Exploring Major Flavonoid Phytochemicals from Nelumbo nucifera Gaertn. as Potential Skin Anti-Aging Agents: In Silico and In Vitro Evaluations
Abstract
:1. Introduction
2. Results and Discussion
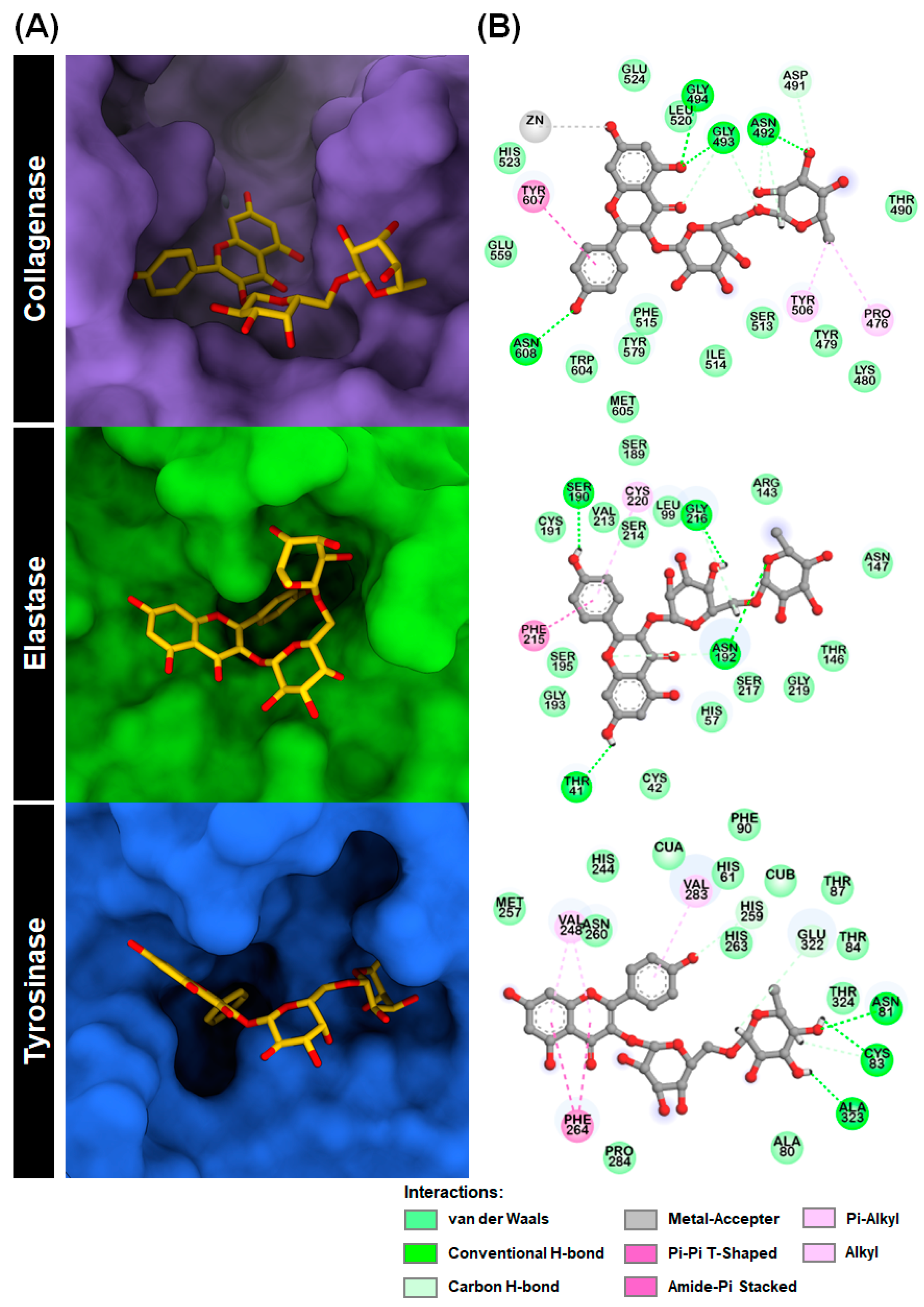
2.1. Molecular Docking
2.2. In Vitro Assay of Kae-3-Rob toward Aging-Related Enzymes Inhibition
2.3. Molecular Dynamics of Kae-3-Rob Bound to Aging-Related Enzymes
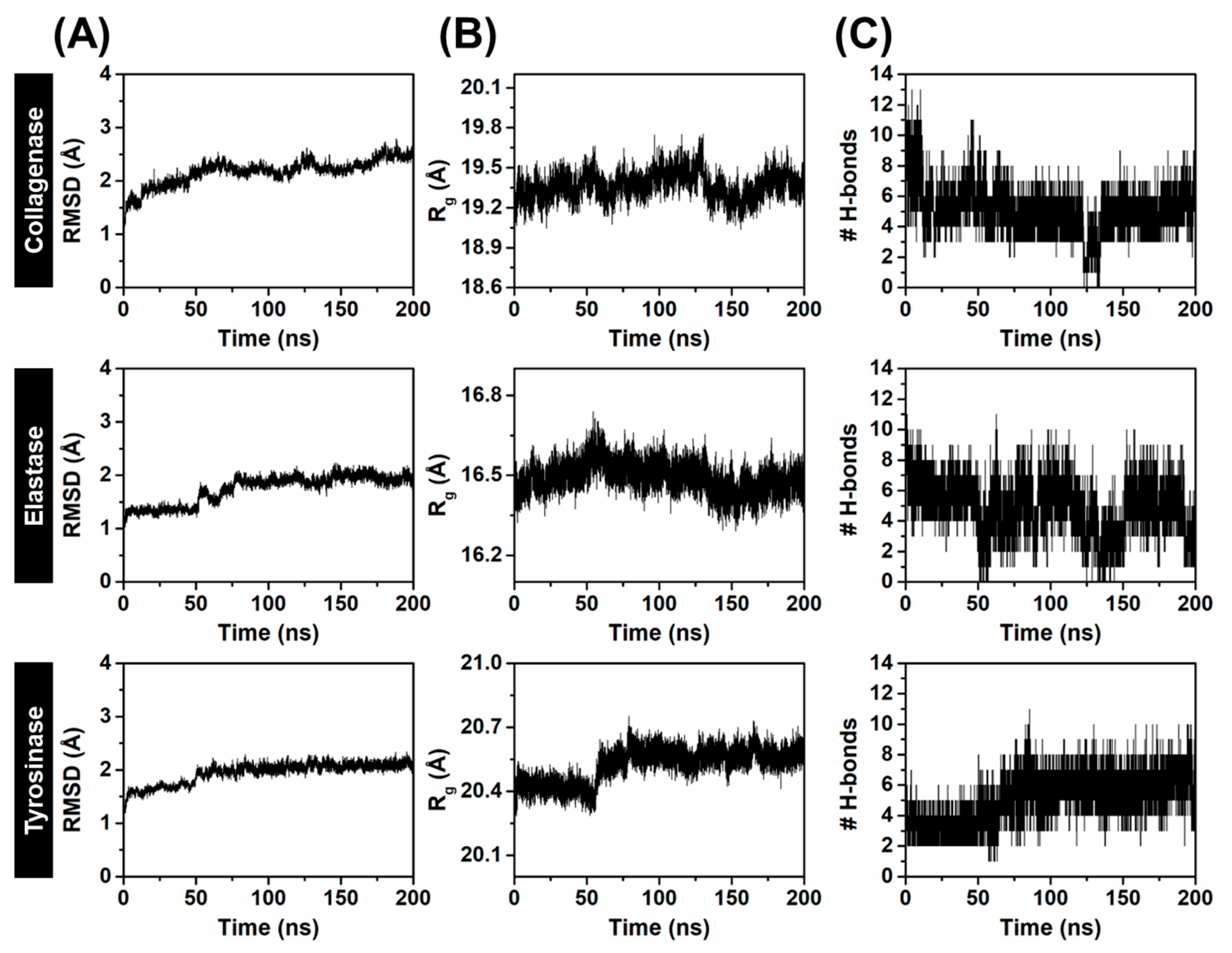
2.3.1. System Stability
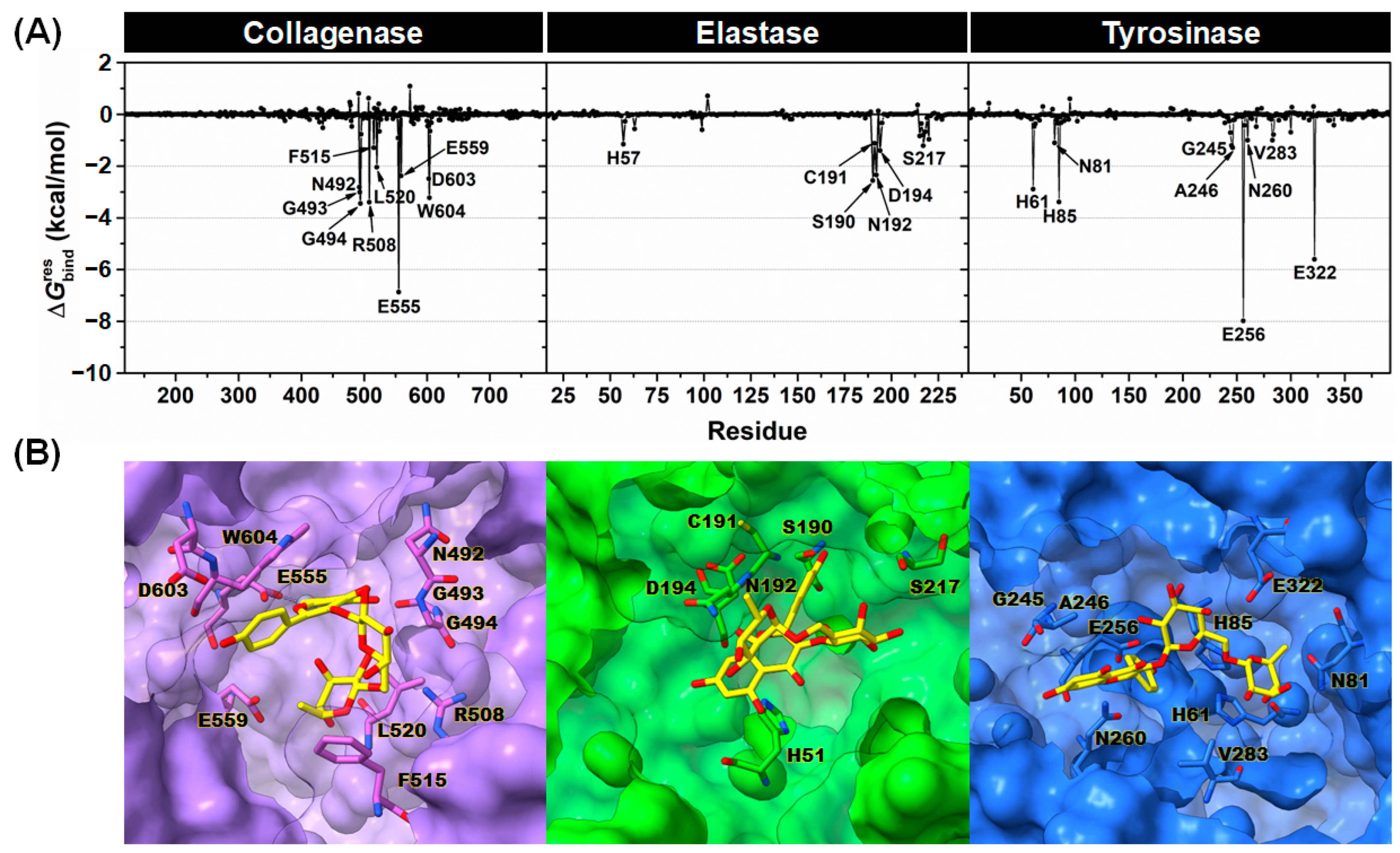
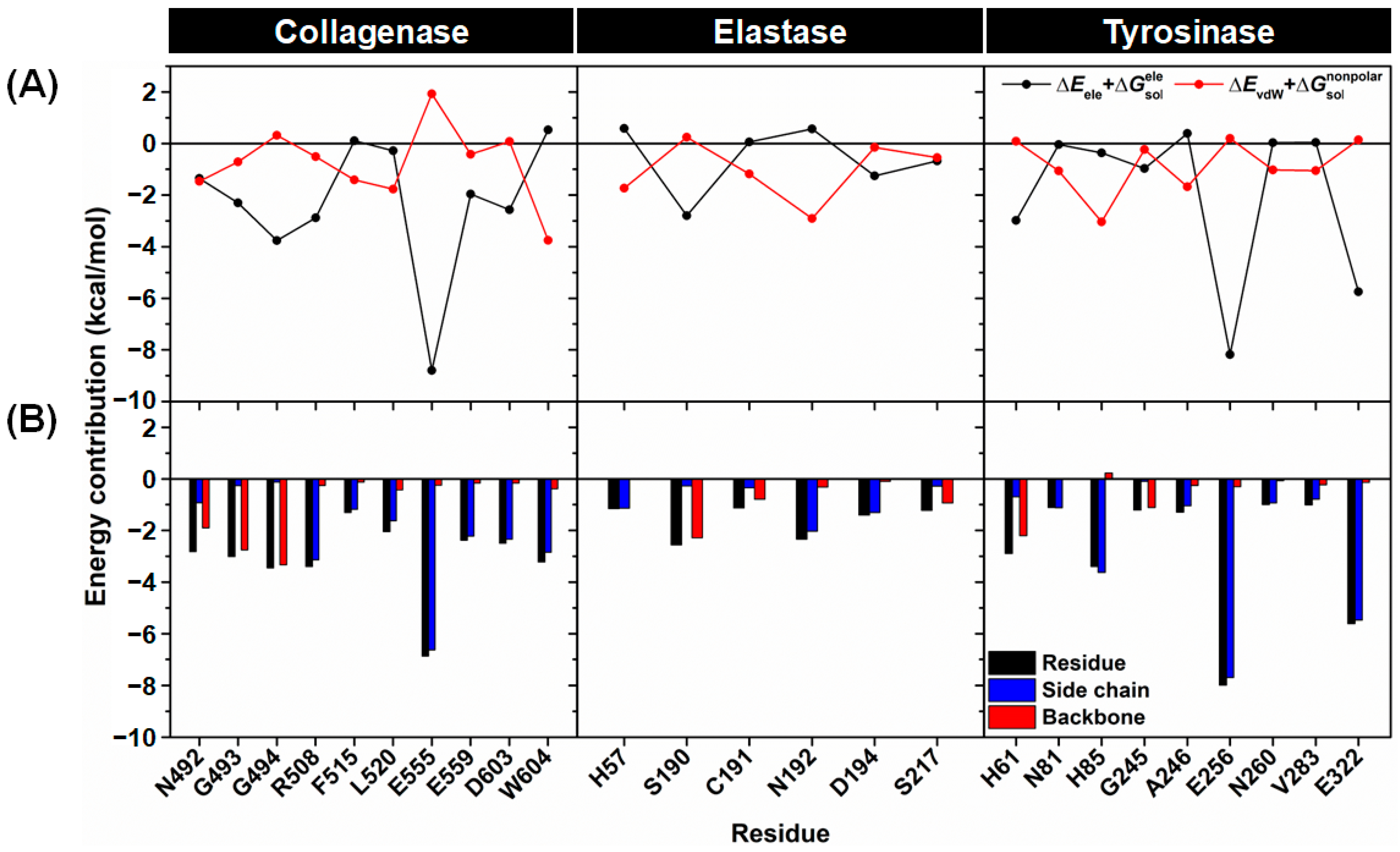
2.3.2. Binding Affinity
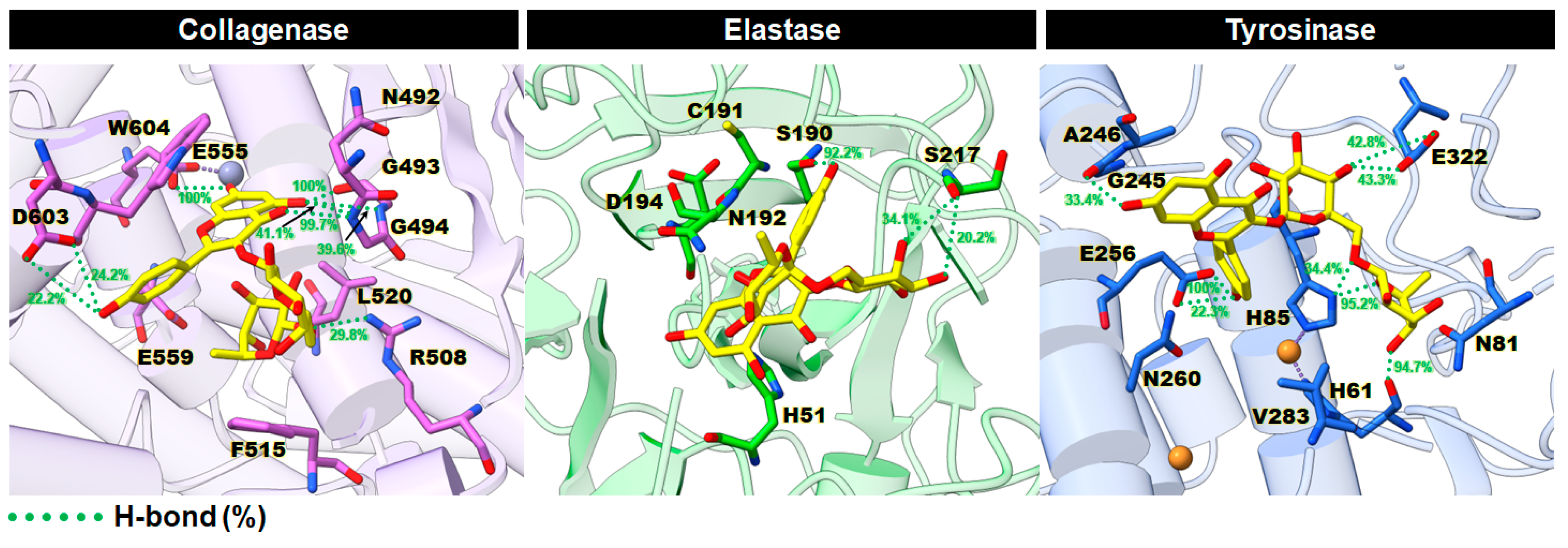
2.3.3. Key Binding Residues
2.3.4. Intermolecular Hydrogen Bonds (H-Bonds)
3. Materials and Methods
3.1. Computational Studies
3.1.1. System Preparation and Molecular Docking
3.1.2. Molecular Dynamic (MD) Simulations
3.1.3. Post-Dynamic Trajectories Analyses
3.2. In Vitro Anti-Aging Activity
3.2.1. Chemicals
3.2.2. Collagenase Assay
3.2.3. Elastase Assay
3.2.4. Tyrosinase Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farage, M.A.; Miller, K.W.; Elsner, P.; Maibach, H.I. Intrinsic and extrinsic factors in skin ageing: A review. Int. J. Cosmet. Sci. 2008, 30, 87–95. [Google Scholar] [CrossRef]
- Russell-Goldman, E.; Murphy, G.F. The pathobiology of skin aging: New insights into an old dilemma. Am. J. Pathol. 2020, 190, 1356–1369. [Google Scholar] [CrossRef]
- Pientaweeratch, S.; Panapisal, V.; Tansirikongkol, A. Antioxidant, anti-collagenase and anti-elastase activities of Phyllanthus emblica, Manilkara zapota and silymarin: An in vitro comparative study for anti-aging applications. Pharm. Biol. 2016, 54, 1865–1872. [Google Scholar] [CrossRef]
- Thring, T.S.A.; Hili, P.; Naughton, D.P. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement. Altern. Med. 2009, 9, 27. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin whitening agents: Medicinal chemistry perspective of tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 403–425. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Maity, N.; Nema, N.K.; Sarkar, B.K. Bioactive compounds from natural resources against skin aging. Phytomedicine 2011, 19, 64–73. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Drouet, S.; Hano, C. Flavonoids from sacred lotus stamen extract slows chronological aging in yeast model by reducing oxidative stress and maintaining cellular metabolism. Cells 2022, 11, 599. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Pinthong, D.; Hano, C. Flavonoids from Nelumbo nucifera Gaertn. a medicinal plant: Uses in traditional medicine, phytochemistry and pharmacological activities. Medicines 2018, 5, 127. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Drouet, S.; Hano, C. Validation of a high-performance liquid chromatography with photodiode array detection method for the separation and quantification of antioxidant and skin anti-aging flavonoids from Nelumbo nucifera Gaertn. stamen extract. Molecules 2022, 27, 1102. [Google Scholar] [CrossRef]
- Li, S.Y.; Lo, A.C.Y. Lutein protects RGC-5 cells against hypoxia and oxidative stress. Int. J. Mol. Sci. 2010, 11, 2109–2117. [Google Scholar] [CrossRef]
- Battaglia, R.; Caponnetto, A.; Caringella, A.M.; Cortone, A.; Ferrara, C.; Smirni, S.; Iannitti, R.; Purrello, M.; D’amato, G.; Fioretti, B.; et al. Resveratrol treatment induces mito-miRNome modification in follicular fluid from aged women with a poor prognosis for in vitro fertilization cycles. Antioxidants 2022, 11, 1019. [Google Scholar] [CrossRef]
- Chen, T.Y.; Chen, Y.L.; Chiu, W.C.; Yeh, C.L.; Tung, Y.T.; Shirakawa, H.; Liao, W.T.; Yang, S.C. Effects of the water extract of fermented rice bran on liver damage and intestinal injury in aged rats with high-fat diet feeding. Plants 2022, 11, 607. [Google Scholar] [CrossRef]
- Dezhi, F.; Wiersema, J.H. Nelumbo nucifera. In Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2001; p. 1. [Google Scholar]
- Chayamarit, K.; Balslav, H.; Esser, H.J. Flora of Thailand; Chayamarit, K., Balslav, H., Eds.; 14/4.; The Forest Herbarium, Royal Forest Department: Bangkok, Thailand, 2020; ISBN 9786163165923. [Google Scholar]
- Sikarwar, R.L.S. Angiosperm diversity assessment of Chitrakootthe legendary place of Vindhyan range. India J. Econ. Taxon. Bot. 2014, 38, 563–619. [Google Scholar]
- Tungmunnithum, D.; Renouard, S.; Drouet, S.; Blondeau, J.P.; Hano, C. A critical cross-species comparison of pollen from Nelumbo nucifera gaertn. vs. Nymphaea lotus L. for authentication of thai medicinal herbal tea. Plants 2020, 9, 921. [Google Scholar] [CrossRef]
- Lee, J.S.; Shukla, S.; Kim, J.A.; Kim, M. Anti-angiogenic effect of Nelumbo nucifera leaf extracts in human umbilical vein endothelial cells with antioxidant potential. PLoS ONE 2015, 10, e0118552. [Google Scholar] [CrossRef]
- Deng, J.; Chen, S.; Yin, X.; Wang, K.; Liu, Y.; Li, S.; Yang, P. Systematic qualitative and quantitative assessment of anthocyanins, flavones and flavonols in the petals of 108 lotus (Nelumbo nucifera) cultivars. Food Chem. 2013, 139, 307–312. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Drouet, S.; Hano, C. Phytochemical diversity and antioxidant potential of natural populations of Nelumbo nucifera Gaertn. throughout the floristic regions in Thailand. Molecules 2022, 27, 681. [Google Scholar] [CrossRef]
- Sheikh, S.A. Ethno-medicinal uses and pharmacological activities of lotus (Nelumbo nucifera). J. Med. Plants Stud. 2014, 2, 42–46. [Google Scholar]
- Zhu, M.Z.; Wu, W.; Jiao, L.L.; Yang, P.F.; Guo, M.Q. Analysis of flavonoids in lotus (Nelumbo nucifera) leaves and their antioxidant activity using macroporous resin chromatography coupled with LC-MS/MS and antioxidant biochemical assays. Molecules 2015, 20, 10553–10565. [Google Scholar] [CrossRef]
- Lin, H.Y.; Kuo, Y.H.; Lin, Y.L.; Chiang, W. Antioxidative effect and active components from leaves of lotus (Nelumbo nucifera). J. Agric. Food Chem. 2009, 57, 6623–6629. [Google Scholar] [CrossRef]
- Rai, S.; Wahile, A.; Mukherjee, K.; Saha, B.P.; Mukherjee, P.K. Antioxidant activity of Nelumbo nucifera (sacred lotus) seeds. J. Ethnopharmacol. 2006, 104, 322–327. [Google Scholar] [CrossRef]
- Yang, M.Y.; Chang, Y.C.; Chan, K.C.; Lee, Y.J.; Wang, C.J. Flavonoid-enriched extracts from Nelumbo nucifera leaves inhibits proliferation of breast cancer in vitro and in vivo. Eur. J. Integr. Med. 2011, 3, e153–e163. [Google Scholar] [CrossRef]
- Huang, C.F.; Chen, Y.W.; Yang, C.Y.; Lin, H.Y.; Way, T.D.; Chiang, W.; Liu, S.H. Extract of lotus leaf (Nelumbo nucifera) and its active constituent catechin with insulin secretagogue activity. J. Agric. Food Chem. 2011, 59, 1087–1094. [Google Scholar] [CrossRef]
- Ho, H.H.; Hsu, L.S.; Chan, K.C.; Chen, H.M.; Wu, C.H.; Wang, C.J. Extract from the leaf of nucifera reduced the development of atherosclerosis via inhibition of vascular smooth muscle cell proliferation and migration. Food Chem. Toxicol. 2010, 48, 159–168. [Google Scholar] [CrossRef]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kim, E.S.; Lee, C.; Kim, S.; Cho, S.H.; Hwang, B.Y.; Lee, M.K. Chemical constituents from Nelumbo nucifera leaves and their anti-obesity effects. Bioorg. Med. Chem. Lett. 2013, 23, 3604–3608. [Google Scholar] [CrossRef]
- Chen, S.; Wu, B.H.; Fang, J.B.; Liu, Y.L.; Zhang, H.H.; Fang, L.C.; Guan, L.; Li, S.H. Analysis of flavonoids from lotus (Nelumbo nucifera) leaves using high performance liquid chromatography/photodiode array detector tandem electrospray ionization mass spectrometry and an extraction method optimized by orthogonal design. J. Chromatogr. A 2012, 1227, 145–153. [Google Scholar] [CrossRef]
- Temviriyanukul, P.; Sritalahareuthai, V.; Promyos, N.; Thangsiri, S.; Pruesapan, K.; Srinuanchai, W.; Nuchuchua, O.; Siriwan, D.; On-Nom, N.; Suttisansanee, U. The effect of sacred lotus (Nelumbo nucifera) and its mixtures on phenolic profiles, antioxidant activities, and inhibitions of the key enzymes relevant to Alzheimer’s disease. Molecules 2020, 25, 3713. [Google Scholar] [CrossRef]
- Choi, S.J.; Lee, S.N.; Kim, K.; Joo, D.H.; Shin, S.; Lee, J.; Lee, H.K.; Kim, J.; Kwon, S.B.; Kim, M.J.; et al. Biological effects of rutin on skin aging. Int. J. Mol. Med. 2016, 38, 357–363. [Google Scholar] [CrossRef]
- Eckhard, U.; Schönauer, E.; Nüss, D.; Brandstetter, H. Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat. Struct. Mol. Biol. 2010, 18, 1109–1114. [Google Scholar] [CrossRef]
- Nakanishi, I.; Kinoshita, T.; Sato, A.; Tada, T. Structure of porcine pancreatic elastase complexed with FR901277, a novel macrocyclic inhibitor of elastases, at 1.6 Å resolution. Biopolymers 2000, 53, 434–445. [Google Scholar] [CrossRef]
- Altyar, A.E.; Ashour, M.L.; Youssef, F.S. Premna odorata: Seasonal metabolic variation in the essential oil composition of its leaf and verification of its anti-ageing potential via in vitro assays and molecular modelling. Biomolecules 2020, 10, 879. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Hassan Khan, M.T.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef]
- Yepes, A.; Ochoa-Bautista, D.; Murillo-Arango, W.; Quintero-Saumeth, J.; Bravo, K.; Osorio, E. Purple passion fruit seeds (Passiflora edulis f. edulis Sims) as a promising source of skin anti-aging agents: Enzymatic, antioxidant and multi-level computational studies. Arab. J. Chem. 2021, 14, 102905. [Google Scholar]
- Ma, S.; Zheng, X.; Zhang, Y.; Zhao, S.; Yi, J.; Cai, S. Exploring the promotive effects and mechanisms of different polyphenolic extracts from Prinsepia utilis royle seed shell on tyrosinase. Foods 2022, 11, 4015. [Google Scholar] [CrossRef]
- Wagle, A.; Seong, S.H.; Joung, E.J.; Kim, H.R.; Jung, H.A.; Choi, J.S. Discovery of a highly potent tyrosinase inhibitor, luteolin 5-O-β-d-glucopyranoside, isolated from Cirsium japonicum var. maackii (Maxim.) Matsum. Korean thistle: Kinetics and computational molecular docking simulation. ACS Omega 2018, 3, 17236–17245. [Google Scholar]
- Garros, L.; Drouet, S.; Corbin, C.; Decourtil, C.; Fidel, T.; De Lacour, J.L.; Leclerc, E.A.; Renouard, S.; Tungmunnithum, D.; Doussot, J.; et al. Insight into the influence of cultivar type, cultivation year, and site on the lignans and related phenolic profiles, and the health-promoting antioxidant potential of flax (Linum usitatissimum L.) seeds. Molecules 2018, 23, 2636. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Drouet, S.; Lorenzo, J.M.; Hano, C. Characterization of bioactive phenolics and antioxidant capacity of edible bean extracts of 50 fabaceae populations grown in Thailand. Foods 2021, 10, 3118. [Google Scholar] [CrossRef]
- Poli, G.; Granchi, C.; Rizzolio, F.; Tuccinardi, T. Application of MM-PBSA methods in virtual screening. Molecules 2020, 25, 1971. [Google Scholar] [CrossRef]
- Brooks, B.; Karplus, M. Harmonic dynamics of proteins: Normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc. Natl. Acad. Sci. USA 1983, 80, 6571–6575. [Google Scholar] [CrossRef]
- Nutho, B.; Wilasluck, P.; Deetanya, P.; Wangkanont, K.; Arsakhant, P.; Saeeng, R.; Rungrotmongkol, T. Discovery of C-12 dithiocarbamate andrographolide analogues as inhibitors of SARS-CoV-2 main protease: In vitro and in silico studies. Comput. Struct. Biotechnol. J. 2022, 20, 2784–2797. [Google Scholar] [CrossRef]
- Nutho, B.; Pengthaisong, S.; Tankrathok, A.; Lee, V.S.; Cairns, J.R.K.; Rungrotmongkol, T.; Hannongbua, S. Structural basis of specific glucoimidazole and mannoimidazole binding by os3bglu7. Biomolecules 2020, 10, 907. [Google Scholar] [CrossRef]
- Boonma, T.; Soikudrua, N.; Nutho, B.; Rungrotmongkol, T.; Nunthaboot, N. Insights into binding molecular mechanism of hemagglutinin H3N2 of influenza virus complexed with arbidol and its derivative: A molecular dynamics simulation perspective. Comput. Biol. Chem. 2022, 101, 107764. [Google Scholar] [CrossRef]
- Marques, R.V.; Guillaumin, A.; Abdelwahab, A.B.; Salwinski, A.; Gotfredsen, C.H.; Bourgaud, F.; Enemark-rasmussen, K.; Miguel, S.; Simonsen, H.T. Collagenase and tyrosinase inhibitory effect of isolated constituents from the moss Polytrichum formosum. Plants 2021, 10, 1271. [Google Scholar] [CrossRef]
- Saechan, C.; Nguyen, U.H.; Wang, Z.; Sugimoto, S.; Yamano, Y.; Matsunami, K.; Otsuka, H.; Phan, G.M.; Pham, V.H.; Tipmanee, V.; et al. Potency of bisresorcinol from Heliciopsis terminalis on skin aging: In vitro bioactivities and molecular interactions. PeerJ 2021, 9, e11618. [Google Scholar] [CrossRef]
- Jugreet, B.S.; Lall, N.; Anina Lambrechts, I.; Reid, A.M.; Maphutha, J.; Nel, M.; Hassan, A.H.; Khalid, A.; Abdalla, A.N.; Van, B.L.; et al. In Vitro and in silico pharmacological and cosmeceutical potential of ten essential oils from aromatic medicinal plants from the Mascarene islands. Molecules 2022, 27, 8705. [Google Scholar] [CrossRef]
- Eun Lee, K.; Bharadwaj, S.; Yadava, U.; Gu Kang, S. Evaluation of caffeine as inhibitor against collagenase, elastase and tyrosinase using in silico and in vitro approach. J. Enzyme Inhib. Med. Chem. 2019, 34, 927–936. [Google Scholar] [CrossRef]
- Paudel, P.; Wagle, A.; Seong, S.H.; Park, H.J.; Jung, H.A.; Choi, J.S. A new tyrosinase inhibitor from the red alga Symphyocladia latiuscula (harvey) Yamada (rhodomelaceae). Mar. Drugs 2019, 17, 295. [Google Scholar] [CrossRef]
- Ashraf, Z.; Rafiq, M.; Nadeem, H.; Hassan, M.; Afzal, S.; Waseem, M.; Afzal, K.; Latip, J. Carvacrol derivatives as mushroom tyrosinase inhibitors; synthesis, kinetics mechanism and molecular docking studies. PLoS ONE 2017, 12, e0178069. [Google Scholar] [CrossRef]
- Ferreira De Freitas, R.; Schapira, M. A systematic analysis of atomic protein-ligand interactions in the PDB. Med. Chem. Comm. 2017, 8, 1970–1981. [Google Scholar] [CrossRef]
- Structure of Porcine Pancreatic Elastase Complexed with the Elastase Inhibitor GR143783. Available online: https://www.rcsb.org/structure/1BRU (accessed on 9 August 2023).
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Li, P.; Merz, K.M., Jr. Taking into account the ion-induced dipole interaction in the nonbonded model of ions. J. Chem. Theory Comput. 2014, 10, 289–297. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Uberuaga, B.P.; Anghel, M.; Voter, A.F. Synchronization of trajectories in canonical molecular-dynamics simulations: Observation, explanation, and exploitation. J. Chem. Phys. 2004, 120, 6363–6374. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: Strategies and applications in drug design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller Iii, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Wittenauer, J.; MäcKle, S.; Sußmann, D.; Schweiggert-Weisz, U.; Carle, R. Inhibitory effects of polyphenols from grape pomace extract on collagenase and elastase activity. Fitoterapia 2015, 101, 179–187. [Google Scholar] [CrossRef]
- Chai, W.M.; Huang, Q.; Lin, M.Z.; Ou-Yang, C.; Huang, W.Y.; Wang, Y.X.; Xu, K.L.; Feng, H.L. Condensed tannins from longan bark as inhibitor of tyrosinase: Structure, activity, and mechanism. J. Agric. Food Chem. 2018, 66, 908–917. [Google Scholar] [CrossRef]


| Anti-Aging Activity | % of Enzyme Inhibition a |
|---|---|
| Tyrosinase | 69.84 ± 6.07 |
| Collagenase | 58.24 ± 8.27 |
| Elastase | 26.29 ± 7.16 |
| Collagenase | Elastase | Tyrosinase | |
|---|---|---|---|
| −28.96 ± 0.27 | −35.38 ± 0.14 | −32.02 ± 0.18 | |
| −46.08 ± 0.25 | −13.23 ± 0.17 | −35.34 ± 0.36 | |
| −75.04 ± 0.27 | −48.61 ± 0.21 | −67.36 ± 0.29 | |
| 22.57 ± 2.02 | 21.71 ± 1.03 | 21.72 ± 1.84 | |
| 50.61 ± 0.22 | 28.25 ± 0.18 | 41.36 ± 0.24 | |
| −4.44 ± 0.01 | −4.06 ± 0.01 | −4.64 ± 0.01 | |
| 46.17 ± 0.21 | 24.19 ± 0.17 | 36.71 ± 0.24 | |
| −28.87 ± 0.20 | −24.42 ± 0.12 | −30.65 ± 0.14 | |
| −6.30 | −2.71 | −8.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nutho, B.; Tungmunnithum, D. Exploring Major Flavonoid Phytochemicals from Nelumbo nucifera Gaertn. as Potential Skin Anti-Aging Agents: In Silico and In Vitro Evaluations. Int. J. Mol. Sci. 2023, 24, 16571. https://doi.org/10.3390/ijms242316571
Nutho B, Tungmunnithum D. Exploring Major Flavonoid Phytochemicals from Nelumbo nucifera Gaertn. as Potential Skin Anti-Aging Agents: In Silico and In Vitro Evaluations. International Journal of Molecular Sciences. 2023; 24(23):16571. https://doi.org/10.3390/ijms242316571
Chicago/Turabian StyleNutho, Bodee, and Duangjai Tungmunnithum. 2023. "Exploring Major Flavonoid Phytochemicals from Nelumbo nucifera Gaertn. as Potential Skin Anti-Aging Agents: In Silico and In Vitro Evaluations" International Journal of Molecular Sciences 24, no. 23: 16571. https://doi.org/10.3390/ijms242316571
APA StyleNutho, B., & Tungmunnithum, D. (2023). Exploring Major Flavonoid Phytochemicals from Nelumbo nucifera Gaertn. as Potential Skin Anti-Aging Agents: In Silico and In Vitro Evaluations. International Journal of Molecular Sciences, 24(23), 16571. https://doi.org/10.3390/ijms242316571








