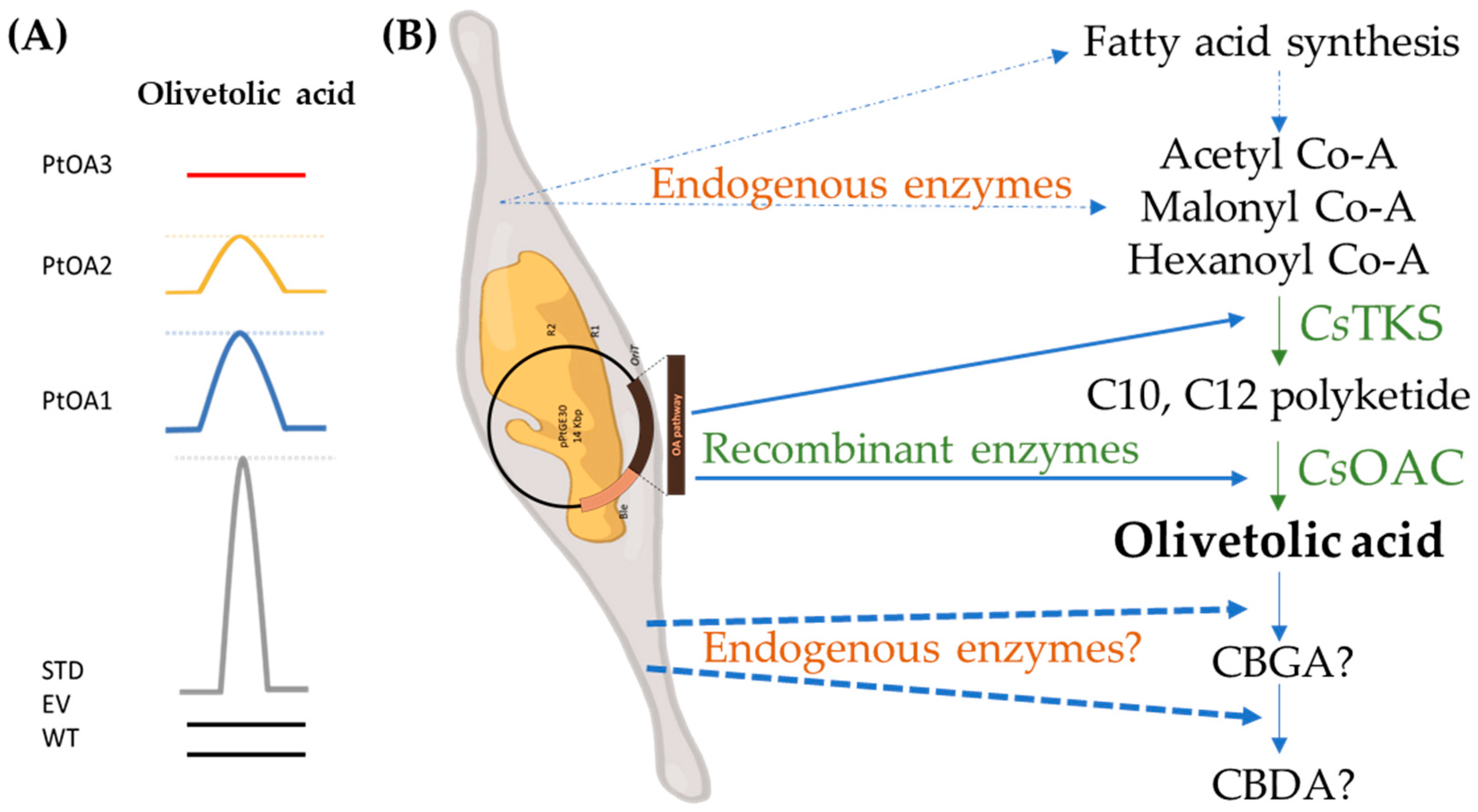
2.2. Temporal Metabolite Production
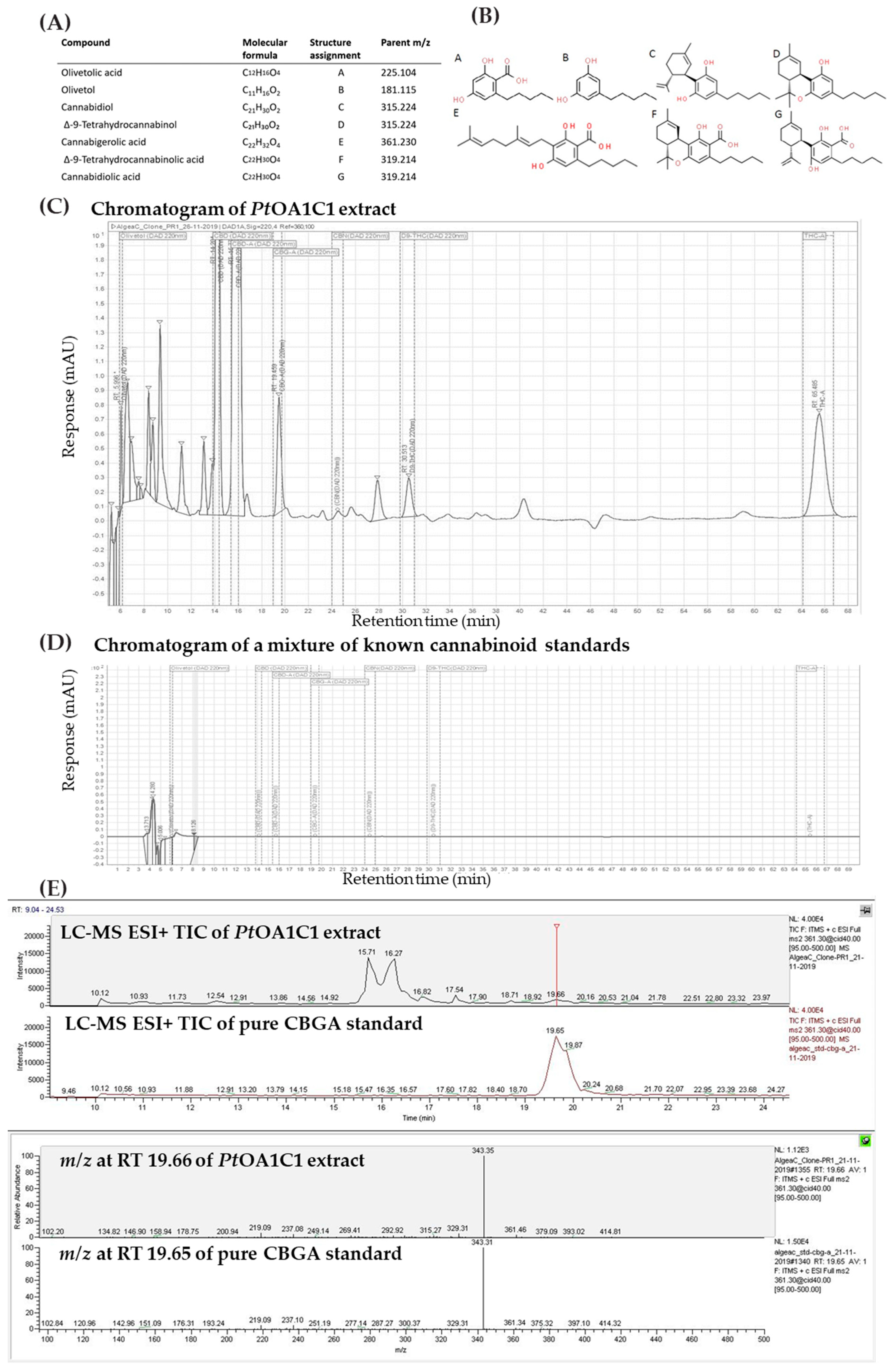
2.2.1. Metabolite Detection and Identification
2.2.2. Metabolite Production Timeline
3. Discussion
4. Materials and Methods
4.1. Microbial Strains and Growth Conditions
4.2. Diatom Transformation, Selection, and Subculturing
4.3. Transgene Sequences Validation
4.4. Heterologous Protein Detection
4.5. Subcellular Localization of YFP
4.6. Flow Cytometry Analysis
4.7. Metabolite Extraction Method
4.8. HPLC Detection Method for Cannabinoid Precursors
4.9. Mass Spectrum Validation
4.10. Precursor Supplementation Assay
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Au | Absorbance units |
| CB | Cannabinoid |
| CBD | Cannabidiol |
| CBDA | Cannabidiolic acid |
| CBGA | Cannabigerolic acid |
| cPCR | Colony polymerase chain reaction |
| FACS | Fluorescence-activated cell sorting |
| Gly | Glycine |
| GPP | Geranyl pyrophosphate |
| His | Histidine |
| OA | Olivetolic acid |
| OAC | Olivetolic acid cyclase |
| OD | Optical density |
| Ser | Serine |
| T2A | Thosea asigna virus self-cleavable sequence |
| THC | Δ-9-tetrahydrocannabinol |
| THCA | Delta (9)-tetrahydrocannabinolic acid |
| TKS | Tetraketide synthase |
| YFP | Yellow fluorescent protein |
Appendix A
| Sequence Name | Amino Acid Sequence | Length (a.a.) | Accession Number (CDS) | Accession Number AA Seq |
|---|---|---|---|---|
| C. sativa type 3 tetraketide synthase (TKS) | MNHLRAEGPASVLAIGTANPENILLQDEFPDYYFRVTKSEHMTQLKEKFRKICDK SMIRKRNCFLNEEHLKQNPRLVEHEMQTLDARQDMLVVEVPKLGKDACAKAIKEWGQPKS KITHLIFTSASTTDMPGADYHCAKLLGLSPSVKRVMMYQLGCYGGGTVLRIAKDIAENN KGARVLAVCCDIMACLFRGPSESDLELLVGQAIFGDGAAAVIVGAEPDESVGERPIFELVSTG QTILPNSEGTIGGHIREAGLIFDLHKDVPMLISNNIEKCLIEAFTPIGISDWNSIFWITH PGGKAILDKVEEKLHLKSDKFVDSRHVLSEHGNMSSSTVLFVMDELRKRSLEEGKSTTGDGFE WGVLFGFGPGLTVERVVVRSVPIKY | 385 | EU551165.1 | ACD76855.1 |
| C. sativa olivetolic acid cyclase (OAC) | MAVKHLIVLKFKDEITEAQKEEFFKTYVNLVNIIPAMKDVYWGKDVTQKNKEEGYTHIVEV TFESVETIQDYIIHPAHVGFGDVYRSFWEKLLIFDYTPRK | 101 | NM_001397939.1 | NP_001384868.1 |
| Name | Nucleotide Sequence (Codon Optimized) | Description and/or References |
|---|---|---|
| pPtGE30-part 1 | CAGGGTAATATAGATCTTCCGCTGCATAACCCTGCTTCGGGGTCATTATAGCGATTTTTTCGGTATATCCATCCTTTTTCGCA CGATATACAGGATTTTGCCAAAGGGTTCGTGTAGACTTTCCTTGGTGTATCCAACGGCGTCAGCCGGGCAGGATAGGTGAAGTAG GCCCACCCGCGAGCGGGTGTTCCTTCTTCACTGTCCCTTATTCGCACCTGGCGGTGCTCAACGGGAATCCTGCTCTGCGAGGCTG GCCGGCTACCGCCGGCGTAACAGATGAGGGCAAGCGGATGGCTGATGAAACCAAGCCAACCAGGAAGGGCAGCCCACCTATCAAG GTGTACTGCCTTCCAGACGAACGAAGAGCGATTGAGGAAAAGGCGGCGGCGGCCGGCATGAGCCTGTCGGCCTACCTGCTGGCCG TCGGCCAGGGCTACAAAATCACGGGCGTCGTGGACTATGAGCACGTCCGCGAGCTGGCCCGCATCAATGGCGACCTGGGCCGCCT GGGCGGCCTGCTGAAACTCTGGCTCACCGACGACCCGCGCACGGCGCGGTTCGGTGATGCCACGATCCTCGCCCTGCTGGCGAAG ATCGAAGAGAAGCAGGACGAGCTTGGCAAGGTCATGATGGGCGTGGTCCGCCCGAGGGCAGAGCCATGACTTTTTTAGCCGCTAA AACGGCCGGGGGGTGCGCGTGATTGCCAAGCACGTCCCCATGCGCTCCATCAAGAAGAGGCACTTCGAGCTGTAAGTACATCACC GACGAGCAAGGCAAGACGATCCGCGCCTGTTTTATTGAGAACGTTGTTCGTGTTGGCCTCAATGGTAGCGATGCGTCATTCAGCG AAGTTCTGGTGCTGATGATGTGGTTCGCTTTGCCACTGGTCAATGTGGTAAGCCCGTGTAATGTCAGTAACCTTTTTACTGATCT CAGCTTGAGCACGGTCGCTGATGAGCTTATCCATGGCCCCACGGTAACGGATATGATCCTCTAGGGCGTTGACAAATTCTTTGTC GGTTTTCATGGGGTAGACGTCAGTGACCAGGGAACGTCTCCCTACAAAAAGTTGCGCATACTTTGCTCCGTTGTCGACGGCAGGG GTGTCGGACCAGATTGTATCTGTGGCAACGGCCTCATTGCGTCAATGGACCTTTAAAGCCGGGAAACGAGACTTGAAATGTTTAC GTAGAGGTGCATTATAGACCTCACGCGCATATTGAGTGGTTGCAAAAATGGTTTTGCGAACGGTATCACTGGAGACCCATGCCAG GCAAGGACGGAGGGCATCATAATCATGTTCATTGCGTTGGACAGCATGCTTGTTACACGTAAGTATACGGTCGAGAGCATCGTGA CGTTGAGAATTACAGATGGCAACGTGTCGGTGCATGTATTGGCCGAGATTGTCAAATCGTGGCTCAATATATGGCATGCCAGGTA AGTCATGGATGTCGGTATGCCATTCAGCCTTCATGTCGATTTCGTTGTCGAGAACTGATGGGTCCCAGTCAACGTCAGAAGTAAG AACAACATGGGGAAGGGAATGAAGTTCATGACTGGTGGGTGCGCGCATATCCATGTACGGAAGACCCTGTCGTATGTTTAGTGGG ATGATGTAATCATCCAGGGTAACAATACGTTGGTTGCCGCCCACGGTACGAGACCGATCTTGAACACAATTGTGGCAATGTTCAA GTTGGGCGCTGGAATGGATGGTTTTACCTTTACCAAGATGCGCGTATTGATGCATGATAACAACAATGGGCCCTCGTTGCGATTC GACAAGACCGGCAGCCGTGACAATGTCTAGATTGGACAAGGTATGTTCATTTATACCGGTTATGTTGGCTGAACGTCCAGTTTTG TGCAGTACGGTAACATCACTCCCCGCAAGTCCACCGTTGGCACCGCGATCGACAAGTGCAGATGTGGTATTTTGTACTGTGTGTCG GGAGACTTGGTACTGTAACACATTTGACTGAAGTGACTTGGTGGTACCATGAGGACGGAGGGAACCCGTTGATACGGTGTTGCGT GACGCTGCAAGTACCTTACGTATGTCGCCTGGATCCATACGTCCAACCCGATCAGAAAGATGTGCTAGGAGTTCCGTTTCTGGTG CACAATCATGAAAGGTATCCACGGGAGTTCCATTGTTGTCGTCCGGGGCTGCGTCGCGACTGCCAGTTGCATGTACATGCGCCAT CAAGGGACGTGCCAATCCATTACTAACCGGGAGAACCTTAGCTGGGGCAGCCAATCCCTGAAGTATAGCCTTAGCATCATCTGAG AGCTGGTTCCACATATCTCTAGGGATATAAGGTCGTTCACGGTTAGTGGGCGTACTATGATGTCGGTTATTAGTACAATTCTCCC TGCGGGCATGCGCATTGGCTTCATAGAGTACGGAAGGTGACAAGTCAATATTGTAGTTAACATCCGGATCAGTGTCAAAGTCTGT AGGATGGTAAGAAAGATCAGTAGAATGAATACTACGCTTTTCCTTGGGACTACGGGAATTAGAGAAGTTGTTTCCTTTATTGTAG AGTGACGCTGAAGCAAGTAGAAGACTAAGGTAACTCTCGTAGCTAATAGGATTACCTCCTTTGGCTAGGTCAAGAGTGGCTGTGA TCTTCACTTGACAAAGTTCCGGTACATTGTGGACAGCATTCTCCAAAAGACTAAGACACAGTTGCTTTGGGAGTTGCTCAGCCAT TGGTACAGTATTGTGGTAGATACAAAGGTGGTTCTTCCAATGAAGGATAAATCCTTCTGCTGTGCCTGTCCATGAGGATCCATAT TTCGCCGTAGTTAGGTAACCAAGCGTGGTGGCTGAACTGATCTTCGCACTTGCTGATTCCGTATAGTGTTTGACAACTTTACAAA ACACTTCTTGCGCAGTTCGCTCTAGCCTGGACAGAGAAGGGACAATCTTCGTCGTTAAGACTCGTGCGAATAGCAAAAGATCACA AAATAGCACATCGGCACCGACCAACGATTATTTCCAAGGAAAAAAAGAATGCTTCACTACAAGAAATTGTGTCATCCCTATACAG AGTCTTGTTACTGTGACAGAAAATTGATGGAAGATGTGGCGGATTGCCTTTACACTAGCCAACTTGTTCGACTAATTGCAGCTTCT TCTGAGAGGCTTCACCGAGTAACGCGAAGAACACCGGTGTCTCGTACATGCTCGTCGGTGAACGCTCGTCCAATGACACCCCCCA CTTTGTATCAATATCCCAACTTGGTAGTGAACTGGAATGATACATGCAATTTCGCGCCGCATCAACAGCCACGGGCACCATCGACG AATAGACTCGGTCGAGCTGGTTGCCCTCGCCGCTGGGCTGGCGGCCGTCTATGGCCCTGCAAACGCGCCAGAAACGCCGTCGAAGC CGTGTGCGAGACACCGCGGCCGGCCGCCGGCGTTGTGGATACCTCGCGGAAAACTTGGCCCTCACTGACAGATGAGGGGCGGACGT TGACACTTGAGGGGCCGACTCACCCGGCGCGGCGTTGACAGATGAGGGGCAGGCTCGATTTCGGCCGGCGACGTGGAGCTGGCCAG CCTCGCAAATCGGCGAAAACGCCTGATTTTACGCGAGTTTCCCACAGATGATGTGGACAAGCCTGGGGATAAGTGCCCTGCGGTAT TGACACTTGAGGGGCGCGACTACTGACAGATGAGGGGCGCGATCCTTGACACTTGAGGGGCAGAGTGCTGACAGATGAGGGGCGCA CCTATTGACATTTGAGGGGCTGTCCACAGGCAGAAAATCCAGCATTTGCAAGGGTTTCCGCCCGTTTTTCGGCCACCGCTAACCTG TCTTTTAACCTGCTTTTAAACCAATATTTATAAACCTTGTTTTTAACCAGGGCTGCGCCCTGTGCGCGTGACCGCGCACGCCGAAG GGGGGTGCCCCCCCTTCTCGAACCCTCCCGGTCGAGTGAGCGAGGAAGCACCAGGGAACAGCACTTATATATTCTGCTTACACACG ATGCCTGAAAAAACTTCCCTTGGGGTTATCCACTTATCCACGGGGATATTTTTATAATTATTTTTTTTATAGTTTTTAGATCTTCT TTTTTAGAGCGCCTTGTAGGCCTTTATCCATGCTGGTTCTAGAGAAGGTGTTGTGACAAATTGCCCTTTCAGTGTGACAAATCACC CTCAAATGACAGTCCTGTCTGTGACAAATTGCCCTTAACCCTGTGACAAATTGCCCTCAGAAGAAGCTGTTTTTTCACAAAGTTAT CCCTGCTTATTGACTCTTTTTTATTTAGTGTGACAATCTAAAAACTTGTCACACTTCACATGGATCTGTCATGGCGGAAACAGCGG TTATCAATCACAAGAAACGTAAAAATAGCCCGCGAATCGTCCAGTCAAACGACCTCACTGAGGCGGCATATAGTCTCTCCCGGGAT CAAAAACGTATGCTGTATCTGTTCGTTGACCAGATCAGAAAATCTGATGGCACCCTACAGGAACATGACGGTATCTGCGAGATCCA TGTTGCTAAATATGCTGAAATATTCGGATTGACCTCTGCGGAAGCCAGTAAGGATATACGGCAGGCATTGAAGAGTTTCGCGGGGA AGGAAGTGGTTTTTTATCGCCCTGAAGAGGATGCCGGCGATGAAAAAGGCTATGAATCTTTTCCTTGGTTTATCAAACGTGCGCAC AGTCCATCCAGAGGGCTTTACAGTGTACATATCAACCCATATCTCATTCCCTTCTTTATCGGGTTACAGAACCGGTTTACGCAGTTT CGGCTTAGTGAAACAAAAGAAATCACCAATCCGTATGCCATGCGTTTATACGAATCCCTGTGTCAGTATCGTAAGCCGGATGGCTCA GGCATCGTCTCTCTGAAAATCGACTGGATCATAGAGCGTTACCAGCTGCCTCAAAGTTACCAGCGTATGCCTGACTTCCGCCGCCG CTTCCTGCAGGTCTGTGTTAATGAGATCAACAGCAGAACTCCAATGCGCCTCTCATACATTGAGAAAAAGAAAGGCCGCCAGACGA CTCATATCGTATTTTCCTTCCGCGATATCACTTCCATGACGACAGGATAGTCTGAGGGTTATCTGTCACAGATTTGAGGGTGGTTCG TCACATTTGTTCTGACCTACTGAGGGTAATTTGTCACAGTTTTGCTGTTTCCTTCAGCCTGCATGGATTTTCTCATACTTTTTGAA CTGTAATTTTTAAGGAAGCCAAATTTGAGGGCAGTTTGTCACAGTTGATTTCCTTCTCTTTCCCTTCGTCATGTGACCTGATATC GGGGGTTAGTTCGTCATCATTGATGAGGGTTGATTATCACAGTTTATTACTCTGAATTGGCTATCCGCGTGTGTACCTCTACCTGG AGTTTTTCCCACGGTGGATATTTCTTCTTGCGCTGAGCGTAAGAGCTATCTGACAGAACAGTTCTTCTTTGCTTCCTCGCCAGTT CGCTCGCTATGCTCGGTTACACGGCTGCGGCGAGCATCACGTGCTATAAAAATAATTATAATTTAAATTTTTTAATATAAATATA TAAATTAAAAATAGAAAGTAAAAAAAGAAATTAAAGAAAAAATAGTTTTTGTTTTCCGAAGATGTAAAAGACTCTAGGGGGATCG CCAACAAATACTACCTTTTACCTTGCTCTTCCTGCTCTCAGGTATTAATGCCGAATTGTTTCATCTTGTCTGTGTAGAAGACCAC ACACGAAAATCCTGTGATTTTACATTTTACTTATCGTTAATCGAATGTATATCTATTTAATCTGCTTTTCTTGTCTAATAAATATA TATGTAAAGTACGCTTTTTGTTGAAATTTTTTAAACCTTTGTTTATTTTTTTTTCTTCATTCCGTAACTCTTCTACCTTCTTTATT TACTTTCTAAAATCCAAATACAAAACATAAAAATAAATAAACACAGAGTAAATTCCCAAATTATTCCATCATTAAAAGATACGAGG CGCGTGTAAGTTACAGGCAAGCGATCCTAGTACACTCTATATTTTTTTATGCCTCGGTAATGATTTTCATTTTTTTTTTCCACCTA GCGGATGACTCTTTTTTTTTCTTAGCGATTGGCATTATCACATAATGAATTATACATTATATAAAGTAATGTGATTTCTTCGAA GAATATACTAAAAAATGAGCAGGCAAGATAAACGAAGGCAAAGATGACAGAGCAGAAAGCCCTAGTAAAGCGTATTACAAATGA AACCAAGATTCAGATTGCGATCTCTTTAAAGGGTGGTCCCCTAGCGATAGAGCACTCGATCTTCCCAGAAAAAGAGGCAGAAGCA GTAGCAGAACAGGCCACACAATCGCAAGTGATTAACGTCCACACAGGTATAGGGTTTCTGGACCATATGATACATGCTCTGGCCA AGCATTCCGGCTGGTCGCTAATCGTTGAGTGCATTGGTGACTTACACATAGACGACCATCACACCACTGAAGACTGCGGGATTGC TCTCGGTCAAGCTTTTAAAGAGGCCCTAG | Slattery et al., 2018 [61] |
| pPtGE30-part 2 | TCGAGCTGGTTGCCCTCGCCGCTGGGCTGGCGGCCGTCTATGGCCCTGCAAACGCGCCAGAAACGCCGTCGAAGCCGTGTGCGA GACACCGCGGCCGGCCGCCGGCGTTGTGGATACCTCGCGGAAAACTTGGCCCTCACTGACAGATGAGGGGCGGACGTTGACACTT GAGGGGCCGACTCACCCGGCGCGGCGTTGACAGATGAGGGGCAGGCTCGATTTCGGCCGGCGACGTGGAGCTGGCCAGCCTCGCA AATCGGCGAAAACGCCTGATTTTACGCGAGTTTCCCACAGATGATGTGGACAAGCCTGGGGATAAGTGCCCTGCGGTATTGACAC TTGAGGGGCGCGACTACTGACAGATGAGGGGCGCGATCCTTGACACTTGAGGGGCAGAGTGCTGACAGATGAGGGGCGCACCTAT TGACATTTGAGGGGCTGTCCACAGGCAGAAAATCCAGCATTTGCAAGGGTTTCCGCCCGTTTTTCGGCCACCGCTAACCTGTCTT TTAACCTGCTTTTAAACCAATATTTATAAACCTTGTTTTTAACCAGGGCTGCGCCCTGTGCGCGTGACCGCGCACGCCGAAGGGG GGTGCCCCCCCTTCTCGAACCCTCCCGGTCGAGTGAGCGAGGAAGCACCAGGGAACAGCACTTATATATTCTGCTTACACACGAT GCCTGAAAAAACTTCCCTTGGGGTTATCCACTTATCCACGGGGATATTTTTATAATTATTTTTTTTATAGTTTTTAGATCTTCTT TTTTAGAGCGCCTTGTAGGCCTTTATCCATGCTGGTTCTAGAGAAGGTGTTGTGACAAATTGCCCTTTCAGTGTGACAAATCACC CTCAAATGACAGTCCTGTCTGTGACAAATTGCCCTTAACCCTGTGACAAATTGCCCTCAGAAGAAGCTGTTTTTTCACAAAGTTA TCCCTGCTTATTGACTCTTTTTTATTTAGTGTGACAATCTAAAAACTTGTCACACTTCACATGGATCTGTCATGGCGGAAACAGC GGTTATCAATCACAAGAAACGTAAAAATAGCCCGCGAATCGTCCAGTCAAACGACCTCACTGAGGCGGCATATAGTCTCTCCCGG GATCAAAAACGTATGCTGTATCTGTTCGTTGACCAGATCAGAAAATCTGATGGCACCCTACAGGAACATGACGGTATCTGCGAGA TCCATGTTGCTAAATATGCTGAAATATTCGGATTGACCTCTGCGGAAGCCAGTAAGGATATACGGCAGGCATTGAAGAGTTTCGC GGGGAAGGAAGTGGTTTTTTATCGCCCTGAAGAGGATGCCGGCGATGAAAAAGGCTATGAATCTTTTCCTTGGTTTATCAAACGT GCGCACAGTCCATCCAGAGGGCTTTACAGTGTACATATCAACCCATATCTCATTCCCTTCTTTATCGGGTTACAGAACCGGTTTA CGCAGTTTCGGCTTAGTGAAACAAAAGAAATCACCAATCCGTATGCCATGCGTTTATACGAATCCCTGTGTCAGTATCGTAAGCC GGATGGCTCAGGCATCGTCTCTCTGAAAATCGACTGGATCATAGAGCGTTACCAGCTGCCTCAAAGTTACCAGCGTATGCCTGAC TTCCGCCGCCGCTTCCTGCAGGTCTGTGTTAATGAGATCAACAGCAGAACTCCAATGCGCCTCTCATACATTGAGAAAAAGAAAG GCCGCCAGACGACTCATATCGTATTTTCCTTCCGCGATATCACTTCCATGACGACAGGATAGTCTGAGGGTTATCTGTCACAGAT TTGAGGGTGGTTCGTCACATTTGTTCTGACCTACTGAGGGTAATTTGTCACAGTTTTGCTGTTTCCTTCAGCCTGCATGGATTTT CTCATACTTTTTGAACTGTAATTTTTAAGGAAGCCAAATTTGAGGGCAGTTTGTCACAGTTGATTTCCTTCTCTTTCCCTTCGTC ATGTGACCTGATATCGGGGGTTAGTTCGTCATCATTGATGAGGGTTGATTATCACAGTTTATTACTCTGAATTGGCTATCCGCGT GTGTACCTCTACCTGGAGTTTTTCCCACGGTGGATATTTCTTCTTGCGCTGAGCGTAAGAGCTATCTGACAGAACAGTTCTTCTT TGCTTCCTCGCCAGTTCGCTCGCTATGCTCGGTTACACGGCTGCGGCGAGCATCACGTGCTATAAAAATAATTATAATTTAAATT TTTTAATATAAATATATAAATTAAAAATAGAAAGTAAAAAAAGAAATTAAAGAAAAAATAGTTTTTGTTTTCCGAAGATGTAAAA GACTCTAGGGGGATCGCCAACAAATACTACCTTTTACCTTGCTCTTCCTGCTCTCAGGTATTAATGCCGAATTGTTTCATCTTGT CTGTGTAGAAGACCACACACGAAAATCCTGTGATTTTACATTTTACTTATCGTTAATCGAATGTATATCTATTTAATCTGCTTTT CTTGTCTAATAAATATATATGTAAAGTACGCTTTTTGTTGAAATTTTTTAAACCTTTGTTTATTTTTTTTTCTTCATTCCGTAACT CTTCTACCTTCTTTATTTACTTTCTAAAATCCAAATACAAAACATAAAAATAAATAAACACAGAGTAAATTCCCAAATTATTCCA TCATTAAAAGATACGAGGCGCGTGTAAGTTACAGGCAAGCGATCCTAGTACACTCTATATTTTTTTATGCCTCGGTAATGATTTT CATTTTTTTTTTCCACCTAGCGGATGACTCTTTTTTTTTCTTAGCGATTGGCATTATCACATAATGAATTATACATTATATAAAG TAATGTGATTTCTTCGAAGAATATACTAAAAAATGAGCAGGCAAGATAAACGAAGGCAAAGATGACAGAGCAGAAAGCCCTAGTA AAGCGTATTACAAATGAAACCAAGATTCAGATTGCGATCTCTTTAAAGGGTGGTCCCCTAGCGATAGAGCACTCGATCTTCCCAG AAAAAGAGGCAGAAGCAGTAGCAGAACAGGCCACACAATCGCAAGTGATTAACGTCCACACAGGTATAGGGTTTCTGGACCATAT GATACATGCTCTGGCCAAGCATTCCGGCTGGTCGCTAATCGTTGAGTGCATTGGTGACTTACACATAGACGACCATCACACCACTG AAGACTGCGGGATTGCTCTCGGTCAAGCTTTTAAAGAGGCCCTAGGGGCCGTGCGTGGAGTAAAAAGGTTTGGATCAGGATTTGC GCCTTTGGATGAGGCACTTTCCAGAGCGGTGGTAGATCTTTCGAACAGGCCGTACGCAGTTGTCGAACTTGGTTTGCAAAGGGAG AAAGTAGGAGATCTCTCTTGCGAGATGATCCCGCATTTTCTTGAAAGCTTTGCAGAGGCTAGCAGAATTACCCTCCACGTTGATT GTCTGCGAGGCAAGAATGATCATCACCGTAGTGAGAGTGCGTTCAAGGCTCTTGCGGTTGCCATAAGAGAAGCCACCTCGCCCAA TGGTACCAACGATGTTCCCTCCACCAAAGGTGTTCTTATGTAGTTTTACACAGGAGTCTGGACTTGACGCTAGTGATAATAAGTG ACTGAGGTATGTGCTCTTCTTATCTCCTTTTGTAGTGTTGCTCTTATTTTAAACAACTTTGCGGTTTTTTGATGACTTTGCGATTT TGTTGTTGCTTTGCAGTAAATTGCAAGATTTAATAAAAAAACGCAAAGCAATGATTAAAGGATGTTCAGAATGAAACTCATGGAA ACACTTAACCAGTGCATAAACGCTGGTCATGAAATGACGAAGGCTATCGCCATTGCACAGTTTAATGATGACAGCCCGGAAGCGA GGAAAATAACCCGGCGCTGGAGAATAGGTGAAGCAGCGGATTTAGTTGGGGTTTCTTCTCAGGCTATCAGAGATGCCGAGAAAGCA GGGCGACTACCGCACCCGGATATGGAAATTCGAGGACGGGTTGAGCAACGTGTTGGTTATACAATTGAACAAATTAATCATATGCG TGATGTGTTTGGTACGCGATTGCGACGTGCTGAAGACGTATTTCCACCGGTGATCGGGGTTGCTGCCCATAAAGGTGGCGTTTACA AAACCTCAGTTTCTGTTCATCTTGCTCAGGATCTGGCTCTGAAGGGGCTACGTGTTTTGCTCGTGGAAGGTAACGACCCCCAGGGA ACAGCCTCAATGTATCACGGATGGGTACCAGATCTTCATATTCATGCAGAAGACACTCTCCTGCCTTTCTATCTTGGGGAAAAGG ACGATGTCACTTATGCAATAAAGCCCACTTGCTGGCCGGGGCTTGACATTATTCCTTCCTGTCTGGCTCTGCACCGTATTGAAACT GAGTTAATGGGCAAATTTGATGAAGGTAAACTGCCCACCGATCCACACCTGATGCTCCGACTGGCCATTGAAACTGTTGCTCATG ACTATGATGTCATAGTTATTGACAGCGCGCCTAACCTGGGTATCGGCACGATTAATGTCGTATGTGCTGCTGATGTGCTGATTGT TCCCACGCCTGCTGAGTTGTTTGACTACACCTCCGCACTGCAGTTTTTCGATATGCTTCGTGATCTGCTCAAGAACGTTGATCTTA AAGGGTTCGAGCCTGATGTACGTATTTTGCTTACCAAATACAGCAATAGCAATGGCTCTCAGTCCCCGTGGATGGAGGAGCAAAT TCGGGATGCCTGGGGAAGCATGGTTCTAAAAAATGTTGTACGTGAAACGGATGAAGTTGGTAAAGGTCAGATCCGGATGAGAACTG TTTTTGAACAGGCCATTGATCAACGCTCTTCAACTGGTGCCTGGAGAAATGCTCTTTCTATTTGGGAACCTGTCTGCAATGAAATT TTCGATCGTCTGATTAAACCACGCTGGGAGATTAGATAATGAAGCGTGCGCCTGTTATTCCAAAACATACGCTCAATACTCAACCG GTTGAAGATACTTCGTTATCGACACCAGCTGCCCCGATGGTGGATTCGTTAATTGCGCGCGTAGGAGTAATGGCTCGCGGTAATGC CATTACTTTGCCTGTATGTGGTCGGGATGTGAAGTTTACTCTTGAAGTGCTCCGGGGTGATAGTGTTGAGAAGACCTCTCGGGTAT GGTCAGGTAATGAACGTGACCAGGAGCTGCTTACTGAGGACGCACTGGATGATCTCATCCCTTCTTTTCTACTGACTGGTCAACAG ACACCGGCGTTCGGTCGAAGAGTATCTGGTGTCATAGAAATTGCCGATGGGAGTCGCCGTCGTAAAGCTGCTGCACTTACCGAAAG TGATTATCGTGTTCTGGTTGGCGAGCTGGATGATGAGCAGATGGCTGCATTATCCAGATTGGGTAACGATTATCGCCCAACAAGTG CTTATGAACGTGGTCAGCGTTATGCAAGCCGATTGCAGAATGAATTTGCTGGAAATATTTCTGCGCTGGCTGATGCGGAAAATATT TCACGTAAGATTATTACCCGCTGTATCAACACCGCCAAATTGCCTAAATCAGTTGTTGCTCTTTTTTCTCACCCCGGTGAACTATC TGCCCGGTCAGGTGATGCACTTCAAAAAGCCTTTACAGATAAAGAGGAATTACTTAAGCAGCAGGCATCTAACCTTCATGAGCAGA AAAAAGCTGGGGTGATATTTGAAGCTGAAGAAGTTATCACTCTTTTAACTTCTGTGCTTAAAACGTCATCTGCATCAAGAACTAGT TTAAGCTCACGACATCAGTTTGCTCCTGGAGCGACAGTATTGTATAAGGGCGATAAAATGGTGCTTAACCTGGACAGGTCTCGTGTT CCAACTGAGTGTATAGAGAAAATTGAGGCCATTCTTAAGGAACTTGAAAAGCCAGCACCCTGATGCGACCTCGTTTTAGTCTACGT TTATCTGTCTTTACTTAATGTCCTTTGTTACAGGCCAGAAAGCATAACTGGCCTGAATATTCTCTCTGGGCCCACTGTTCCACTTG TATCGTCGGTCTGATAATCAGACTGGGACCACGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCACGGTCCCACTCG TATCGTCGGTCTGATTATTAGTCTGGGACCACGGTCCCACTCGTATCGTCGGTCTGATAATCAGACTGGGACCACGGTCCCACTCG TATCGTCGGTCTGATTATTAGTCTGGGACCATGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCACGGTCCCACTCG TATCGTCGGTCTGATTATTAGTCTGGAACCACGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCACGGTCCCACTCG TATCGTCGGTCTGATTATTAGTCTGGGACCACGATCCCACTCGTGTTGTCGGTCTGATTATCGGTCTGGGACCACGGTCCCACTTG TATTGTCGATCAGACTATCAGCGTGAGACTACGATTCCATCAATGCCTGTCAAGGGCAAGTATTGACATGTCGTCGTAACCTGTAG AACGGAGTAACCTCGGTGTGCGGTTGTATGCCTGCTGTGGATTGCTGCTGTGTCCTGCTTATCCACAACATTTTGCGCACGGTTAT GTGGACAAAATACCTGGTTACCCAGGCCGTGCCGGCACGTTAACCGGGCTGCATCCGATGCAAGTGTGTCGCTGTCGACGAGCTCG CGAGCTCGGACATGAGGTTGCCCCGTATTCAGTGTCGCTGATTTGTATTGTCTGAAGTTGTTTTTACGTTAAGTTGATGCAGATCA ATTAATACGATACCTGCGTCATAATTGATTATTTGACGTGGTTTGATGGCCTCCACGCACGTTGTGATATGTAGATGATAATCATT ATCACTTTACGGGTCCTTTCCGGTGATCCGACAGGTTACGGGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTT TAAGGCGTTTCCGTTCTTCTTCGTCATAACTTAATGTTTTTATTTAAAATACCCTCTGAAAAGAAAGGAAACGACAGGTGCTGAAA GCGAGCTTTTTGGCCTCTGTCGTTTCCTTTCTCTGTTTTTGTCCGTGGAATGAACAATGGAAGTCCGAGCTCATCGCTAATAACTT CGTATAGCATACATTATACGAAGTTATATTCGATGCGGCCGCAAGGGGTTCGCGTCAGCGGGTGTTGGCGGGTGTCGGGGCTGGCT TAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCACACAGATGCGTAAGGAGAAAATACC GCATCAGGCGCCATTCGCCATTCAGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAA AGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTGT AATACGACTCACTATAGGGCGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGAGTATTCT ATAGTCTCACCTAAATAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACA TACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCT TTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGAACCCCTTGCGGCCGCCCGGGCCGTCGACCAATT CTCATGTTTGACAGCTTATCATCGAATTTCTGCCATTCATCCGCTTATTATCACTTATTCAGGCGTAGCAACCAGGCGTTTAAGGG CACCAATAACTGCCTTAAAAAAATTACGCCCCGCCCTGCCACTCATCGCAGTACTGTTGTAATTCATTAAGCATTCTGCCGACATGG AAGCCATCACAAACGGCATGATGAACCTGAATCGCCAGCGGCATCAGCACCTTGTCGCCTTGCGTATAATATTTGCCCATGGTGAA AACGGGGGCGAAGAAGTTGTCCATATTGGCCACGTTTAAATCAAAACTGGTGAAACTCACCCAGGGATTGGCTGAGACGAAAAACA TATTCTCAATAAACCCTTTAGGGAAATAGGCCAGGTTTTCACCGTAACACGCCACATCTTGCGAATATATGTGTAGAAACTGCCGGA AATCGTCGTGGTATTCACTCCAGAGCGATGAAAACGTTTCAGTTTGCTCATGGAAAACGGTGTAACAAGGGTGAACACTATCCCAT ATCACCAGCTCACCGTCTTTCATTGCCATACGAAATTCCGGATGAGCATTCATCAGGCGGGCAAGAATGTGAATAAAGGCCGGATA AAACTTGTGCTTATTTTTCTTTACGGTCTTTAAAAAGGCCGTAATATCCAGCTGAACGGTCTGGTTATAGGTACATTGAGCAACT GACTGAAATGCCTCAAAATGTTCTTTACGATGCCATTGGGATATATCAACGGTGGTATATCCAGTGATTTTTTTCTCCATTTTAGC TTCCTTAGCTCCTGAAAATCTCGATAACTCAAAAAATACGCCCGGTAGTGATCTTATTTCATTATGGTGAAAGTTGGAACCTCTT ACGTGCCGATCAACGTCTCATTTTCGCCAAAAGTTGGCCCAGGGCTTCCCGGTATCAACAGGGACACCAGGATTTATTTATTCTGC GAAGTGATCTTCCGTCACAGGTATTTATTCGCGATAAGCTCATGGAGCGGCGTAACCGTCGCACAGGAAGGACAGAGAAAGCGCGGA TCTGGGAAGTGACGGACAGAACGGTCAGGACCTGGATTGGGGAGGCGGTTGCCGCCGCTGCTGCTGACGGTGTGACGTTCTCTGTT CCGGTCACACCACATACGTTCCGCCATTCCTATGCGATGCACATGCTGTATGCCGGTATACCGCTGAAAGTTCTGCAAAGCCTGAT GGGACATAAGTCCATCAGTTCAACGGAAGTCTACACGAAGGTTTTTGCGCTGGATGTGGCTGCCCGGCACCGGGTGCAGTTTGCGA TGCCGGAGTCTGATGCGGTTGCGATGCTGAAACAATTATCCTGAGAATAAATGCCTTGGCCTTTATATGGAAATGTGGAACTGAGTG GATATGCTGTTTTTGTCTGTTAAACAGAGAAGCTGGCTGTTATCCACTGAGAAGCGAACGAAACAGTCGGGAAAATCTCCCATTAT CGTAGAGATCCGCATTATTAATCTCAGGAGCCTGTGTAGCGTTTATAGGAAGTAGTGTTCTGTCATGATGCCTGCAAGCGGTAACGA AAACGATTTGAATATGCCTTCAGGAACAATAGAAATCTTCGTGCGGTGTTACGTTGAAGTGGAGCGGATTATGTCAGCAATGGACA GAACAACCTAATGAACACAGAACCATGATGTGGTCTGTCCTTTTACAGCCAGTAGTGCTCGCCGCAGTCGAGCGACAGGGCGAAGCCC | Slattery et al., 2018 [61] |
| ShBle cassette | TCGAGCTGGTTGCCCTCGCCGCTGGGCTGGCGGCCGTCTATGGCCCTGCAAACGCGCCAGAAACGCCGTCGAAGCCGTGTGCGAGA CACCGCGGCCGGCCGCCGGCGTTGTGGATACCTCGCGGAAAACTTGGCCCTCACTGACAGATGAGGGGCGGACGTTGACACTTGAG GGGCCGACTCACCCGGCGCGGCGTTGACAGATGAGGGGCAGGCTCGATTTCGGCCGGCGACGTGGAGCTGGCCAGCCTCGCAAATC GGCGAAAACGCCTGATTTTACGCGAGTTTCCCACAGATGATGTGGACAAGCCTGGGGATAAGTGCCCTGCGGTATTGACACTTGAG GGGCGCGACTACTGACAGATGAGGGGCGCGATCCTTGACACTTGAGGGGCAGAGTGCTGACAGATGAGGGGCGCACCTATTGACAT TTGAGGGGCTGTCCACAGGCAGAAAATCCAGCATTTGCAAGGGTTTCCGCCCGTTTTTCGGCCACCGCTAACCTGTCTTTTAACCT GCTTTTAAACCAATATTTATAAACCTTGTTTTTAACCAGGGCTGCGCCCTGTGCGCGTGACCGCGCACGCCGAAGGGGGGTGCCCC CCCTTCTCGAACCCTCCCGGTCGAGTGAGCGAGGAAGCACCAGGGAACAGCACTTATATATTCTGCTTACACACGATGCCTGAAAA AACTTCCCTTGGGGTTATCCACTTATCCACGGGGATATTTTTATAATTATTTTTTTTATAGTTTTTAGATCTTCTTTTTTAGAGCG CCTTGTAGGCCTTTATCCATGCTGGTTCTAGAGAAGGTGTTGTGACAAATTGCCCTTTCAGTGTGACAAATCACCCTCAAATGACA GTCCTGTCTGTGACAAATTGCCCTTAACCCTGTGACAAATTGCCCTCAGAAGAAGCTGTTTTTTCACAAAGTTATCCCTGCTTATT GACTCTTTTTTATTTAGTGTGACAATCTAAAAACTTGTCACACTTCACATGGATCTGTCATGGCGGAAACAGCGGTTATCAATCAC AAGAAACGTAAAAATAGCCCGCGAATCGTCCAGTCAAACGACCTCACTGAGGCGGCATATAGTCTCTCCCGGGATCAAAAACGTAT GCTGTATCTGTTCGTTGACCAGATCAGAAAATCTGATGGCACCCTACAGGAACATGACGGTATCTGCGAGATCCATGTTGCTAAAT ATGCTGAAATATTCGGATTGACCTCTGCGGAAGCCAGTAAGGATATACGGCAGGCATTGAAGAGTTTCGCGGGGAAGGAAGTGGTT TTTTATCGCCCTGAAGAGGATGCCGGCGATGAAAAAGGCTATGAATCTTTTCCTTGGTTTATCAAACGTGCGCACAGTCCATCCAG AGGGCTTTACAGTGTACATATCAACCCATATCTCATTCCCTTCTTTATCGGGTTACAGAACCGGTTTACGCAGTTTCGGCTTAGTG AAACAAAAGAAATCACCAATCCGTATGCCATGCGTTTATACGAATCCCTGTGTCAGTATCGTAAGCCGGATGGCTCAGGCATCGTC TCTCTGAAAATCGACTGGATCATAGAGCGTTACCAGCTGCCTCAAAGTTACCAGCGTATGCCTGACTTCCGCCGCCGCTTCCTGCA GGTCTGTGTTAATGAGATCAACAGCAGAACTCCAATGCGCCTCTCATACATTGAGAAAAAGAAAGGCCGCCAGACGACTCATATCG TATTTTCCTTCCGCGATATCACTTCCATGACGACAGGATAGTCTGAGGGTTATCTGTCACAGATTTGAGGGTGGTTCGTCACATTT GTTCTGACCTACTGAGGGTAATTTGTCACAGTTTTGCTGTTTCCTTCAGCCTGCATGGATTTTCTCATACTTTTTGAACTGTAATT TTTAAGGAAGCCAAATTTGAGGGCAGTTTGTCACAGTTGATTTCCTTCTCTTTCCCTTCGTCATGTGACCTGATATCGGGGGTTAG TTCGTCATCATTGATGAGGGTTGATTATCACAGTTTATTACTCTGAATTGGCTATCCGCGTGTGTACCTCTACCTGGAGTTTTTCC CACGGTGGATATTTCTTCTTGCGCTGAGCGTAAGAGCTATCTGACAGAACAGTTCTTCTTTGCTTCCTCGCCAGTTCGCTCGCTAT GCTCGGTTACACGGCTGCGGCGAGCATCACGTGCTATAAAAATAATTATAATTTAAATTTTTTAATATAAATATATAAATTAAAAA TAGAAAGTAAAAAAAGAAATTAAAGAAAAAATAGTTTTTGTTTTCCGAAGATGTAAAAGACTCTAGGGGGATCGCCAACAAATACT ACCTTTTACCTTGCTCTTCCTGCTCTCAGGTATTAATGCCGAATTGTTTCATCTTGTCTGTGTAGAAGACCACACACGAAAATCCT GTGATTTTACATTTTACTTATCGTTAATCGAATGTATATCTATTTAATCTGCTTTTCTTGTCTAATAAATATATATGTAAAGTACGC TTTTTGTTGAAATTTTTTAAACCTTTGTTTATTTTTTTTTCTTCATTCCGTAACTCTTCTACCTTCTTTATTTACTTTCTAAAATC CAAATACAAAACATAAAAATAAATAAACACAGAGTAAATTCCCAAATTATTCCATCATTAAAAGATACGAGGCGCGTGTAAGTTAC AGGCAAGCGATCCTAGTACACTCTATATTTTTTTATGCCTCGGTAATGATTTTCATTTTTTTTTTCCACCTAGCGGATGACTCTTT TTTTTTCTTAGCGATTGGCATTATCACATAATGAATTATACATTATATAAAGTAATGTGATTTCTTCGAAGAATATACTAAAAAAT GAGCAGGCAAGATAAACGAAGGCAAAGATGACAGAGCAGAAAGCCCTAGTAAAGCGTATTACAAATGAAACCAAGATTCAGATTGC GATCTCTTTAAAGGGTGGTCCCCTAGCGATAGAGCACTCGATCTTCCCAGAAAAAGAGGCAGAAGCAGTAGCAGAACAGGCCACAC AATCGCAAGTGATTAACGTCCACACAGGTATAGGGTTTCTGGACCATATGATACATGCTCTGGCCAAGCATTCCGGCTGGTCGCTA ATCGTTGAGTGCATTGGTGACTTACACATAGACGACCATCACACCACTGAAGACTGCGGGATTGCTCTCGGTCAAGCTTTTAAAGA GGCCCTAGGGGCCGTGCGTGGAGTAAAAAGGTTTGGATCAGGATTTGCGCCTTTGGATGAGGCACTTTCCAGAGCGGTGGTAGATC TTTCGAACAGGCCGTACGCAGTTGTCGAACTTGGTTTGCAAAGGGAGAAAGTAGGAGATCTCTCTTGCGAGATGATCCCGCATTTT CTTGAAAGCTTTGCAGAGGCTAGCAGAATTACCCTCCACGTTGATTGTCTGCGAGGCAAGAATGATCATCACCGTAGTGAGAGTGC GTTCAAGGCTCTTGCGGTTGCCATAAGAGAAGCCACCTCGCCCAATGGTACCAACGATGTTCCCTCCACCAAAGGTGTTCTTATGT AGTTTTACACAGGAGTCTGGACTTGACGCTAGTGATAATAAGTGACTGAGGTATGTGCTCTTCTTATCTCCTTTTGTAGTGTTGCT CTTATTTTAAACAACTTTGCGGTTTTTTGATGACTTTGCGATTTTGTTGTTGCTTTGCAGTAAATTGCAAGATTTAATAAAAAAAC GCAAAGCAATGATTAAAGGATGTTCAGAATGAAACTCATGGAAACACTTAACCAGTGCATAAACGCTGGTCATGAAATGACGAAGG CTATCGCCATTGCACAGTTTAATGATGACAGCCCGGAAGCGAGGAAAATAACCCGGCGCTGGAGAATAGGTGAAGCAGCGGATTTA GTTGGGGTTTCTTCTCAGGCTATCAGAGATGCCGAGAAAGCAGGGCGACTACCGCACCCGGATATGGAAATTCGAGGACGGGTTGA GCAACGTGTTGGTTATACAATTGAACAAATTAATCATATGCGTGATGTGTTTGGTACGCGATTGCGACGTGCTGAAGACGTATTTC CACCGGTGATCGGGGTTGCTGCCCATAAAGGTGGCGTTTACAAAACCTCAGTTTCTGTTCATCTTGCTCAGGATCTGGCTCTGAAG GGGCTACGTGTTTTGCTCGTGGAAGGTAACGACCCCCAGGGAACAGCCTCAATGTATCACGGATGGGTACCAGATCTTCATATTCAT GCAGAAGACACTCTCCTGCCTTTCTATCTTGGGGAAAAGGACGATGTCACTTATGCAATAAAGCCCACTTGCTGGCCGGGGCTTGAC ATTATTCCTTCCTGTCTGGCTCTGCACCGTATTGAAACTGAGTTAATGGGCAAATTTGATGAAGGTAAACTGCCCACCGATCCACA CCTGATGCTCCGACTGGCCATTGAAACTGTTGCTCATGACTATGATGTCATAGTTATTGACAGCGCGCCTAACCTGGGTATCGGCAC GATTAATGTCGTATGTGCTGCTGATGTGCTGATTGTTCCCACGCCTGCTGAGTTGTTTGACTACACCTCCGCACTGCAGTTTTTCG ATATGCTTCGTGATCTGCTCAAGAACGTTGATCTTAAAGGGTTCGAGCCTGATGTACGTATTTTGCTTACCAAATACAGCAATAGC AATGGCTCTCAGTCCCCGTGGATGGAGGAGCAAATTCGGGATGCCTGGGGAAGCATGGTTCTAAAAAATGTTGTACGTGAAACGGA TGAAGTTGGTAAAGGTCAGATCCGGATGAGAACTGTTTTTGAACAGGCCATTGATCAACGCTCTTCAACTGGTGCCTGGAGAAATGC TCTTTCTATTTGGGAACCTGTCTGCAATGAAATTTTCGATCGTCTGATTAAACCACGCTGGGAGATTAGATAATGAAGCGTGCGCCT GTTATTCCAAAACATACGCTCAATACTCAACCGGTTGAAGATACTTCGTTATCGACACCAGCTGCCCCGATGGTGGATTCGTTAATT GCGCGCGTAGGAGTAATGGCTCGCGGTAATGCCATTACTTTGCCTGTATGTGGTCGGGATGTGAAGTTTACTCTTGAAGTGCTCCGG GGTGATAGTGTTGAGAAGACCTCTCGGGTATGGTCAGGTAATGAACGTGACCAGGAGCTGCTTACTGAGGACGCACTGGATGATCTC ATCCCTTCTTTTCTACTGACTGGTCAACAGACACCGGCGTTCGGTCGAAGAGTATCTGGTGTCATAGAAATTGCCGATGGGAGTCGC CGTCGTAAAGCTGCTGCACTTACCGAAAGTGATTATCGTGTTCTGGTTGGCGAGCTGGATGATGAGCAGATGGCTGCATTATCCAGA TTGGGTAACGATTATCGCCCAACAAGTGCTTATGAACGTGGTCAGCGTTATGCAAGCCGATTGCAGAATGAATTTGCTGGAAATATT TCTGCGCTGGCTGATGCGGAAAATATTTCACGTAAGATTATTACCCGCTGTATCAACACCGCCAAATTGCCTAAATCAGTTGTTGCTC TTTTTTCTCACCCCGGTGAACTATCTGCCCGGTCAGGTGATGCACTTCAAAAAGCCTTTACAGATAAAGAGGAATTACTTAAGCAGCA GGCATCTAACCTTCATGAGCAGAAAAAAGCTGGGGTGATATTTGAAGCTGAAGAAGTTATCACTCTTTTAACTTCTGTGCTTAAAACG TCATCTGCATCAAGAACTAGTTTAAGCTCACGACATCAGTTTGCTCCTGGAGCGACAGTATTGTATAAGGGCGATAAAATGGTGCTTA ACCTGGACAGGTCTCGTGTTCCAACTGAGTGTATAGAGAAAATTGAGGCCATTCTTAAGGAACTTGAAAAGCCAGCACCCTGATGCGAC CTCGTTTTAGTCTACGTTTATCTGTCTTTACTTAATGTCCTTTGTTACAGGCCAGAAAGCATAACTGGCCTGAATATTCTCTCTGGGC CCACTGTTCCACTTGTATCGTCGGTCTGATAATCAGACTGGGACCACGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGAC CACGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCACGGTCCCACTCGTATCGTCGGTCTGATAATCAGACTGGGACC ACGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCATGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCA CGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGAACCACGGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCAC GGTCCCACTCGTATCGTCGGTCTGATTATTAGTCTGGGACCACGATCCCACTCGTGTTGTCGGTCTGATTATCGGTCTGGGACCACG GTCCCACTTGTATTGTCGATCAGACTATCAGCGTGAGACTACGATTCCATCAATGCCTGTCAAGGGCAAGTATTGACATGTCGTCGT AACCTGTAGAACGGAGTAACCTCGGTGTGCGGTTGTATGCCTGCTGTGGATTGCTGCTGTGTCCTGCTTATCCACAACATTTTGCGC ACGGTTATGTGGACAAAATACCTGGTTACCCAGGCCGTGCCGGCACGTTAACCGGGCTGCATCCGATGCAAGTGTGTCGCTGTCGAC GAGCTCGCGAGCTCGGACATGAGGTTGCCCCGTATTCAGTGTCGCTGATTTGTATTGTCTGAAGTTGTTTTTACGTTAAGTTGATGC AGATCAATTAATACGATACCTGCGTCATAATTGATTATTTGACGTGGTTTGATGGCCTCCACGCACGTTGTGATATGTAGATGATAA TCATTATCACTTTACGGGTCCTTTCCGGTGATCCGACAGGTTACGGGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCC GGTTTAAGGCGTTTCCGTTCTTCTTCGTCATAACTTAATGTTTTTATTTAAAATACCCTCTGAAAAGAAAGGAAACGACAGGTGCTGA AAGCGAGCTTTTTGGCCTCTGTCGTTTCCTTTCTCTGTTTTTGTCCGTGGAATGAACAATGGAAGTCCGAGCTCATCGCTAATAAC TTCGTATAGCATACATTATACGAAGTTATATTCGATGCGGCCGCAAGGGGTTCGCGTCAGCGGGTGTTGGCGGGTGTCGGGGCTGGC TTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCACACAGATGCGTAAGGAGAAAATAC CGCATCAGGCGCCATTCGCCATTCAGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCG AAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATT GTAATACGACTCACTATAGGGCGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGAGTATT CTATAGTCTCACCTAAATAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAA CATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCG CTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGAACCCCTTGCGGCCGCCCGGGCCGTCGACCAA TTCTCATGTTTGACAGCTTATCATCGAATTTCTGCCATTCATCCGCTTATTATCACTTATTCAGGCGTAGCAACCAGGCGTTTAAG GGCACCAATAACTGCCTTAAAAAAATTACGCCCCGCCCTGCCACTCATCGCAGTACTGTTGTAATTCATTAAGCATTCTGCCGACA TGGAAGCCATCACAAACGGCATGATGAACCTGAATCGCCAGCGGCATCAGCACCTTGTCGCCTTGCGTATAATATTTGCCCATGGT GAAAACGGGGGCGAAGAAGTTGTCCATATTGGCCACGTTTAAATCAAAACTGGTGAAACTCACCCAGGGATTGGCTGAGACGAAAA ACATATTCTCAATAAACCCTTTAGGGAAATAGGCCAGGTTTTCACCGTAACACGCCACATCTTGCGAATATATGTGTAGAAACTGCC GGAAATCGTCGTGGTATTCACTCCAGAGCGATGAAAACGTTTCAGTTTGCTCATGGAAAACGGTGTAACAAGGGTGAACACTATCCC ATATCACCAGCTCACCGTCTTTCATTGCCATACGAAATTCCGGATGAGCATTCATCAGGCGGGCAAGAATGTGAATAAAGGCCGGA TAAAACTTGTGCTTATTTTTCTTTACGGTCTTTAAAAAGGCCGTAATATCCAGCTGAACGGTCTGGTTATAGGTACATTGAGCAACT GACTGAAATGCCTCAAAATGTTCTTTACGATGCCATTGGGATATATCAACGGTGGTATATCCAGTGATTTTTTTCTCCATTTTAGCT TCCTTAGCTCCTGAAAATCTCGATAACTCAAAAAATACGCCCGGTAGTGATCTTATTTCATTATGGTGAAAGTTGGAACCTCTTACG TGCCGATCAACGTCTCATTTTCGCCAAAAGTTGGCCCAGGGCTTCCCGGTATCAACAGGGACACCAGGATTTATTTATTCTGCGAAG TGATCTTCCGTCACAGGTATTTATTCGCGATAAGCTCATGGAGCGGCGTAACCGTCGCACAGGAAGGACAGAGAAAGCGCGGATCTG GGAAGTGACGGACAGAACGGTCAGGACCTGGATTGGGGAGGCGGTTGCCGCCGCTGCTGCTGACGGTGTGACGTTCTCTGTTCCGGT CACACCACATACGTTCCGCCATTCCTATGCGATGCACATGCTGTATGCCGGTATACCGCTGAAAGTTCTGCAAAGCCTGATGGGACA TAAGTCCATCAGTTCAACGGAAGTCTACACGAAGGTTTTTGCGCTGGATGTGGCTGCCCGGCACCGGGTGCAGTTTGCGATGCCGGA GTCTGATGCGGTTGCGATGCTGAAACAATTATCCTGAGAATAAATGCCTTGGCCTTTATATGGAAATGTGGAACTGAGTGGATATGC TGTTTTTGTCTGTTAAACAGAGAAGCTGGCTGTTATCCACTGAGAAGCGAACGAAACAGTCGGGAAAATCTCCCATTATCGTAGAGA TCCGCATTATTAATCTCAGGAGCCTGTGTAGCGTTTATAGGAAGTAGTGTTCTGTCATGATGCCTGCAAGCGGTAACGAAAACGATT TGAATATGCCTTCAGGAACAATAGAAATCTTCGTGCGGTGTTACGTTGAAGTGGAGCGGATTATGTCAGCAATGGACAGAACAACCT AATGAACACAGAACCATGATGTGGTCTGTCCTTTTACAGCCAGTAGTGCTCGCCGCAGTCGAGCGACAGGGCGAAGCCC | Karas et al., 2015 [38] |
| 40SRPS8 promoter | CCCTGCGATAGACCTTTTCCAAACTCACGCAGTCCAAGAAAACAAAGGGGTGAGAAGTATACGCACCTTTCGGTTTCGGCATAATTCT TAAACTCTTGTGGTCACTTTCTTGTGAAGAAGCTAGGGGCACTCGTTTTCCCTCAGAGCCTGCAAACACAAAATTCCTGCAGTCAAT TGTCCCAACACTCGGCAAACCGTATGCGCAAGCAACGATGCGCAGAAGGCCGTGGATGGATGGCGACTCGCGATATGGCTTCTTGGG TCGCCAGTGTGGTACGTCCGGCGTATGTCAATACGCGAATTCGGACGACTGGCATCTCTAGGAGGAGGATTCCTTCTTTTATGACAT GTTTATTTTATATACATTGATGCTTTCCGACAGTCGGAAGTAATAAATGAATTTATTTCAAGACTACCTATACTCCTTTGACTTGTT CGACTAATCTTACCGCTTACTAAAATCTCGAAATCACGCTTGACCTCTCGCACGCAAATTTTTGCTGCTGGACGCTACGCACTCGGC CCAATTCTTCTCGGTCCTCGTCGTCGCAATTGTCGTTGCGTTGATCTTGCACCGAAGGAATCAGAGAATAGAATACC | Slattery et al., 2018 [61] |
| FcpA terminator | CCGCAACAACTACCTCGACTTTGGCTGGGACACTTTCAGTGAGGACAAGAAGCTTCAGAAGCGTGCTATCGAACTCAACCAGGGACGTG CGGCACAAATGGGCATCCTTGCTCTCATGGTGCACGAACAGTTGGGAGTCTCTATCCTTCCTTAAAAATTTAATTTTCATTAGTTGCAGTCACTCCGCTTTGGTTT | Niu et al., 2013 [63] |
| OA1 | ATGAACCACCTCCGTGCCGAAGGACCCGCCTCCGTCCTCGCCATTGGAACCGCCAACCCCGAAAACATTCTCCTCCAGGACGAATTCCC GGACTACTACTTCCGCGTCACCAAGTCGGAACACATGACCCAGCTCAAGGAAAAGTTCCGTAAGATCTGTGACAAGTCGATGATTCG TAAGCGCAACTGCTTCCTCAACGAAGAACACTTGAAGCAGAACCCCCGTCTCGTCGAACACGAAATGCAGACCCTCGACGCCCGTCA GGACATGCTCGTCGTCGAAGTCCCCAAGCTCGGAAAGGATGCCTGCGCCAAGGCCATCAAGGAATGGGGTCAGCCCAAGTCCAAGAT CACCCACCTCATTTTCACCTCCGCCTCCACCACCGACATGCCCGGAGCTGACTACCACTGCGCCAAGTTGCTCGGCCTCTCCCCCTC CGTCAAGCGCGTCATGATGTACCAGCTTGGCTGCTACGGCGGAGGTACCGTCTTGCGTATTGCCAAGGACATCGCCGAAAACAACAA GGGAGCCCGTGTCTTGGCCGTCTGCTGTGACATTATGGCCTGCCTCTTCCGTGGCCCCTCCGAATCCGACCTCGAACTCTTGGTCGGT CAGGCCATCTTTGGCGATGGAGCCGCCGCCGTCATTGTCGGTGCCGAACCCGACGAATCCGTCGGAGAACGTCCCATTTTTGAACTT GTCTCCACCGGACAGACCATTCTCCCCAACTCCGAAGGAACCATTGGCGGCCACATTCGTGAAGCCGGACTCATCTTTGACCTCCAC AAGGACGTCCCGATGCTCATCTCCAACAACATCGAAAAGTGCCTCATTGAAGCCTTCACTCCGATCGGAATTTCCGACTGGAACTCC ATCTTCTGGATTACCCACCCCGGCGGAAAGGCCATTTTGGACAAGGTCGAAGAAAAGCTCCACCTCAAGTCCGATAAGTTTGTCGAC TCCCGTCACGTCCTCTCCGAACACGGTAACATGTCCTCCTCCACCGTCCTCTTTGTCATGGACGAACTCCGTAAGCGTTCCTTGGAA GAAGGCAAGTCGACCACCGGCGACGGCTTCGAATGGGGAGTCTTGTTTGGCTTCGGTCCCGGATTGACCGTCGAACGTGTCGTCGTC CGTTCCGTTCCCATCAAGTACGGATCCGGAGAAGGACGCGGTTCGTTGCTCACCTGCGGAGACGTCGAAGAAAACCCCGGACCCATGG CCGTCAAGCACCTCATTGTCTTGAAGTTCAAGGATGAAATCACCGAAGCTCAGAAGGAAGAATTCTTCAAGACCTACGTCAACCTCG TCAACATCATTCCCGCCATGAAGGACGTCTACTGGGGAAAGGACGTCACCCAGAAGAACAAGGAAGAAGGATACACCCACATTGTCG AAGTCACCTTTGAATCCGTCGAAACCATCCAGGACTACATCATCCACCCCGCCCACGTCGGATTCGGTGACGTCTACCGCTCCTTCT GGGAAAAGCTCTTGATTTTCGACTACACCCCCCGTAAATAA | This study |
| OA2 | ATGAACCACCTCCGTGCCGAAGGTCCCGCCTCCGTCCTCGCCATTGGAACCGCCAACCCCGAAAACATTCTCCTCCAAGACGAATTCCC TGACTACTACTTCCGCGTGACTAAGAGTGAGCACATGACGCAATTGAAAGAAAAGTTCCGTAAGATCTGTGATAAGTCCATGATCCG TAAGCGTAACTGTTTCCTGAATGAGGAACACCTTAAGCAGAACCCCCGCTTGGTCGAACATGAAATGCAGACCTTGGATGCCCGCCA GGACATGCTCGTCGTTGAAGTCCCTAAGTTGGGTAAAGACGCTTGCGCCAAGGCCATTAAGGAATGGGGACAGCCCAAGAGTAAGAT CACCCATTTGATTTTCACATCCGCCTCTACGACCGATATGCCTGGAGCCGATTACCACTGCGCCAAGCTTCTGGGACTGTCCCCCTC CGTTAAGCGCGTCATGATGTACCAGCTTGGATGTTACGGAGGTGGCACCGTCCTCCGTATTGCCAAGGACATTGCCGAAAACAACAA GGGTGCCCGTGTCCTCGCCGTCTGCTGCGACATCATGGCCTGCCTCTTCCGCGGACCCTCCGAATCCGACTTGGAACTCCTCGTGGG ACAGGCCATTTTTGGTGACGGCGCCGCCGCCGTCATTGTCGGAGCTGAACCCGACGAATCCGTCGGAGAAAGGCCCATTTTTGAATT GGTCTCCACCGGACAGACCATTCTCCCGAACTCCGAAGGAACCATCGGCGGACACATTCGTGAAGCCGGTCTCATCTTTGACTTGCA CAAGGACGTTCCCATGCTCATTTCCAACAACATTGAAAAGTGCCTCATTGAAGCCTTCACCCCCATTGGAATCTCCGACTGGAATAG TATCTTTTGGATTACCCACCCCGGAGGAAAGGCCATCTTGGATAAGGTGGAAGAAAAGCTCCACCTCAAGTCCGACAAGTTCGTCGA CTCCCGTCACGTCTTGTCCGAACACGGAAACATGTCGTCCTCCACCGTCCTCTTTGTCATGGATGAATTGCGTAAGCGCTCGCTCGA AGAAGGAAAGTCCACCACCGGCGATGGTTTCGAATGGGGCGTTCTCTTCGGATTTGGACCCGGTCTCACCGTCGAACGTGTCGTCGTC CGTTCTGTCCCGATTAAGTACCACCATCACCACCACCACGGATCGGGCGAAGGACGTGGTTCCCTCCTTACGTGCGGAGACGTCGAA GAAAACCCCGGTCCCATGGCCGTCAAGCACTTGATCGTTCTCAAGTTCAAGGACGAAATCACCGAAGCCCAGAAGGAAGAATTCTTC AAGACCTACGTCAACCTCGTCAACATCATTCCCGCCATGAAGGATGTTTACTGGGGAAAGGACGTCACCCAGAAGAACAAGGAAGAA GGTTACACCCACATCGTCGAAGTCACCTTTGAATCCGTTGAAACCATCCAGGACTACATTATCCACCCGGCCCACGTCGGCTTCGGA GACGTCTACCGTTCCTTTTGGGAAAAGTTGTTGATTTTCGACTACACCCCCCGTAAGGAACAGAAGTTGATTTCCGAAGAAGACCTCTAA | This study |
| OA3 | ATGCGTAAGGGTGAAGAACTCTTCACCGGAGTCGTCCCCATCTTGGTGGAATTGGACGGAGACGTCAACGGCCATAAGTTCTCGGTT TCCGGTGAAGGTGAAGGTGACGCCACTAACGGAAAGCTCACCCTCAAGTTCATTTGCACCACCGGCAAGCTCCCGGTCCCCTGGCCC ACCCTCGTCACCACCTTGACTTACGGTGTCCAGTGCTTCGCCCGTTACCCCGACCACATGAAGCAGCACGACTTCTTTAAGTCCGCC ATGCCCGAAGGATACGTCCAGGAACGCACCATTTCCTTCAAGGACGACGGAACCTACAAGACCCGTGCCGAAGTCAAGTTTGAAGGAG ATACCCTCGTCAACCGCATTGAACTCAAGGGCATTGATTTCAAAGAAGATGGCAACATTCTCGGACACAAGCTCGAATACAACTTCAA CTCCCACAACGTCTACATCACCGCCGATAAGCAAAAGAACGGTATTAAGGCCAACTTCAAGATTCGCCACAACGTCGAAGACGGTTC CGTCCAGTTGGCCGACCACTACCAGCAGAACACCCCGATTGGTGACGGTCCGGTCCTCCTCCCCGACAACCACTACCTCTCGTACCAG TCCGCCCTCTCCAAGGATCCGAACGAGAAGCGTGACCACATGGTCCTCTTGGAATTCGTCACCGCCGCCGGAATCACCCACGGCATGG ACGAACTCTACAAGCGTCCCGCCGCCAACGACGAGAACTACGCCGCCTCCGTCGGTATGAACCACTTGCGCGCCGAAGGACCGGCTTC CGTCTTGGCCATTGGTACCGCCAACCCGGAAAACATCCTCCTCCAGGACGAATTCCCCGACTACTACTTCCGTGTCACCAAGTCCGAA CACATGACCCAGTTGAAGGAAAAATTCCGTAAGATTTGTGATAAGTCCATGATTCGTAAGCGTAACTGCTTCTTGAACGAAGAACACC TCAAGCAAAACCCCCGTCTCGTCGAACACGAAATGCAAACCCTCGACGCCCGTCAGGACATGCTCGTTGTCGAAGTCCCGAAGCTTGG CAAGGACGCCTGCGCCAAGGCCATTAAGGAATGGGGCCAGCCCAAGAGCAAGATTACCCACTTGATTTTCACCTCCGCCTCCACCACC GATATGCCCGGCGCTGACTACCACTGCGCTAAGCTCCTCGGTCTCTCCCCCTCCGTCAAGCGTGTCATGATGTACCAGCTCGGATGCT ACGGAGGTGGAACCGTCCTCCGCATTGCCAAGGACATTGCTGAAAACAACAAGGGCGCCCGTGTCCTCGCCGTCTGCTGTGACATTAT GGCCTGCTTGTTTCGTGGTCCCTCCGAATCCGATTTGGAACTCCTCGTCGGCCAGGCCATTTTTGGTGACGGTGCCGCCGCCGTCATC GTCGGCGCCGAACCGGACGAATCCGTCGGTGAACGTCCGATTTTTGAACTCGTCTCCACCGGTCAAACCATTCTCCCCAACTCCGAAG GAACCATCGGAGGACACATCCGTGAAGCCGGACTCATTTTTGACCTCCACAAGGATGTCCCCATGCTCATCTCCAACAACATCGAAAA GTGCCTCATCGAAGCCTTCACCCCCATTGGCATCTCCGACTGGAACTCCATTTTCTGGATTACCCACCCCGGTGGAAAGGCCATTCTC GACAAGGTCGAAGAAAAGCTCCACTTGAAGTCCGATAAGTTCGTCGACTCCCGCCACGTCCTCTCCGAACACGGAAACATGTCCTCCT CCACCGTCCTCTTCGTCATGGACGAACTCCGTAAGCGTTCCCTCGAAGAAGGAAAGTCCACGACGGGTGACGGATTCGAGTGGGGAGT CCTCTTCGGCTTCGGTCCCGGACTCACCGTCGAACGTGTCGTCGTCCGTTCCGTGCCCATCAAGTACGGAGGAGGAGGATCCGGTGGT GGCGGATCCGGAGGTGGTGGATCGATGGCCGTCAAGCACCTCATTGTCTTGAAGTTTAAGGACGAAATCACCGAAGCCCAGAAGGAAG AATTCTTCAAGACTTACGTCAACTTGGTCAACATCATCCCCGCCATGAAGGACGTCTACTGGGGAAAGGACGTCACCCAAAAGAACAA GGAAGAAGGATACACCCACATTGTCGAAGTCACCTTTGAATCCGTCGAAACCATTCAGGACTACATTATTCACCCCGCCCACGTCGGTT TCGGTGACGTCTACCGTTCCTTCTGGGAAAAGCTCTTGATTTTTGACTACACCCCCCGTAAGGAACAGAAGTTGATTTCCGAAGAAGACTTGTAA | This study |
| Primer Name | Sequence (5′ to 3′) | Amplicon Size (bp) | Description of Product |
|---|---|---|---|
| bk1-FP | CAGGGTAATATAGATCTTCCGCTGCATAACCCTGCTTCGGGGTCATTATAGCGATTTTTT | 6158 | pPTGE30 backbone part 1 |
| bk1-RP | TTTGCAAACCAAGTTCGACAACTGCGTACGGCCTGTTCGAAAGATCTACCACCGCTCTGG | ||
| bk1-FP | GGGCCGTGCGTGGAGTAAAAAGGTTTGGATCAGGATTTGCGCCTTTGGATGAGGCACTTT | 6318 | pPTGE30 backbone part 2 |
| bk2-RP | ATGGGCTTCGCCCTGTCGCTCGACTGCGGCGAGCACTACTGGCTGTAAAAGGACAGACCA | ||
| ShBle-FP | TGGTCTGTCCTTTTACAGCCAGTAGTGCTCGCCGCAGTCGAGCGACAGGGCGAAGCCCATGAGCACAAGAGGTGACAAAA | 1466 | P. t selection marker region |
| ShBle-RP | ATACTTCTCACCCCTTTGTTTTCTTGGACTGCGTGAGTTTGGAAAAGGTCTATCGCAGGGATTGCAGCTTGTTGCAGAAG | ||
| 40SRPS8-FP | TTGCATGGTTAGACCTCCTTGACGACTGTGAGCCTACATCCTTCTGCAACAAGCTGCAATCCCTGCGATAGACCTTTTCC | 700 | 40SRPS8 Promoter region |
| 40SRPS8-RP | CGAGGACGGAGGCGGGTCCTTCGGCACGGAGGTGGTTCATGGTATTCTATTCTCTGATTC | ||
| FcpA terminator-FP | GGAAAAGCTCTTGATTTTCGACTACACCCCCCGTAAATAACCGCAACAACTACCTCGACT | 275 | FcpA Terminator region |
| FcpA terminator-RP | CCGAAGCAGGGTTATGCAGCGGAAGATCTATATTACCCTGAAACCAAAGCGGAGTGACTG | ||
| OA1-FP | GCGTTGATCTTGCACCGAAGGAATCAGAGAATAGAATACCATGAACCACCTCCGTGCCGA | 1524 | OA1 pathway genes |
| OA1-RP | ACTGAAAGTGTCCCAGCCAAAGTCGAGGTAGTTGTTGCGGTTATTTACGGGGGGTGTAGT | ||
| OA2-FP | GCGTTGATCTTGCACCGAAGGAATCAGAGAATAGAATACCATGAACCACCTCCGTGCCGA | 1572 | OA2 pathway genes |
| OA2-RP | AGTCGAGGTAGTTGTTGCGGTTAGAGGTCTTCTTCGGAAATCAACTTCTGTTCCTTACGG | ||
| OA3-FP | GCGTTGATCTTGCACCGAAGGAATCAGAGAATAGAATACCATGCGTAAGGGTGAAGAACT | 2352 | OA3 pathway genes |
| OA3-RP | AGTCGAGGTAGTTGTTGCGGTTACAAGTCTTCTTCGGAAATCAACTTCTGTTCCTTACGG | ||
| Q-TKS1-Gn2-F | CGA CTG GAA CTC CAT CTT CTG | 527 | Pt OA1 colony PCR |
| Q-OAC1-Gn2-R | GGGTGTATCCTTCTTCCTTGTT | ||
| Q-TKS3-Gn2-F | ATTGGAATCTCCGACTGGAATAG | 594 | Pt OA2 colony PCR |
| Q-OAC3-Gn2-R | GATGGTTTCAACGGATTCAAAGG | ||
| Q-TKS7-Gn2-F | GTCAGGACATGCTCGTTGT | 1211 | Pt OA3 colony PCR |
| Q-OAC7-Gn2-R | TTCCCAGAAGGAACGGTAGA |
| Sequence ID | Identity | Mismatch Position (ON-SN) | Effect on Protein | Number of Insertion |
|---|---|---|---|---|
| PtOA1 | >99% | 9305 (T-C) | No | 60 bp insertion (after terminator, no effect) |
| PtOA2 | >99% | 8526 (C-T) 9304 (T-C) | No | 0 |
| PtOA3 | >99% | 8192 (G-A) 9304 (T-C) | No | 0 |

Appendix B. The In Silico Analyses Identified Putative Endogenous P. tricornutum Candidates for CB-like Biosynthesis

References
- Aizpurua-Olaizola, O.; Soydaner, U.; Öztürk, E.; Schibano, D.; Simsir, Y.; Navarro, P.; Etxebarria, N.; Usobiaga, A. Evolution of the Cannabinoid and Terpene Content during the Growth of Cannabis sativa Plants from Different Chemotypes. J. Nat. Prod. 2016, 79, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, F.; Pagano, E.; Romano, B.; Panzera, S.; Maiello, F.; Coppola, D.; De Petrocellis, L.; Buono, L.; Orlando, P.; Izzo, A.A. Colon Carcinogenesis Is Inhibited by the TRPM8 Antagonist Cannabigerol, a Cannabis-Derived Non-Psychotropic Cannabinoid. Carcinogenesis 2014, 35, 2787–2797. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, B.; Ravi, J.; Ganju, R.K. Cannabinoids as Therapeutic Agents in Cancer: Current Status and Future Implications. Oncotarget 2014, 5, 5852–5872. [Google Scholar] [CrossRef] [PubMed]
- Eibach, L.; Scheffel, S.; Cardebring, M.; Lettau, M.; Özgür Celik, M.; Morguet, A.; Roehle, R.; Stein, C. Cannabidivarin for HIV-Associated Neuropathic Pain: A Randomized, Blinded, Controlled Clinical Trial. Clin. Pharmacol. Ther. 2021, 109, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Brunt, T.M.; Bossong, M.G. The Neuropharmacology of Cannabinoid Receptor Ligands in Central Signaling Pathways. Eur. J. Neurosci. 2022, 55, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Schachtsiek, J.; Warzecha, H.; Kayser, O.; Stehle, F. Current Perspectives on Biotechnological Cannabinoid Production in Plants. Planta Medica 2018, 84, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.; Weinstein, A. The Effects of Cannabinoids on Executive Functions: Evidence from Cannabis and Synthetic Cannabinoids—A Systematic Review. Brain Sci. 2018, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Vadivelu, N.; Kai, A.M.; Kodumudi, G.; Sramcik, J.; Kaye, A.D. Medical Marijuana: Current Concepts, Pharmacological Actions of Cannabinoid Receptor Mediated Activation, and Societal Implications. Curr. Pain Headache Rep. 2018, 22, 3. [Google Scholar] [CrossRef]
- Gagne, S.J.; Stout, J.M.; Liu, E.; Boubakir, Z.; Clark, S.M.; Page, J.E. Identification of Olivetolic Acid Cyclase from Cannabis sativa Reveals a Unique Catalytic Route to Plant Polyketides. Proc. Natl. Acad. Sci. USA 2012, 109, 12811–12816. [Google Scholar] [CrossRef]
- Valliere, M.A.; Korman, T.P.; Woodall, N.B.; Khitrov, G.A.; Taylor, R.E.; Baker, D.; Bowie, J.U. A Cell-Free Platform for the Prenylation of Natural Products and Application to Cannabinoid Production. Nat. Commun. 2019, 10, 565. [Google Scholar] [CrossRef]
- Garfinkel, A.R.; Otten, M.; Crawford, S. SNP in Potentially Defunct Tetrahydrocannabinolic Acid Synthase Is a Marker for Cannabigerolic Acid Dominance in Cannabis sativa L. Genes 2021, 12, 228. [Google Scholar] [CrossRef] [PubMed]
- Sirikantaramas, S.; Taura, F.; Tanaka, Y.; Ishikawa, Y.; Morimoto, S.; Shoyama, Y. Tetrahydrocannabinolic Acid Synthase, the Enzyme Controlling Marijuana Psychoactivity, Is Secreted into the Storage Cavity of the Glandular Trichomes. Plant Cell Physiol. 2005, 46, 1578–1582. [Google Scholar] [CrossRef]
- Morita: How Structural Subtleties Lead to Molecular...—Google Scholar. Available online: https://scholar.google.com/scholar_lookup?title=How%20structural%20subtleties%20lead%20to%20molecular%20diversity%20for%20the%20type%20III%20polyketide%20synthases&journal=J%20Biol%20Chem&doi=10.1074%2Fjbc.REV119.006129&volume=294&pages=15121-15136&publication_year=2019&author=Morita%2CH&author=Wong%2CCP&author=Abe%2CI (accessed on 8 August 2023).
- Abe, I. Biosynthesis of Medicinally Important Plant Metabolites by Unusual Type III Polyketide Synthases. J. Nat. Med. 2020, 74, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Pandith, S.A.; Ramazan, S.; Khan, M.I.; Reshi, Z.A.; Shah, M.A. Chalcone Synthases (CHSs): The Symbolic Type III Polyketide Synthases. Planta 2019, 251, 15. [Google Scholar] [CrossRef] [PubMed]
- Nivina, A.; Yuet, K.P.; Hsu, J.; Khosla, C. Evolution and Diversity of Assembly-Line Polyketide Synthases. Chem. Rev. 2019, 119, 12524–12547. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Liu, J. Resveratrol: A Review of Plant Sources, Synthesis, Stability, Modification and Food Application. J. Sci. Food Agric. 2020, 100, 1392–1404. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, F.; Stehle, F.; Kayser, O. The Biosynthesis of Cannabinoids. In Handbook of Cannabis and Related Pathologies; Elsevier: London, UK, 2017; pp. 13–23. [Google Scholar]
- Gaoni, Y.; Mechoulam, R. Isolation, Structure, and Partial Synthesis of an Active Constituent of Hashish. J. Am. Chem. Soc. 1964, 86, 1646–1647. [Google Scholar] [CrossRef]
- Welling, M.T.; Deseo, M.A.; Bacic, A.; Doblin, M.S. Biosynthetic Origins of Unusual Cannabimimetic Phytocannabinoids in Cannabis sativa L: A Review. Phytochemistry 2022, 201, 113282. [Google Scholar] [CrossRef]
- Taura, F.; Tanaya, R.; Sirikantaramas, S. Recent Advances in Cannabinoid Biochemistry and Biotechnology. ScienceAsia 2019, 45, 399. [Google Scholar] [CrossRef]
- Livingston, S.J.; Quilichini, T.D.; Booth, J.K.; Wong, D.C.; Rensing, K.H.; Laflamme-Yonkman, J.; Castellarin, S.D.; Bohlmann, J.; Page, J.E.; Samuels, A.L. Cannabis Glandular Trichomes Alter Morphology and Metabolite Content during Flower Maturation. Plant J. 2020, 101, 37–56. [Google Scholar] [CrossRef]
- Günnewich, N.; Page, J.E.; Köllner, T.G.; Degenhardt, J.; Kutchan, T.M. Functional Expression and Characterization of Trichome-Specific (-)-Limonene Synthase and (+)-α-Pinene Synthase from Cannabis Sativa. Nat. Prod. Commun. 2007, 2, 223–232. [Google Scholar]
- Grof, C.P. Cannabis, from Plant to Pill. Br. J. Clin. Pharmacol. 2018, 84, 2463–2467. [Google Scholar] [CrossRef]
- Fellermeier, M.; Zenk, M.H. Prenylation of Olivetolate by a Hemp Transferase Yields Cannabigerolic Acid, the Precursor of Tetrahydrocannabinol. FEBS Lett. 1998, 427, 283–285. [Google Scholar] [CrossRef]
- Frontiers|Cannabis sativa: The Plant of the Thousand and One Molecules|Plant Science. Available online: https://www.frontiersin.org/articles/10.3389/fpls.2016.00019/full (accessed on 28 January 2022).
- Balthazar, C.; Novinscak, A.; Cantin, G.; Joly, D.L.; Filion, M. Biocontrol Activity of Bacillus spp. and Pseudomonas spp. against Botrytis Cinerea and Other Cannabis Fungal Pathogens. Phytopathology 2022, 112, 549–560. [Google Scholar] [CrossRef]
- Poulos, J.L.; Farnia, A.N. Production of Cannabidiolic Acid in Yeast 2018. U.S. Patent US20180073043A1, 15 March 2018. [Google Scholar]
- Poulos, J.L.; Farnia, A.N. Production of Tetrahydrocannabinolic Acid in Yeast 2019. U.S. Patent US10392635B2, 27 August 2019. [Google Scholar]
- Thomas, F.; Schmidt, C.; Kayser, O. Bioengineering Studies and Pathway Modeling of the Heterologous Biosynthesis of Tetrahydrocannabinolic Acid in Yeast. Appl. Microbiol. Biotechnol. 2020, 104, 9551–9563. [Google Scholar] [CrossRef]
- Luo, X.; Reiter, M.A.; d’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W. Complete Biosynthesis of Cannabinoids and Their Unnatural Analogues in Yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef]
- Kufs, J.E.; Reimer, C.; Steyer, E.; Valiante, V.; Hillmann, F.; Regestein, L. Scale-up of an Amoeba-Based Process for the Production of the Cannabinoid Precursor Olivetolic Acid. Microb. Cell Fact. 2022, 21, 217. [Google Scholar] [CrossRef]
- Tan, Z.; Clomburg, J.M.; Gonzalez, R. Synthetic Pathway for the Production of Olivetolic Acid in Escherichia coli. ACS Synth. Biol. 2018, 7, 1886–1896. [Google Scholar] [CrossRef]
- Zirpel, B.; Degenhardt, F.; Martin, C.; Kayser, O.; Stehle, F. Engineering Yeasts as Platform Organisms for Cannabinoid Biosynthesis. J. Biotechnol. 2017, 259, 204–212. [Google Scholar] [CrossRef]
- Dhaouadi, F.; Awwad, F.; Diamond, A.; Desgagné-Penix, I. Diatoms’ Breakthroughs in Biotechnology: Phaeodactylum tricornutum as a Model for Producing High-Added Value Molecules. Am. J. Plant Sci. 2020, 11, 1632–1670. [Google Scholar] [CrossRef]
- Fabris, M.; George, J.; Kuzhiumparambil, U.; Lawson, C.A.; Jaramillo-Madrid, A.C.; Abbriano, R.M.; Vickers, C.E.; Ralph, P. Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum tricornutum Enables the Heterologous Production of Monoterpenoids. ACS Synth. Biol. 2020, 9, 598–612. [Google Scholar] [CrossRef]
- Kassaw, T.K.; Paton, A.J.; Peers, G. Episome-Based Gene Expression Modulation Platform in the Model Diatom Phaeodactylum tricornutum. ACS Synth. Biol. 2022, 11, 191–204. [Google Scholar] [CrossRef]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J. Designer Diatom Episomes Delivered by Bacterial Conjugation. Nat. Commun. 2015, 6, 6925. [Google Scholar] [CrossRef]
- Karas, B.J.; Molparia, B.; Jablanovic, J.; Hermann, W.J.; Lin, Y.-C.; Dupont, C.L.; Tagwerker, C.; Yonemoto, I.T.; Noskov, V.N.; Chuang, R.-Y. Assembly of Eukaryotic Algal Chromosomes in Yeast. J. Biol. Eng. 2013, 7, 30. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q. The BioCyc Collection of Microbial Genomes and Metabolic Pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Athey, J.; Alexaki, A.; Osipova, E.; Rostovtsev, A.; Santana-Quintero, L.V.; Katneni, U.; Simonyan, V.; Kimchi-Sarfaty, C. A New and Updated Resource for Codon Usage Tables. BMC Bioinform. 2017, 18, 391. [Google Scholar] [CrossRef]
- Diamond, A.; Diaz-Garza, A.M.; Li, J.; Slattery, S.S.; Merindol, N.; Fantino, E.; Meddeb-Mouelhi, F.; Karas, B.J.; Barnabé, S.; Desgagné-Penix, I. Instability of Extrachromosomal DNA Transformed into the Diatom Phaeodactylum tricornutum. Algal Res. 2023, 70, 102998. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The Lipid Biochemistry of Eukaryotic Algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef]
- Liu, B.; Benning, C. Lipid Metabolism in Microalgae Distinguishes Itself. Curr. Opin. Biotechnol. 2013, 24, 300–309. [Google Scholar] [CrossRef]
- Chng, J.; Wang, T.; Nian, R.; Lau, A.; Hoi, K.M.; Ho, S.C.; Gagnon, P.; Bi, X.; Yang, Y. Cleavage Efficient 2A Peptides for High Level Monoclonal Antibody Expression in CHO Cells. MAbs 2015, 7, 403–412. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, O.; Wall, J.B.J.; Zheng, M.; Zhou, Y.; Wang, L.; Ruth Vaseghi, H.; Qian, L.; Liu, J. Systematic Comparison of 2A Peptides for Cloning Multi-Genes in a Polycistronic Vector. Sci. Rep. 2017, 7, 2193. [Google Scholar] [CrossRef]
- Ma, J.; Gu, Y.; Xu, P. Biosynthesis of Cannabinoid Precursor Olivetolic Acid in Genetically Engineered Yarrowia Lipolytica. Commun. Biol. 2022, 5, 1239. [Google Scholar] [CrossRef]
- George, J.; Kahlke, T.; Abbriano, R.M.; Kuzhiumparambil, U.; Ralph, P.J.; Fabris, M. Metabolic Engineering Strategies in Diatoms Reveal Unique Phenotypes and Genetic Configurations with Implications for Algal Genetics and Synthetic Biology. Front. Bioeng. Biotechnol. 2020, 8, 513. [Google Scholar] [CrossRef]
- Munakata, R.; Kitajima, S.; Nuttens, A.; Tatsumi, K.; Takemura, T.; Ichino, T.; Galati, G.; Vautrin, S.; Bergès, H.; Grosjean, J.; et al. Convergent Evolution of the UbiA Prenyltransferase Family Underlies the Independent Acquisition of Furanocoumarins in Plants. New Phytol. 2020, 225, 2166–2182. [Google Scholar] [CrossRef]
- Wiles, D.; Shanbhag, B.K.; O’Brien, M.; Doblin, M.S.; Bacic, A.; Beddoe, T. Heterologous Production of Cannabis sativa-Derived Specialised Metabolites of Medicinal Significance—Insights into Engineering Strategies. Phytochemistry 2022, 203, 113380. [Google Scholar] [CrossRef]
- Chuberre, C.; Chan, P.; Walet-Balieu, M.-L.; Thiébert, F.; Burel, C.; Hardouin, J.; Gügi, B.; Bardor, M. Comparative Proteomic Analysis of the Diatom Phaeodactylum tricornutum Reveals New Insights Into Intra- and Extra-Cellular Protein Contents of Its Oval, Fusiform, and Triradiate Morphotypes. Front. Plant Sci. 2022, 13, 673113. [Google Scholar] [CrossRef]
- Hussain, M.H.; Hong, Q.; Zaman, W.Q.; Mohsin, A.; Wei, Y.; Zhang, N.; Fang, H.; Wang, Z.; Hang, H.; Zhuang, Y.; et al. Rationally Optimized Generation of Integrated Escherichia coli with Stable and High Yield Lycopene Biosynthesis from Heterologous Mevalonate (MVA) and Lycopene Expression Pathways. Synth. Syst. Biotechnol. 2021, 6, 85–94. [Google Scholar] [CrossRef]
- Taylor, G.M.; Hitchcock, A.; Heap, J.T. Combinatorial Assembly Platform Enabling Engineering of Genetically Stable Metabolic Pathways in Cyanobacteria. Nucleic Acids Res. 2021, 49, e123. [Google Scholar] [CrossRef]
- Hensing, M.C.M.; Rouwenhorst, R.J.; Heijnen, J.J.; van Dijken, J.P.; Pronk, J.T. Physiological and Technological Aspects of Large-Scale Heterologous-Protein Production with Yeasts. Antonie Leeuwenhoek 1995, 67, 261–279. [Google Scholar] [CrossRef]
- Okorafor, I.C.; Chen, M.; Tang, Y. High-Titer Production of Olivetolic Acid and Analogs in Engineered Fungal Host Using a Nonplant Biosynthetic Pathway. ACS Synth. Biol. 2021, 10, 2159–2166. [Google Scholar] [CrossRef]
- Trantas, E.; Panopoulos, N.; Ververidis, F. Metabolic Engineering of the Complete Pathway Leading to Heterologous Biosynthesis of Various Flavonoids and Stilbenoids in Saccharomyces Cerevisiae. Metab. Eng. 2009, 11, 355–366. [Google Scholar] [CrossRef]
- Favero, G.R.; de Melo Pereira, G.V.; de Carvalho, J.C.; de Carvalho Neto, D.P.; Soccol, C.R. Converting Sugars into Cannabinoids—The State-of-the-Art of Heterologous Production in Microorganisms. Fermentation 2022, 8, 84. [Google Scholar] [CrossRef]
- Barbosa, M.J.; Janssen, M.; Südfeld, C.; D’Adamo, S.; Wijffels, R.H. Hypes, Hopes, and the Way Forward for Microalgal Biotechnology. Trends Biotechnol. 2023, 41, 452–471. [Google Scholar] [CrossRef]
- Sharma, N.; Fantino, E.I.; Awwad, F.; Mérindol, N.; Augustine, A.; Meddeb, F.; Desgagné-Penix, I. Impact of Different Light Characteristics on the Growth and Lipid Content of Diatom Phaeodactylum tricornutum Transconjugant Strains. Am. J. Plant Sci. 2023, 14, 41–63. [Google Scholar] [CrossRef]
- Kouprina, N.; Larionov, V. Recent Advances in Chromosome Engineering. Chromosome Res. 2015, 23, 1–5. [Google Scholar] [CrossRef][Green Version]
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagné-Penix, I.; Karas, B.J. An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Niu, Y.F.; Zhang, M.H.; Li, D.W.; Yang, W.D.; Liu, J.S.; Bai, W.B.; Li, H.Y. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar Drugs 2013, 11, 4558–4569. [Google Scholar] [CrossRef]
- Shoichet, B.; Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar]
- Kuzuyama, T.; Noel, J.P.; Richard, S.B. Structural Basis for the Promiscuous Biosynthetic Prenylation of Aromatic Natural Products. Nature 2005, 435, 983–987. [Google Scholar] [CrossRef]
| Supplemented mM | Detected | |||||
|---|---|---|---|---|---|---|
| OA | GPP | CBGA | HPLC | MS (m/z) Confirmation | HPLC | MS (m/z) Confirmation |
| 0.45 | 1 | 0 | CBGA | yes | CBDA, CBNA | No |
| 0 | 0 | 0.27 | CBDA | yes | THCA, CBNA | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awwad, F.; Fantino, E.I.; Héneault, M.; Diaz-Garza, A.M.; Merindol, N.; Custeau, A.; Gélinas, S.-E.; Meddeb-Mouelhi, F.; Li, J.; Lemay, J.-F.; et al. Bioengineering of the Marine Diatom Phaeodactylum tricornutum with Cannabis Genes Enables the Production of the Cannabinoid Precursor, Olivetolic Acid. Int. J. Mol. Sci. 2023, 24, 16624. https://doi.org/10.3390/ijms242316624
Awwad F, Fantino EI, Héneault M, Diaz-Garza AM, Merindol N, Custeau A, Gélinas S-E, Meddeb-Mouelhi F, Li J, Lemay J-F, et al. Bioengineering of the Marine Diatom Phaeodactylum tricornutum with Cannabis Genes Enables the Production of the Cannabinoid Precursor, Olivetolic Acid. International Journal of Molecular Sciences. 2023; 24(23):16624. https://doi.org/10.3390/ijms242316624
Chicago/Turabian StyleAwwad, Fatima, Elisa Ines Fantino, Marianne Héneault, Aracely Maribel Diaz-Garza, Natacha Merindol, Alexandre Custeau, Sarah-Eve Gélinas, Fatma Meddeb-Mouelhi, Jessica Li, Jean-François Lemay, and et al. 2023. "Bioengineering of the Marine Diatom Phaeodactylum tricornutum with Cannabis Genes Enables the Production of the Cannabinoid Precursor, Olivetolic Acid" International Journal of Molecular Sciences 24, no. 23: 16624. https://doi.org/10.3390/ijms242316624
APA StyleAwwad, F., Fantino, E. I., Héneault, M., Diaz-Garza, A. M., Merindol, N., Custeau, A., Gélinas, S.-E., Meddeb-Mouelhi, F., Li, J., Lemay, J.-F., Karas, B. J., & Desgagne-Penix, I. (2023). Bioengineering of the Marine Diatom Phaeodactylum tricornutum with Cannabis Genes Enables the Production of the Cannabinoid Precursor, Olivetolic Acid. International Journal of Molecular Sciences, 24(23), 16624. https://doi.org/10.3390/ijms242316624






