Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response
Abstract
:1. Introduction
2. Results
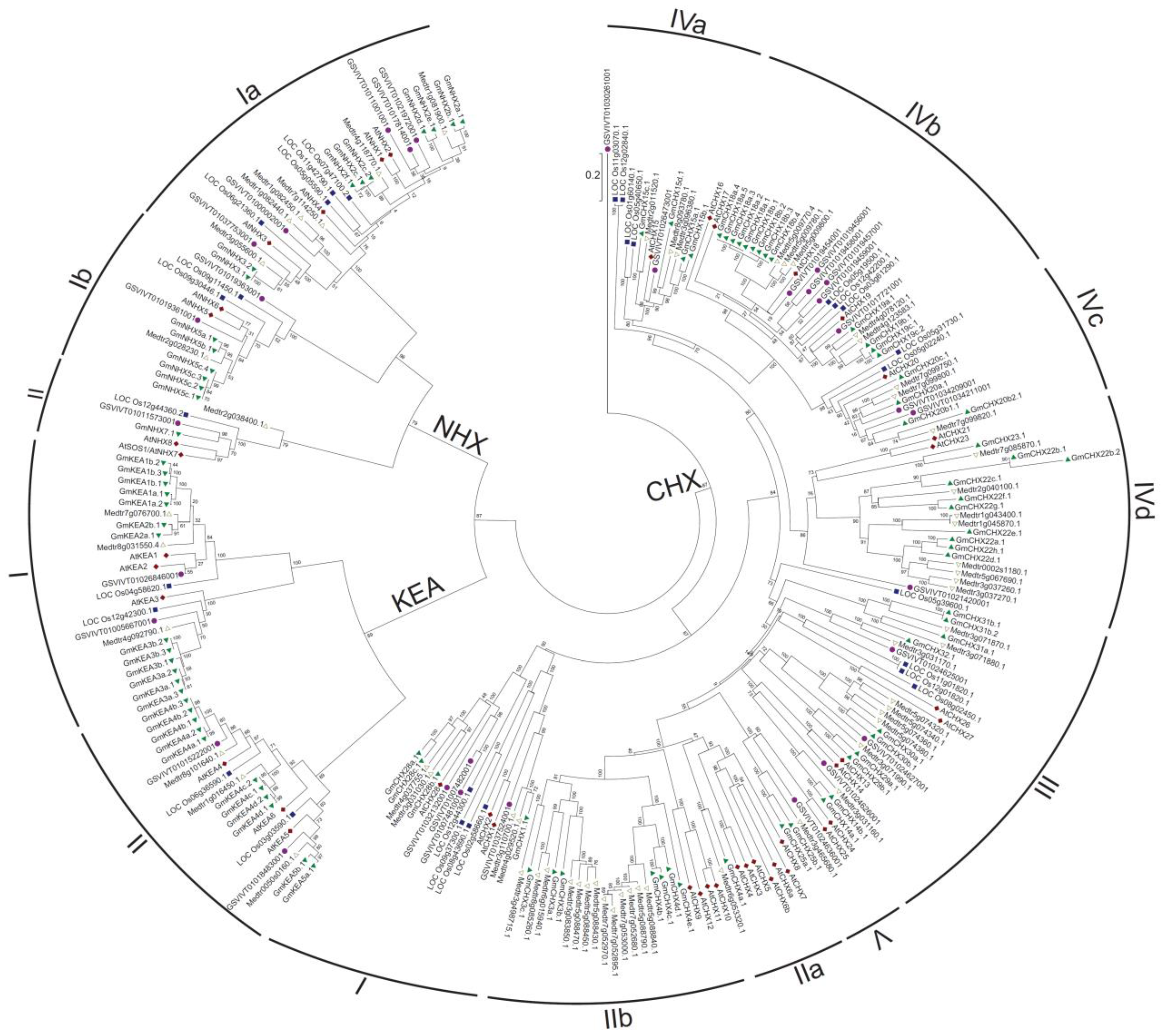
2.1. Identification and Classification of CPA Genes in Soybean
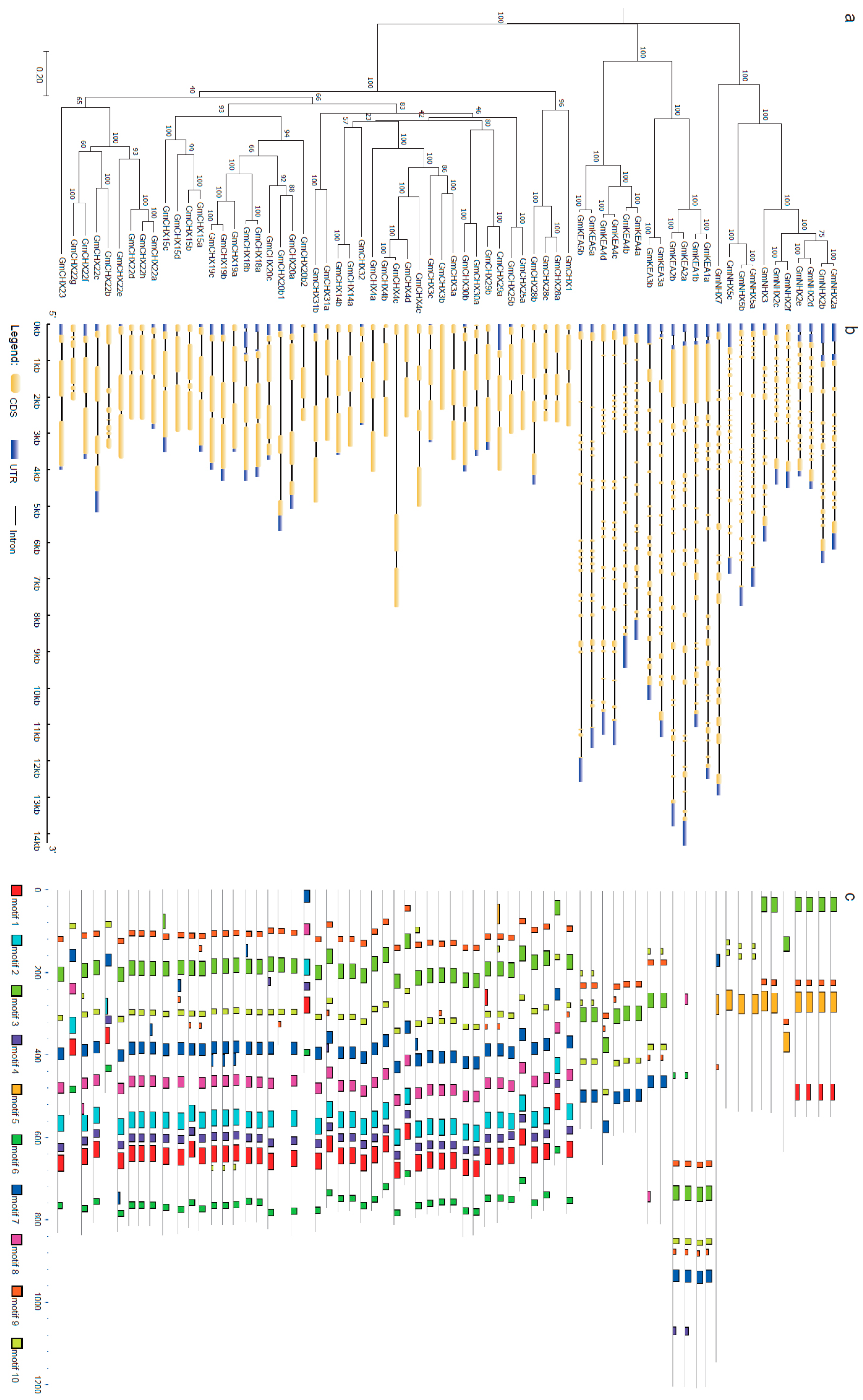
2.2. Gene Structure and Conserved Domain Analysis
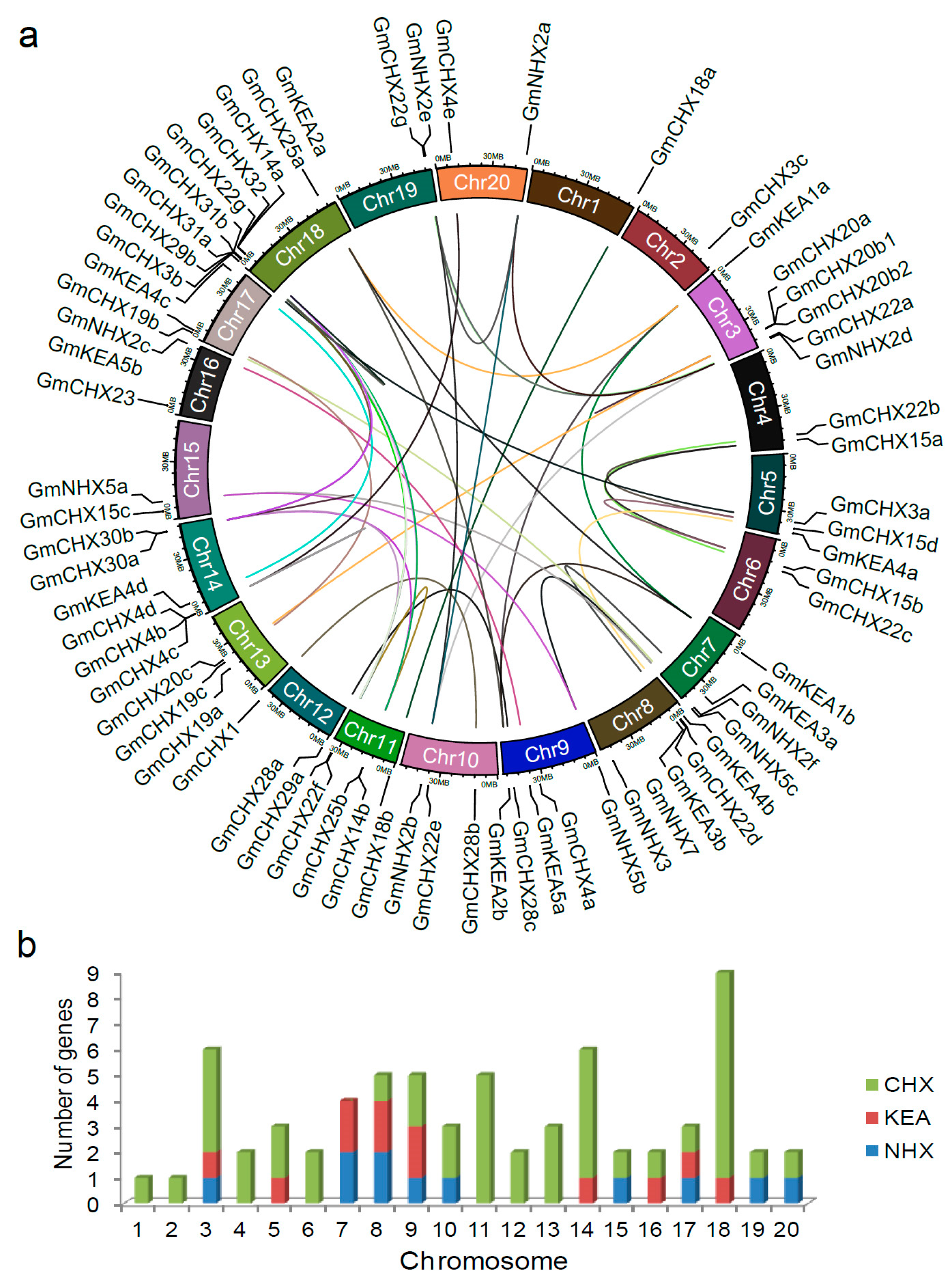
2.3. Chromosomal Locations and Expansion Pattern of GmCPAs
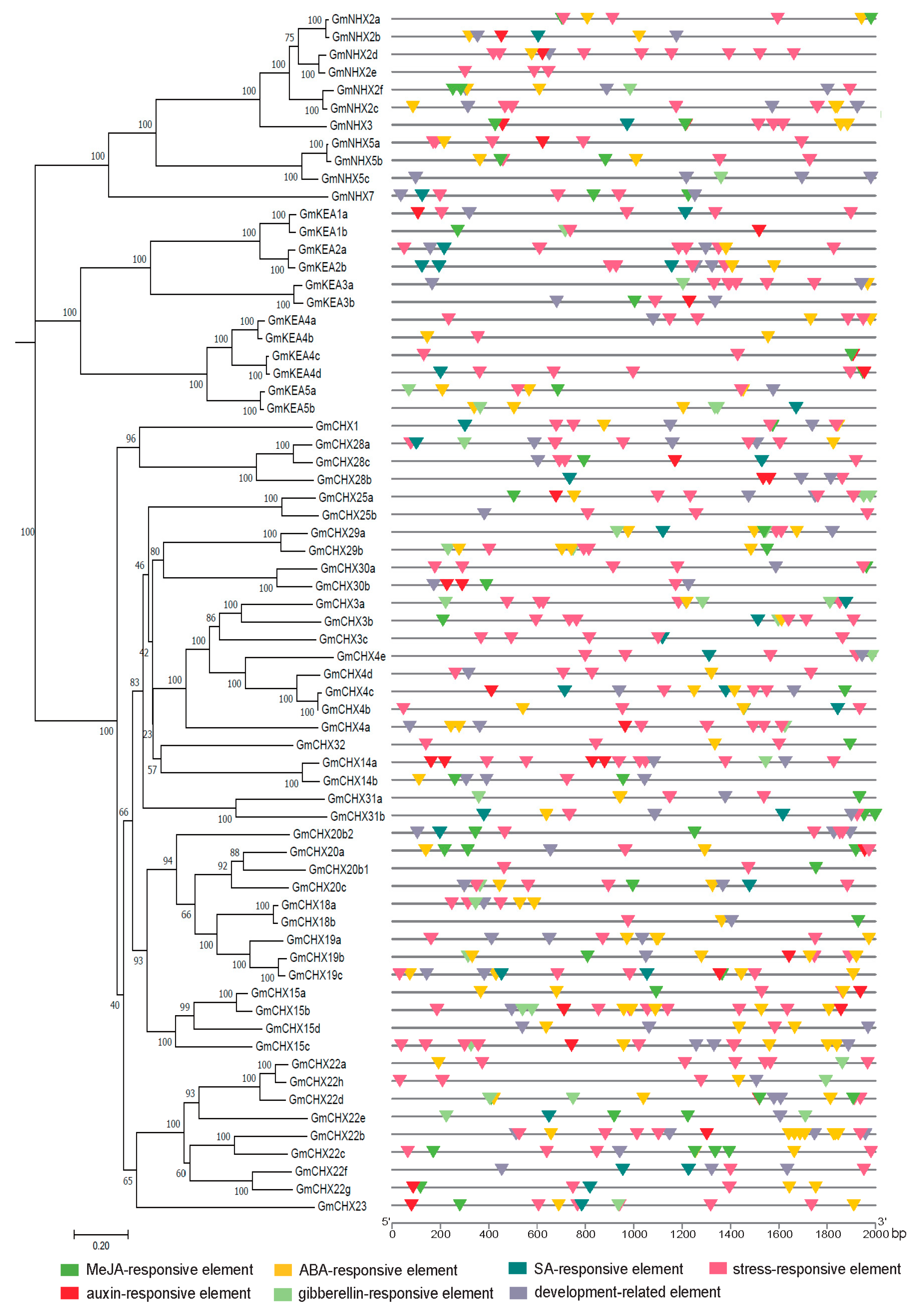
2.4. cis-Elements Analysis in the Promoter of GmCPAs
2.5. Expression Analysis of GmCPAs in Different Tissues
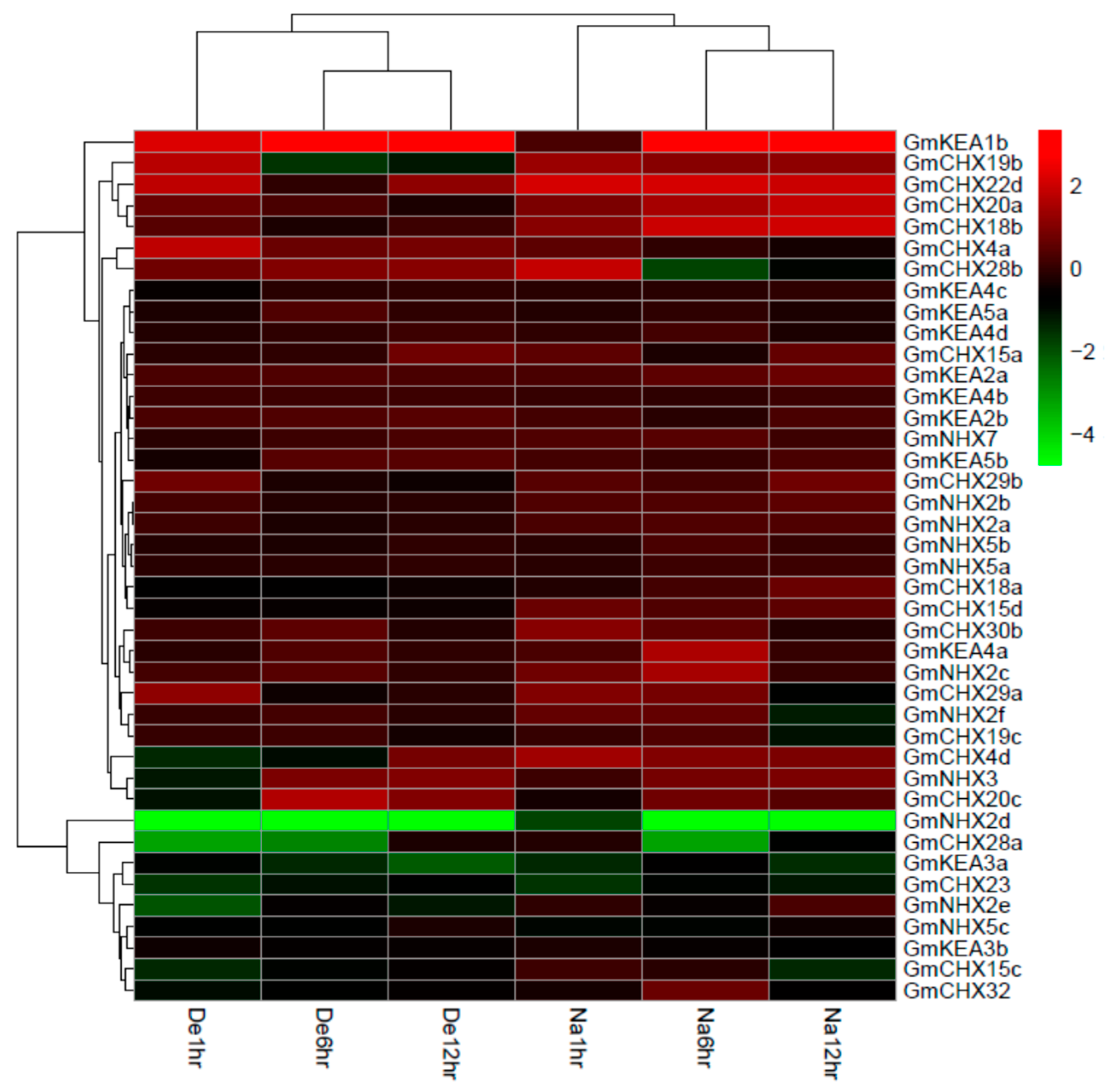
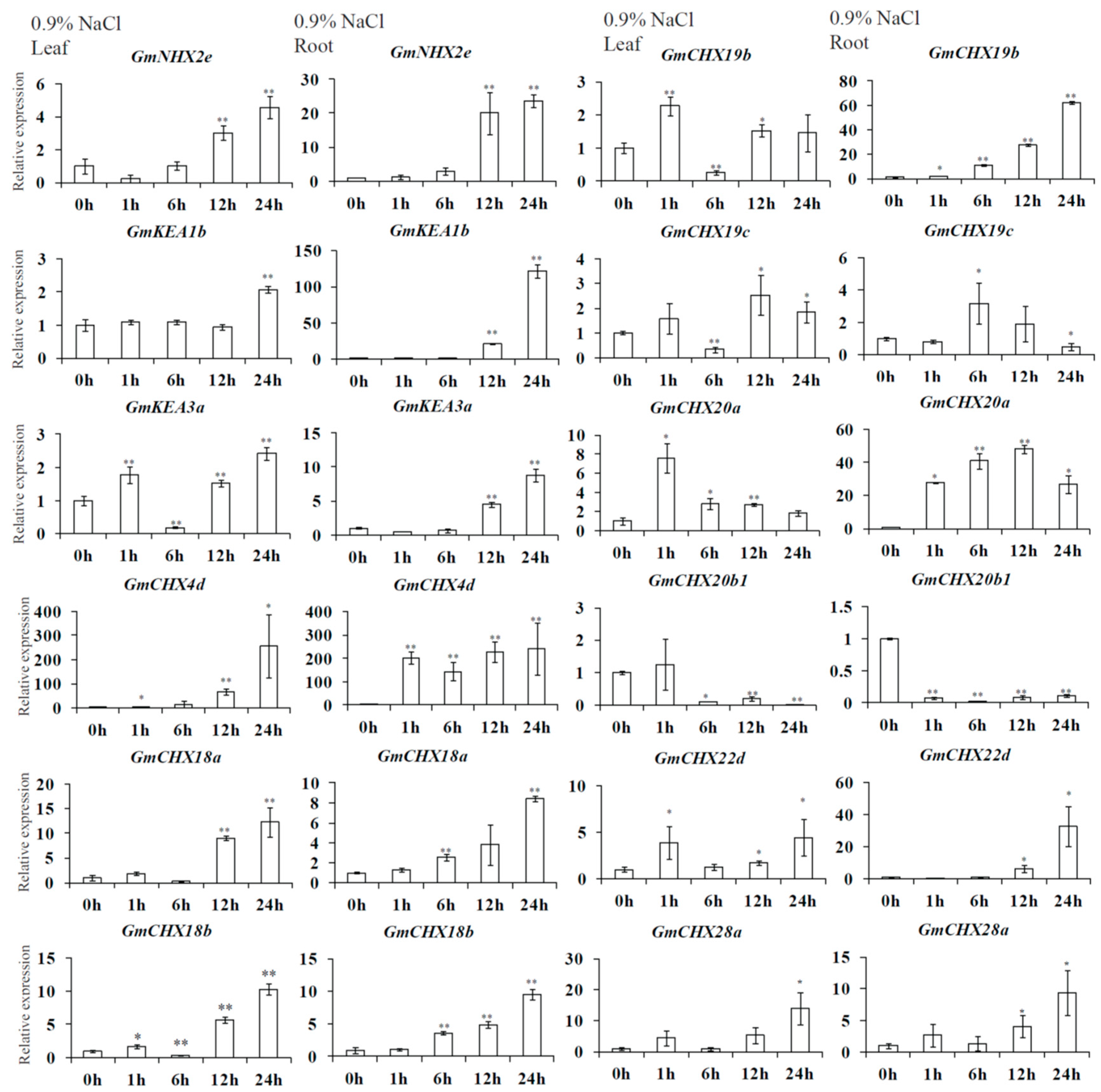
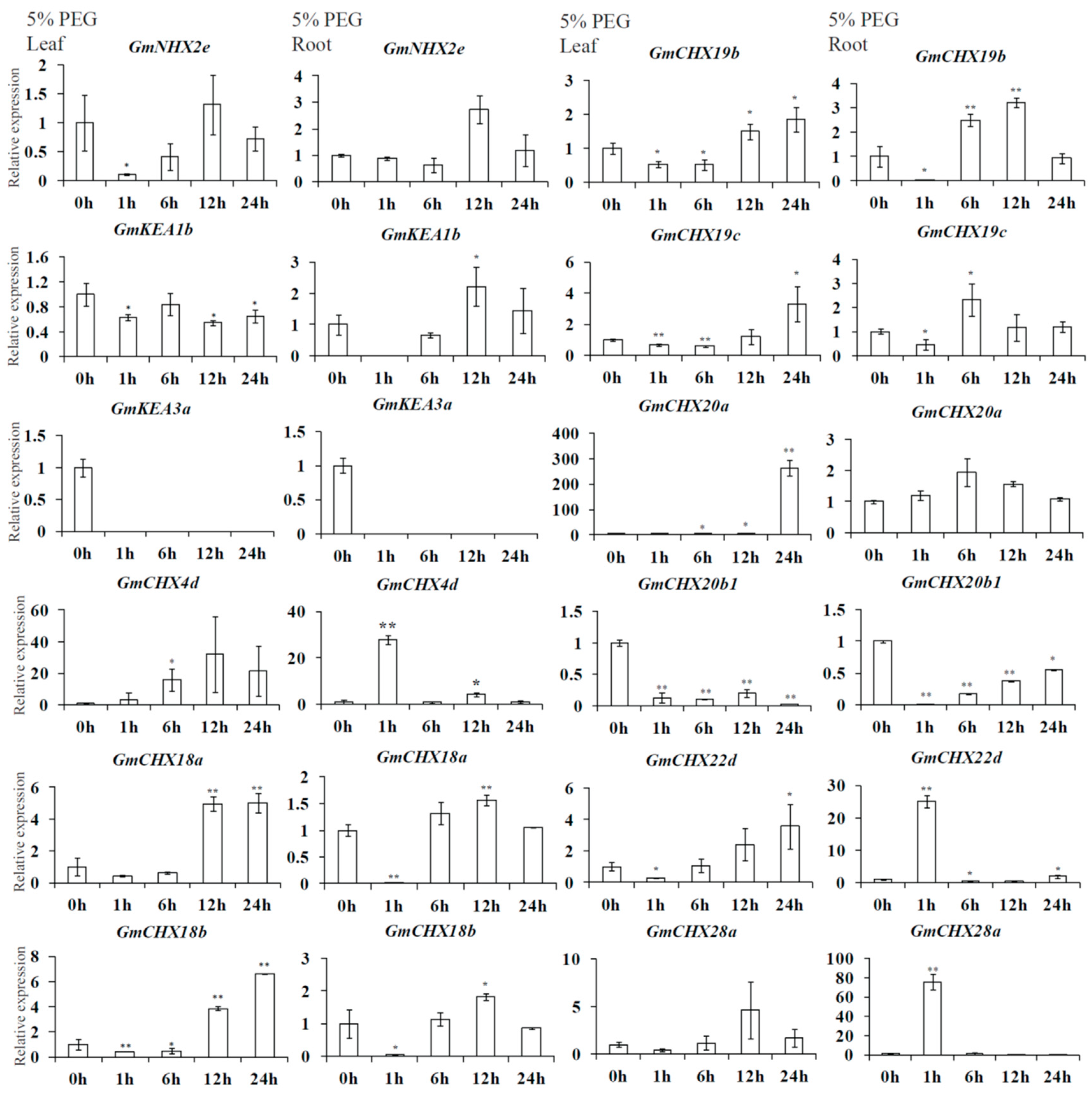
2.6. Expression Profiles of GmCPAs under Salt Stress and Osmotic Stress
2.7. Ectopic Expression of GmCHX20a Increased Salt Sensitivity in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Identification and Characteristics of CPA Genes in Soybean
4.2. Phylogenetic Analysis
4.3. Analysis of Gene Structures and Conserved Domains
4.4. Chromosome Localization and Gene Duplication Analysis
4.5. Analysis of cis-Acting Regulatory Elements in Promoter
4.6. Digital Expression Analysis of GmCPAs in Different Tissues or under Abiotic Stress
4.7. Gene Expression Analysis of GmCPAs under Salt or Osmotic Treatments
4.8. Salt Tolerance Assay on the GmCHX20a-Overexpressing Arabidopsis thaliana Plants
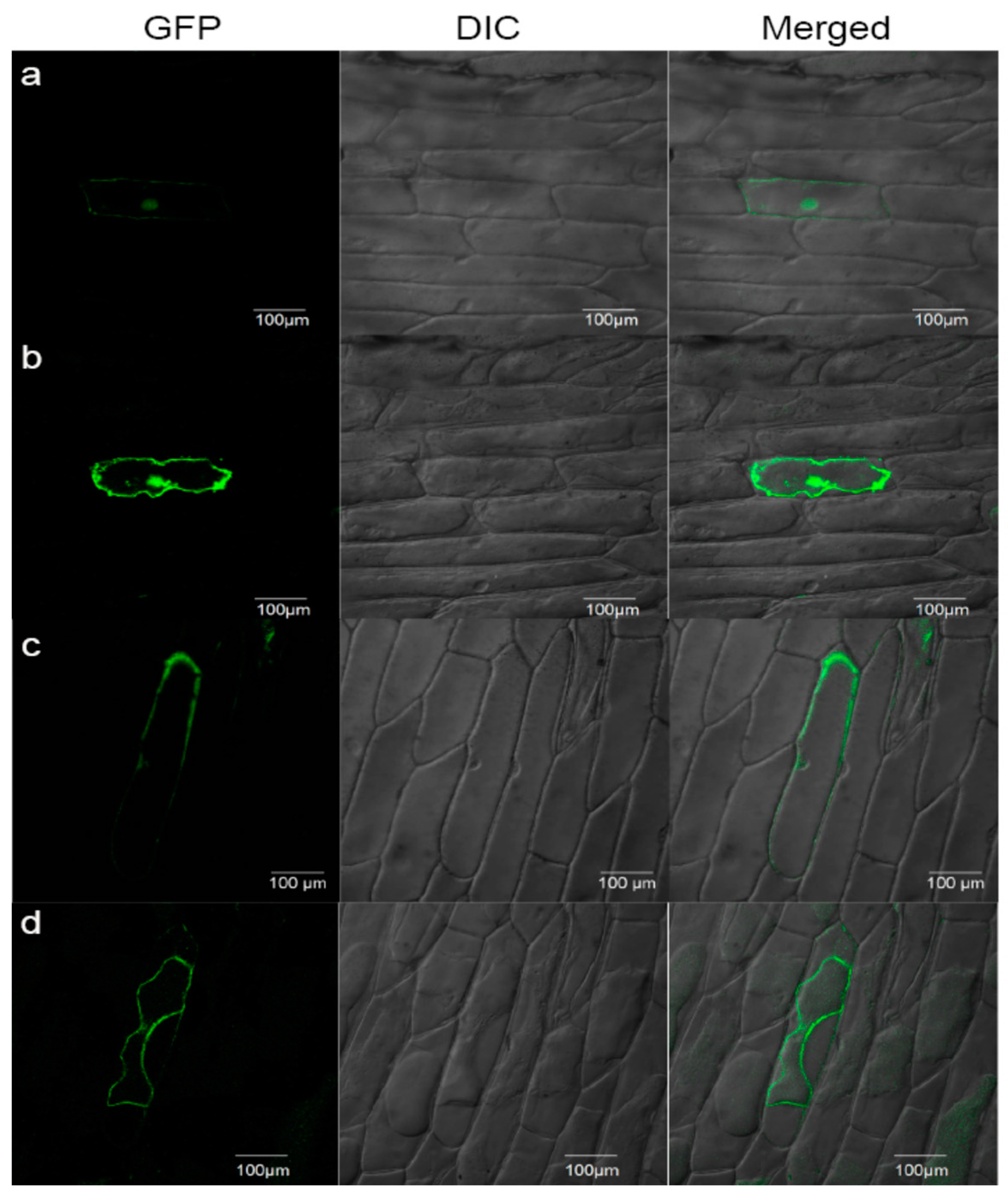
4.9. Subcellular Localization by Transient Expression in Onion Epidermis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sze, H.; Chanroj, S. Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef] [PubMed]
- Masrati, G.; Dwivedi, M.; Rimon, A.; Gluck-Margolin, Y.; Kessel, A.; Ashkenazy, H.; Mayrose, I.; Padan, E.; Ben-Tal, N. Broad phylogenetic analysis of cation/proton antiporters reveals transport determinants. Nat. Commun. 2018, 9, 4205. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Dabravolski, S.A.; Pan, T.; Shabala, S. Phylogenetic diversity and physiological doles of plant monovalent cation/H+ antiporters. Front. Plant Sci. 2020, 11, 573564. [Google Scholar] [CrossRef] [PubMed]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef]
- Chanroj, S.; Wang, G.; Venema, K.; Zhang, M.W.; Delwiche, C.F.; Sze, H. Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 2012, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.-Y.; Yang, X.; Xia, X.; Yin, W. Comparative analysis of cation/proton antiporter superfamily in plants. Gene 2013, 521, 245–251. [Google Scholar] [CrossRef]
- Jia, Q.; Zheng, C.; Sun, S.; Amjad, H.; Liang, K.; Lin, W. The role of plant cation/proton antiporter gene family in salt tolerance. Biol. Plant. 2018, 62, 617–629. [Google Scholar] [CrossRef]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Mishra, A.K.; Kumar, K. Molecular insights into the role of plant transporters in salt stress response. Physiol. Plant. 2021, 173, 1481–1494. [Google Scholar] [CrossRef]
- Bassil, E.; Zhang, S.; Gong, H.; Tajima, H.; Blumwald, E. Cation specificity of vacuolar NHX-Type cation/H+ antiporters. Plant Physiol. 2019, 179, 616–629. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Lee, B.; Wu, S.-J.; Zhu, J.-K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, R.-J.; Yang, X.; Zheng, X.; Shao, Q.; Tang, Q.-L.; Fu, A.; Luan, S. Golgi-localized cation/proton exchangers regulate ionic homeostasis and skotomorphogenesis in Arabidopsis. Plant Cell Environ. 2019, 42, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Pan, T.; Fan, L.; Qiu, Q.-S. A novel AtKEA gene family, homolog of bacterial K+/H+ antiporters, plays potential roles in K+ homeostasis and osmotic adjustment in Arabidopsis. PLoS ONE 2013, 8, e81463. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Pan, T.; Zhang, X.; Fan, L.; Quintero, F.J.; Zhao, H.; Su, X.; Li, X.; Villalta, I.; Mendoza, I.; et al. K+ efflux antiporters 4, 5, and 6 mediate pH and K+ homeostasis in endomembrane compartments. Plant Physiol. 2018, 178, 1657–1678. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, J.; Zhong, Y.; Cramer, G.R.; Cheng, Z. Genome-wide analysis of the cation/proton antiporter (CPA) super family genes in grapevine (Vitis vinifera L.). Plant Omics 2015, 8, 300–311. [Google Scholar]
- Zhou, H.; Qi, K.; Liu, X.; Yin, H.; Wang, P.; Chen, J.; Wu, J.; Zhang, S. Genome-wide identification and comparative analysis of the cation proton antiporters family in pear and four other rosaceae species. Mol. Genet. Genomics 2016, 291, 1727–1742. [Google Scholar] [CrossRef]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, J.; Zhang, Y.; Xu, L.; Zhang, W.; Ni, M.; Zhu, Y.; Liu, L. Genome-wide identification and functional characterization of the cation proton antiporter (CPA) family related to salt stress response in radish (Raphanus sativus L.). Int. J. Mol. Sci. 2020, 21, 8262. [Google Scholar] [CrossRef]
- Kong, M.; Luo, M.; Li, J.; Feng, Z.; Zhang, Y.; Song, W.; Zhang, R.; Wang, R.; Wang, Y.; Zhao, J.; et al. Genome-wide identification, characterization, and expression analysis of the monovalent cation-proton antiporter superfamily in maize, and functional analysis of its role in salt tolerance. Genomics 2021, 113, 1940–1951. [Google Scholar] [CrossRef]
- Wu, L.; Wu, M.; Liu, H.; Gao, Y.; Chen, F.; Xiang, Y. Identification and characterisation of monovalent cation/proton antiporters (CPAs) in phyllostachys edulis and the functional analysis of PheNHX2 in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 164, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Taneja, M.; Upadhyay, S.K. Identification, characterization and expression profiling of cation-proton antiporter superfamily in Triticum aestivum L. and functional analysis of TaNHX4-B. Genomics 2020, 112, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, X.; Wu, B.; Yuan, X.; Zhang, H.; Cui, X.; Liu, X. Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agric. 2015, 14, 1171–1183. [Google Scholar] [CrossRef]
- Hima Kumari, P.; Anil Kumar, S.; Ramesh, K.; Sudhakar Reddy, P.; Nagaraju, M.; Bhanu Prakash, A.; Shah, T.; Henderson, A.; Srivastava, R.K.; Rajasheker, G.; et al. Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in Sorghum bicolor. Genes 2018, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Kaur, K.; Khare, T.; Srivastava, A.K.; Suprasanna, P.; Kumar, V. Genome-wide identification, characterization and transcriptional profiling of NHX-type (Na+/H+) antiporters under salinity stress in soybean. 3 Biotech 2021, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Phang, T.-H.; Shao, G.; Lam, H.-M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, M.-W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Kay-Yuen Yim, A.; Tao, Y.; Wong, F.-L.; et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Qu, Y.; Guo, Y.; Yu, L.; Liu, Y.; Jiang, J.; Chen, J.; Ren, Y.; Liu, G.; Tian, L.; et al. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014, 80, 937–950. [Google Scholar] [CrossRef]
- Do, T.D.; Chen, H.; Hien, V.T.T.; Hamwieh, A.; Yamada, T.; Sato, T.; Yan, Y.; Cong, H.; Shono, M.; Suenaga, K.; et al. Ncl synchronously regulates Na+, K+, and Cl− in soybean and greatly increases the grain yield in saline field conditions. Sci. Rep. 2016, 6, 19147. [Google Scholar] [CrossRef]
- Patil, G.; Do, T.; Vuong, T.D.; Valliyodan, B.; Lee, J.-D.; Chaudhary, J.; Shannon, J.G.; Nguyen, H.T. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 2016, 6, 19199. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, L.; Qu, Y.; Chen, J.; Liu, X.; Hong, H.; Liu, Z.; Chang, R.; Gilliham, M.; Qiu, L.; et al. GmSALT3, which confers improved soybean salt tolerance in the field, increases leaf Cl− exclusion prior to Na+ exclusion but does not improve early vigor under salinity. Front. Plant Sci. 2016, 7, 1485. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Guan, R.; Bose, J.; Henderson, S.W.; Wege, S.; Qiu, L.; Gilliham, M. Soybean CHX-type ion transport protein GmSALT3 confers leaf Na+ exclusion via a root derived mechanism, and Cl− exclusion via a shoot derived process. Plant Cell Environ. 2021, 44, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Guan, R.; Yu, L.; Berkowitz, O.; David, R.; Whelan, J.; Ford, M.; Wege, S.; Qiu, L.; Gilliham, M. Enhanced reactive oxygen detoxification occurs in salt-stressed soybean roots expressing GmSALT3. Physiol. Plant. 2022, 174, e13709. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Lyle, C.; Jiang, G.; Penumala, A. Soybean salt tolerance 1 (GmST1) reduces ROS production, enhances ABA sensitivity, and abiotic stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 445. [Google Scholar] [CrossRef]
- Guan, R.; Yu, L.; Liu, X.; Li, M.; Chang, R.; Gilliham, M.; Qiu, L. Selection of the salt tolerance gene GmSALT3 during six decades of soybean breeding in china. Front. Plant Sci. 2021, 12, 794241. [Google Scholar] [CrossRef]
- Jia, Q.; Li, M.-W.; Zheng, C.; Xu, Y.; Sun, S.; Li, Z.; Wong, F.-L.; Song, J.; Lin, W.-W.; Li, Q.; et al. The soybean plasma membrane-localized cation/H+ exchanger GmCHX20a plays a negative role under salt stress. Physiol. Plant. 2021, 171, 714–727. [Google Scholar] [CrossRef]
- Jia, B.; Sun, M.; DuanMu, H.; Ding, X.; Liu, B.; Zhu, Y.; Sun, X. GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci. Rep. 2017, 7, 9423. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, D.; Bai, Y.; Wang, N.; Wang, Y. Studies on the overexpression of the soybean GmNHX1 in lotus corniculatus: The reduced Na+ level is the basis of the increased salt tolerance. Chin. Sci. Bull. 2006, 51, 1306–1315. [Google Scholar] [CrossRef]
- Li, W.-Y.F.; Wong, F.-L.; Tsai, S.-N.; Phang, T.-H.; Shao, G.; Lam, H.-M. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 2006, 29, 1122–1137. [Google Scholar] [CrossRef]
- Wang, M.; Hou, W.; Wang, Q.; Lam, H.; Han, T. Enhancing salt tolerance of soybean roots by overexpression of GmNHX1. Soybean Sci. 2011, 30, 889–894. [Google Scholar]
- Nie, W.-X.; Xu, L.; Yu, B.-J. A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma 2015, 252, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-J.; Fan, L.; Yang, J.; Cao, R.-Z.; Yang, C.-Y.; Zhang, J.; Wang, D.-M. A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na+ efflux rate and K+/Na+ ratio in Arabidopsis. BMC Plant Biol. 2019, 19, 469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cao, J.; Zhang, T.; Xu, T.; Yang, L.; Li, X.; Ji, F.; Gao, Y.; Ali, S.; Zhang, Q.; et al. A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front. Plant Sci. 2022, 13, 870695. [Google Scholar] [CrossRef]
- Jin, T.; An, J.; Xu, H.; Chen, J.; Pan, L.; Zhao, R.; Wang, N.; Gai, J.; Li, Y. A soybean sodium/hydrogen exchanger GmNHX6 confers plant alkaline salt tolerance by regulating Na+/K+ homeostasis. Front. Plant Sci. 2022, 13, 938635. [Google Scholar] [CrossRef]
- Sun, T.; Ma, N.; Wang, C.; Fan, H.; Wang, M.; Zhang, J.; Cao, J.; Wang, D. A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 2021, 12, 638340. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.X.; Shen, Y.; Zhou, W.H.; Jia, B.W.; Sun, M.Z.; Wang, J.Y.; Yang, J.K.; Li, J.W.; Sun, X.L. Whole genome wide identification of soybean CHX family genes and bioinformatics analysis. Genomics Appl. Biol. 2018, 37, 5360–5369. [Google Scholar]
- Jia, Q.; Xiao, Z.-X.; Wong, F.-L.; Sun, S.; Liang, K.-J.; Lam, H.-M. Genome-wide analyses of the soybean F-Box gene family in response to salt stress. Int. J. Mol. Sci. 2017, 18, 818. [Google Scholar] [CrossRef]
- Filichkin, S.; Priest, H.D.; Megraw, M.; Mockler, T.C. Alternative splicing in plants: Directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 2015, 24, 125–135. [Google Scholar] [CrossRef]
- Sze, H.; Padmanaban, S.; Cellier, F.; Honys, D.; Cheng, N.-H.; Bock, K.W.; Conéjéro, G.; Li, X.; Twell, D.; Ward, J.M.; et al. Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 2004, 136, 2532–2547. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.; Li, Y.; Liu, Y.; Yu, J.; Ma, X.; Sun, S.; Yang, N.; Xia, R.; Lei, T.; et al. Hierarchically aligning 10 legume genomes establishes a family-level genomics platform. Plant Physiol. 2017, 174, 284–300. [Google Scholar] [CrossRef]
- Carmel, L.; Wolf, Y.I.; Rogozin, I.B.; Koonin, E.V. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007, 17, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics 2014, 15, 950. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.-W.; Wong, F.-L.; Yung, W.-S.; Ku, Y.-S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.-Y.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Jia, Q.; Sun, S.; Kong, D.; Song, J.; Wu, L.; Yan, Z.; Zuo, L.; Yang, Y.; Liang, K.; Lin, W.; et al. Ectopic expression of Gs5PTase8, a soybean inositol polyphosphate 5-phosphatase, enhances salt tolerance in plants. Int. J. Mol. Sci. 2020, 21, 1023. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Higgs, D.; Murillo-Amador, B.; Aydemir, S.; Girgin, A.R. Silicon improves salinity tolerance in wheat plants. Environ. Exp. Bot. 2007, 62, 10–16. [Google Scholar] [CrossRef]
- Leng, Z.-X.; Liu, Y.; Chen, Z.-Y.; Guo, J.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; Cui, X.-Y. Genome-side analysis of the DUF4228 family in soybean and functional identification of GmDUF4228 -70 in response to drought and salt stresses. Front. Plant Sci. 2021, 12, 628299. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.-S.; Chan, L.-Y.; Law, C.-H.; Li, M.-W.; Lam, H.-M. Twenty years of mining salt tolerance genes in soybean. Mol. Breed. 2023, 43, 45. [Google Scholar] [CrossRef]
- Zhang, Y.; Berman, A.; Shani, E. Plant hormone transport and localization: Signaling molecules on the move. Annu. Rev. Plant Biol. 2023, 74, 453–479. [Google Scholar] [CrossRef]
- Mulet, J.M.; Porcel, R.; Yenush, L. Modulation of potassium transport to increase abiotic stress tolerance in plants. J. Exp. Bot. 2023, 74, 5989–6005. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H.; Meng, J.L. MapDraw:A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 2003, 25, 317–321. [Google Scholar]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yim, A.K.-Y.; Wong, J.W.-H.; Ku, Y.-S.; Qin, H.; Chan, T.-F.; Lam, H.-M. Using RNA-Seq data to evaluate reference genes suitable for gene expression studies in soybean. PLoS ONE 2015, 10, e0136343. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Qian, X.; Chen, C.; Cheng, S.; Jia, B.; Zhu, Y.; Sun, X. Ectopic expression of GsSRK in Medicago sativa reveals its involvement in plant architecture and salt stress responses. Front. Plant Sci. 2018, 9, 226. [Google Scholar] [CrossRef]
- Wang, K.; Zhong, M.; Wu, Y.-H.; Bai, Z.-Y.; Liang, Q.-Y.; Liu, Q.-L.; Pan, Y.-Z.; Zhang, L.; Jiang, B.-B.; Jia, Y.; et al. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Q.; Song, J.; Zheng, C.; Fu, J.; Qin, B.; Zhang, Y.; Liu, Z.; Jia, K.; Liang, K.; Lin, W.; et al. Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response. Int. J. Mol. Sci. 2023, 24, 16560. https://doi.org/10.3390/ijms242316560
Jia Q, Song J, Zheng C, Fu J, Qin B, Zhang Y, Liu Z, Jia K, Liang K, Lin W, et al. Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response. International Journal of Molecular Sciences. 2023; 24(23):16560. https://doi.org/10.3390/ijms242316560
Chicago/Turabian StyleJia, Qi, Junliang Song, Chengwen Zheng, Jiahui Fu, Bin Qin, Yongqiang Zhang, Zhongjuan Liu, Kunzhi Jia, Kangjing Liang, Wenxiong Lin, and et al. 2023. "Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response" International Journal of Molecular Sciences 24, no. 23: 16560. https://doi.org/10.3390/ijms242316560
APA StyleJia, Q., Song, J., Zheng, C., Fu, J., Qin, B., Zhang, Y., Liu, Z., Jia, K., Liang, K., Lin, W., & Fan, K. (2023). Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response. International Journal of Molecular Sciences, 24(23), 16560. https://doi.org/10.3390/ijms242316560





