The Protective Effects of Reineckia carnea Ether Fraction against Alzheimer’s Disease Pathology: An Exploration in Caenorhabditis elegans Models
Abstract
:1. Introduction
2. Results
2.1. Identification of Bioactive Constituents in R. carnea
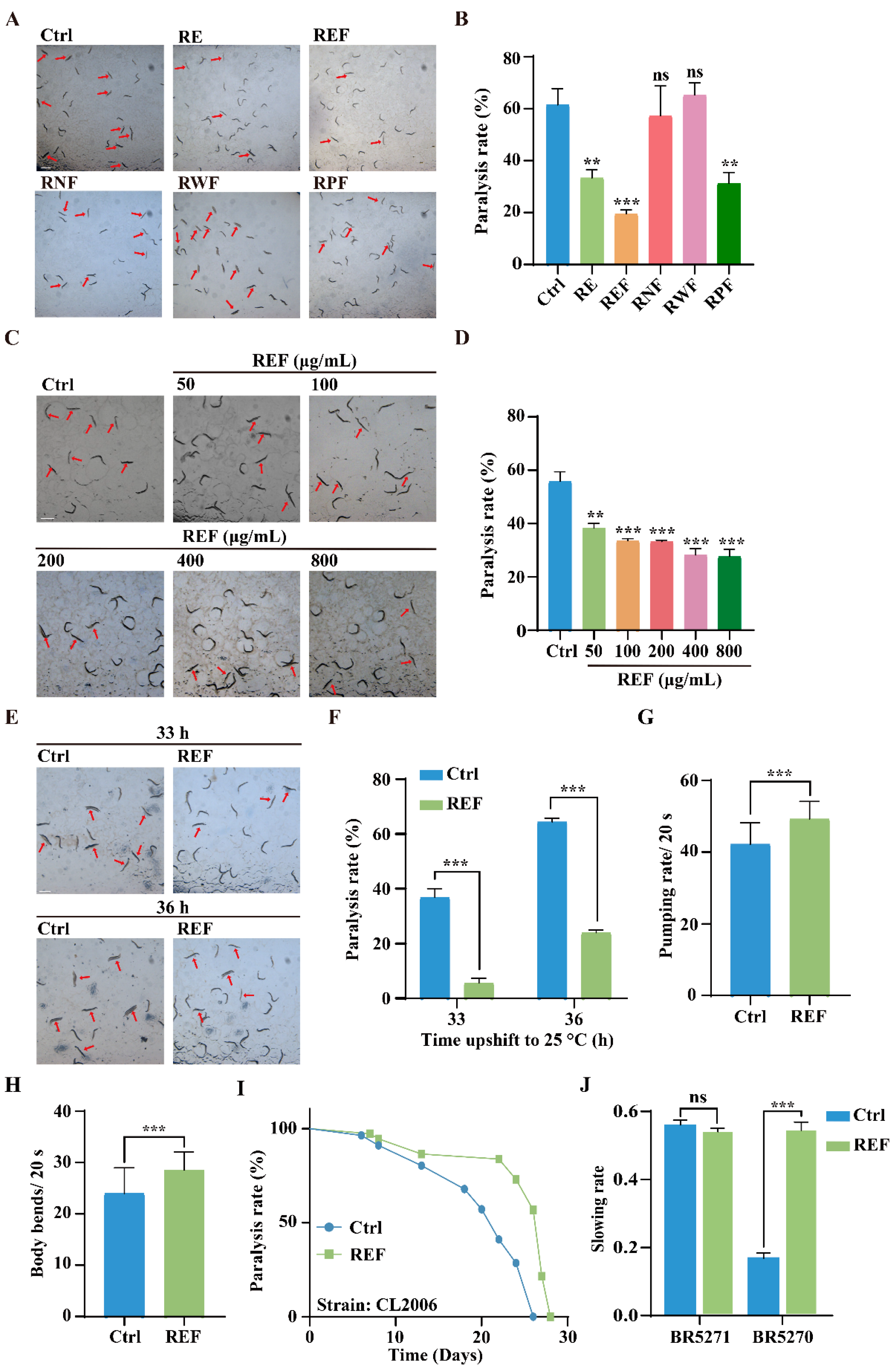
2.2. REF Inhibits Aβ-Induced Paralysis and Tau-Induced Neurotoxicity in C. elegans
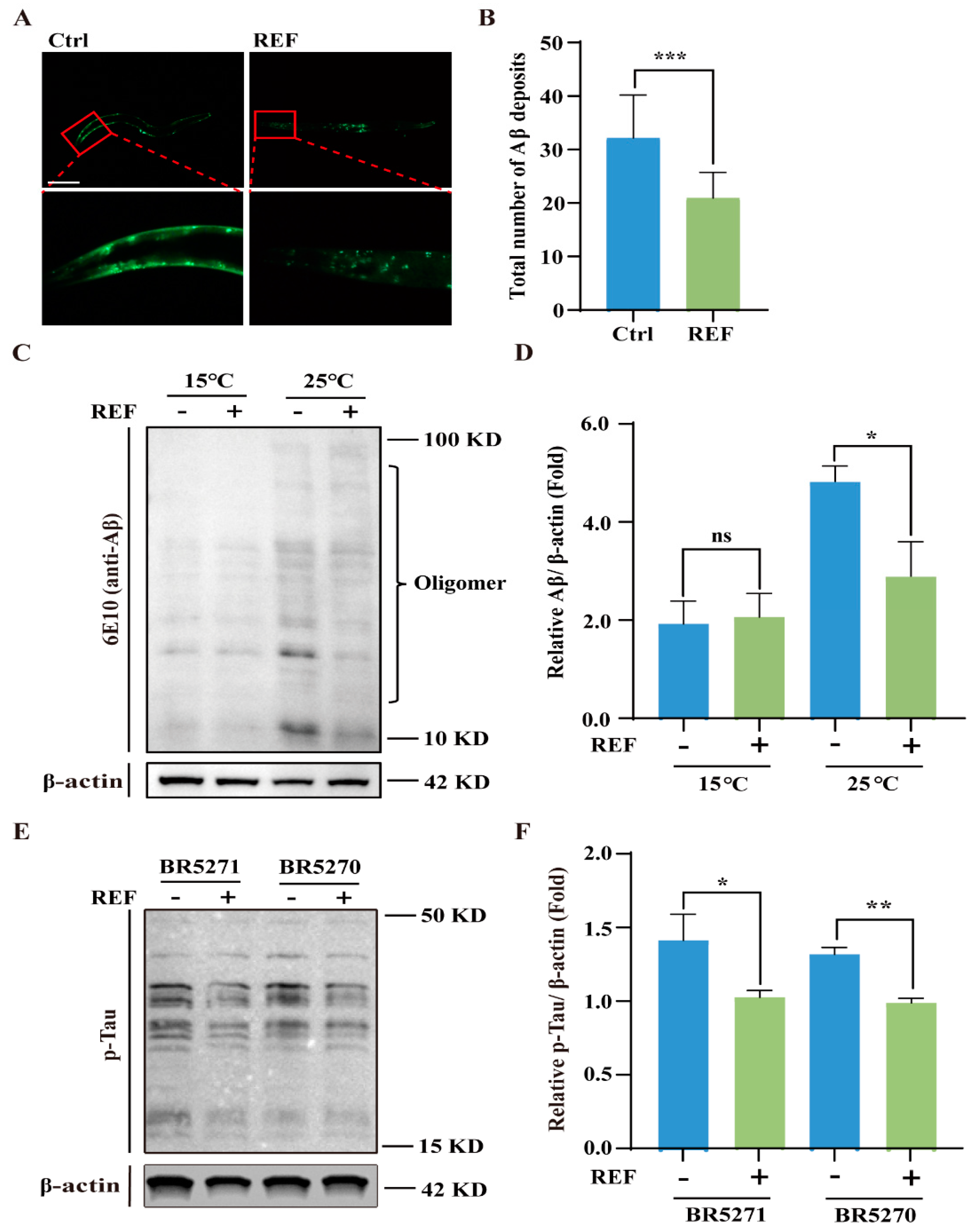
2.3. REF Diminishes the Accumulation of Aβ and pTau Aggregates in C. elegans
2.4. REF Mitigates Oxidative Stress Triggered by Aβ and Tau Aggregates in C. elegans
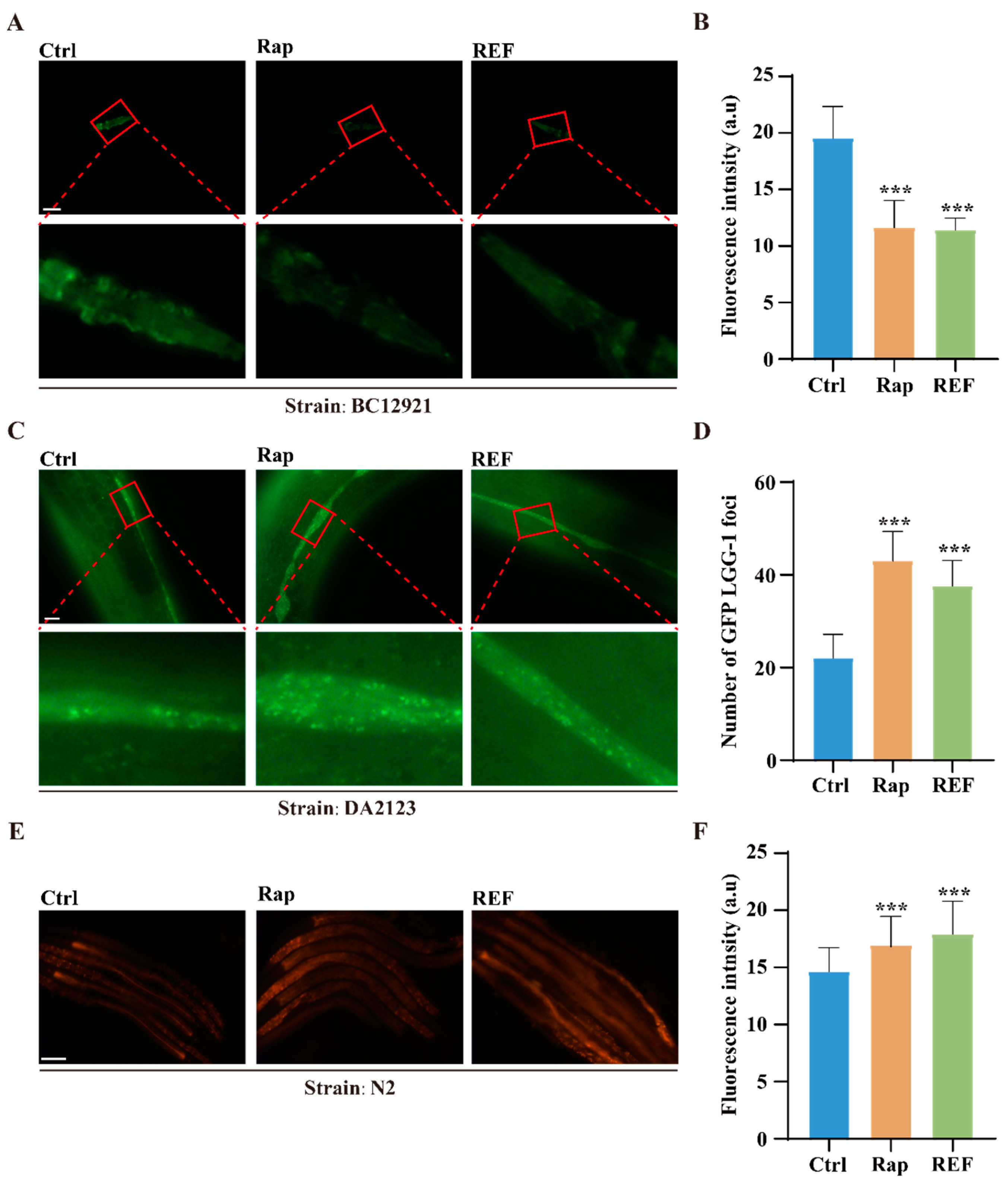
2.5. REF Activates Autophagy in C. elegans and U87 Cells
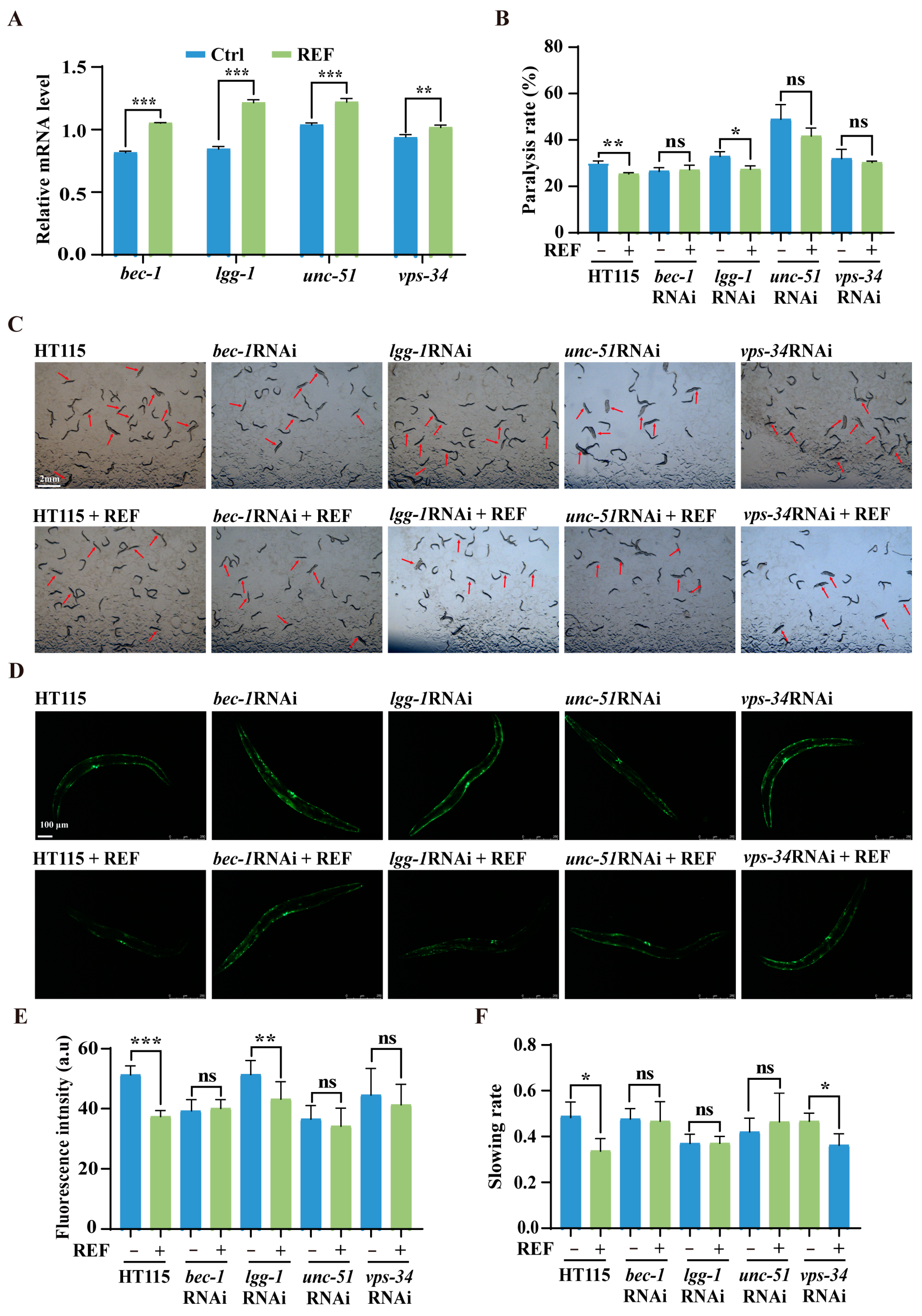
2.6. REF Alleviates Aβ- and Tau-Induced Neurotoxicity via Autophagy Activation in C. elegans
3. Discussion
4. Methods and Materials
4.1. Extraction and Fractionation of R. carnea
4.2. UPLC/Q-TOF-MS/MS Conditions
4.3. Maintenance and Synchronization of C. elegans Strains
4.4. Paralysis Assay
4.5. Measurement of Aβ(3–42) Aggregation in CL2331 Worms
4.6. Western Blot Analysis for Aβ and pTau Levels and Antibodies
4.7. Assessment of Food-Sensing Behavior
4.8. Assessment of ROS Levels and Oxidative Stress in C. elegans
4.9. Gene Silencing through RNAi
4.10. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.11. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| Aβ | β-amyloid |
| pTau | Hyperphosphorylated Tau |
| TCMs | Traditional Chinese medicines |
| C. elegans | Caenorhabditis elegans |
| NDs | Neurodegenerative diseases |
| RE | R. carnea methanol extract |
| REF | R. carnea ether fraction |
| RNAi | RNA interference |
| CGC | Caenorhabditis Genetics Center |
| NGM | Nematode growth medium |
| FUDR | 5-fluoro-2′-deoxyuridine |
| IPTG | Isoprophylthiogalactoside |
| qRT-PCR | Quantitative Real-Time PCR |
| SD | Standard deviation |
| DHE | Dihydroethidium |
References
- Salmon, D.P.; Bondi, M.W. Neuropsychological assessment of dementia. Annu. Rev. Psychol. 2009, 60, 257–282. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Xie, C.; Schenkel, J.A.; Wu, C.; Long, Q.; Cui, H.; Aman, Y.; Frank, J.; Liao, J.; Zou, H. A research agenda for ageing in China in the 21st century: Focusing on basic and translational research, long-term care, policy and social networks. Ageing Res. Rev. 2020, 64, 101174. [Google Scholar] [CrossRef]
- Yiannopoulou, K.G.; Papageorgiou, S.G. Current and future treatments in Alzheimer disease: An update. J. Cent. Nerv. Syst. Dis. 2020, 12, 1179573520907397. [Google Scholar] [CrossRef]
- Joe, E.; Ringman, J.M. Cognitive symptoms of Alzheimer’s disease: Clinical management and prevention. BMJ 2019, 367, l6217. [Google Scholar] [CrossRef]
- Shao, L.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar]
- Long, T.; Tang, Y.; He, Y.-N.; He, C.-L.; Chen, X.; Guo, M.-S.; Wu, J.-M.; Yu, L.; Yu, C.-L.; Yuen-Kwan Law, B. Citri Reticulatae Semen extract promotes healthy aging and neuroprotection via autophagy induction in Caenorhabditis elegans. J. Gerontol. Ser. A 2022, 77, 2186–2194. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.W.; Murugaiyah, V.; Najimudin, N.; Azzam, G. Danshen (Salvia miltiorrhiza) water extract shows potential neuroprotective effects in Caenorhabditis elegans. J. Ethnopharmacol. 2021, 266, 113418. [Google Scholar] [CrossRef]
- Ping-Sheng, H.; Lin-Chu, L. Karyotype Analysis of Reineckia carnea (Andr.) Kunth (Liliaceae). J. Syst. Evol. 1984, 22, 128. [Google Scholar]
- Jian-qiong, Y.; Ye, W.; Chen, Y.; Nan-nan, W.; Xiao-yan, H. Chemical Constituents from Reineckia carnea Kunth. Nat. Prod. Res. Dev. 2010, 22, 245–247. [Google Scholar]
- Wang, J.; Han, N.; Wang, Y.; Wang, Y.; Liu, Z.; Wang, Y.; Yin, J. Three alkaloids from Reineckia carnea herba and their antitussive and expectorant activities. Nat. Prod. Res. 2014, 28, 1306–1309. [Google Scholar] [CrossRef]
- Xu, X.; Tan, T.; Zhang, J.; Li, Z.-F.; Yang, S.-L.; Wen, Q.; Feng, Y.-L. Isolation of chemical constituents with anti-inflammatory activity from Reineckia carnea herbs. J. Asian Nat. Prod. Res. 2020, 22, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, M.; Li, J.; Shi, W.; Wang, H.; Zhao, C. Analysis of fatty acids, aliphatic esters, and in vitro studies of antioxidant and antimicrobial activities for recineckea carnea and tupistra chinensis from the guizhou province. J. Med. Food 2014, 17, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, W.; He, H.; Fu, X.; Wang, J.; Zou, K.; Chen, J. Growth inhibition and apoptosis-inducing effect on human cancer cells by RCE-4, a spirostanol saponin derivative from natural medicines. Int. J. Mol. Med. 2013, 31, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Chen, L.L.; Wang, Y.; Xue, R.; Zou, L.B.; Liu, F.; Yin, J. Steroidal glycosides from Reineckia carnea herba and their antitussive activity. Planta Medica 2013, 79, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Rani, N.; Alam, M.M.; Jamal, A.; Ghaffar, O.B.; Parvez, S. Caenorhabditis elegans: A transgenic model for studying age-associated neurodegenerative diseases. Ageing Res. Rev. 2023, 2023, 102036. [Google Scholar] [CrossRef]
- Meneely, P.M.; Dahlberg, C.L.; Rose, J.K. Working with worms: Caenorhabditis elegans as a model organism. Curr. Protoc. Essent. Lab. Tech. 2019, 19, e35. [Google Scholar] [CrossRef]
- Helmcke, K.J.; Avila, D.S.; Aschner, M. Utility of Caenorhabditis elegans in high throughput neurotoxicological research. Neurotoxicology Teratol. 2010, 32, 62–67. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, Z.; Butko, P.; Christen, Y.; Lambert, M.P.; Klein, W.L.; Link, C.D.; Luo, Y. Amyloid-β-induced pathological behaviors are suppressed by Ginkgo biloba extract EGb 761 and ginkgolides in transgenic Caenorhabditis elegans. J. Neurosci. 2006, 26, 13102–13113. [Google Scholar] [CrossRef]
- Drake, J.; Link, C.D.; Butterfield, D.A. Oxidative stress precedes fibrillar deposition of Alzheimer’s disease amyloid β-peptide (1–42) in a transgenic Caenorhabditis elegans model. Neurobiol. Aging 2003, 24, 415–420. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, H.; Zhao, C.; Gong, X.; Zhang, J. Studies on chemical constituents of herbs of Reineckia carnea. Zhongguo Zhong Yao Za Zhi= Zhongguo Zhongyao Zazhi= China J. Chin. Mater. Medica 2008, 33, 2793–2796. [Google Scholar]
- He, C.-L.; Tang, Y.; Chen, X.; Long, T.; He, Y.-N.; Wei, J.; Wu, J.-M.; Lan, C.; Yu, L.; Huang, F.-H. Folium Hibisci Mutabilis extract, a potent autophagy enhancer, exhibits neuroprotective properties in multiple models of neurodegenerative diseases. Phytomedicine 2023, 109, 154548. [Google Scholar] [CrossRef] [PubMed]
- Panza, F.; Lozupone, M.; Logroscino, G.; Imbimbo, B.P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Matis, I.; Delivoria, D.C.; Mavroidi, B.; Papaevgeniou, N.; Panoutsou, S.; Bellou, S.; Papavasileiou, K.D.; Linardaki, Z.I.; Stavropoulou, A.V.; Vekrellis, K. An integrated bacterial system for the discovery of chemical rescuers of disease-associated protein misfolding. Nat. Biomed. Eng. 2017, 1, 838–852. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.N.; Singh, C.; Singh, A.; Singh, M.; Singh, B.K. Mitochondrial dysfunction: A potential therapeutic target to treat Alzheimer’s disease. Mol. Neurobiol. 2020, 57, 3075–3088. [Google Scholar] [CrossRef]
- Li, Q.; Liu, Y.; Sun, M. Autophagy and Alzheimer’s disease. Cell. Mol. Neurobiol. 2017, 37, 377–388. [Google Scholar] [CrossRef]
- He, C.-L.; Tang, Y.; Wu, J.-M.; Long, T.; Yu, L.; Teng, J.-F.; Qiu, W.-Q.; Pan, R.; Yu, C.-L.; Qin, D.-L. Chlorogenic acid delays the progression of Parkinson’s disease via autophagy induction in Caenorhabditis elegans. Nutr. Neurosci. 2023, 26, 11–24. [Google Scholar] [CrossRef]
- Artal-Sanz, M.; de Jong, L.; Tavernarakis, N. Caenorhabditis elegans: A versatile platform for drug discovery. Biotechnol. J. Healthc. Nutr. Technol. 2006, 1, 1405–1418. [Google Scholar] [CrossRef]
- Giunti, S.; Andersen, N.; Rayes, D.; De Rosa, M.J. Drug discovery: Insights from the invertebrate Caenorhabditis elegans. Pharmacol. Res. Perspect. 2021, 9, e00721. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Locatelli, A.G.; Cenci, S. Autophagy and longevity: Evolutionary hints from hyper-longevous mammals. Front. Endocrinol. 2022, 13, 1085522. [Google Scholar] [CrossRef]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Haas, M.; Yeo, S.K.; Sebti, S.; Fernández, Á.F.; Zou, Z.; Levine, B.; Guan, J.-L. Enhanced autophagy in Becn1F121A/F121A knockin mice counteracts aging-related neural stem cell exhaustion and dysfunction. Autophagy 2022, 18, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wei, Y.; Lautrup, S.; Yang, B.; Wang, Y.; Cordonnier, S.; Mattson, M.P.; Croteau, D.L.; Bohr, V.A. NAD+ supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer’s disease via cGAS–STING. Proc. Natl. Acad. Sci. USA 2021, 118, e2011226118. [Google Scholar] [CrossRef]
- Wang, H.; Fu, J.; Xu, X.; Yang, Z.; Zhang, T. Rapamycin activates mitophagy and alleviates cognitive and synaptic plasticity deficits in a mouse model of Alzheimer’s disease. J. Gerontol. Ser. A 2021, 76, 1707–1713. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, S.; Zhang, X.; Li, T.; Tang, Y.; Liu, H.; Yang, W.; Le, W. Autophagy enhancer carbamazepine alleviates memory deficits and cerebral amyloid-β pathology in a mouse model of Alzheimer’s disease. Curr. Alzheimer Res. 2013, 10, 433–441. [Google Scholar] [CrossRef]
- Long, T.; Chen, X.; Qin, D.L.; Zhu, Y.F.; Zhou, Y.J.; He, Y.N.; Fu, H.J.; Tang, Y.; Yu, L.; Huang, F.H. Ameliorative effect of Luffa cylindrica fruits on Caenorhabditis elegans and cellular models of Alzheimer’s disease-related pathology via autophagy induction. Phytother. Res. 2023, 37, 4639–4654. [Google Scholar] [CrossRef]
- Sangha, J.S.; Sun, X.; Wally, O.S.; Zhang, K.; Ji, X.; Wang, Z.; Wang, Y.; Zidichouski, J.; Prithiviraj, B.; Zhang, J. Liuwei Dihuang (LWDH), a traditional Chinese medicinal formula, protects against β-amyloid toxicity in transgenic Caenorhabditis elegans. PLoS ONE 2012, 7, e43990. [Google Scholar] [CrossRef]
- Long, T.; Chen, X.; Zhang, Y.; Zhou, Y.-J.; He, Y.-N.; Zhu, Y.-F.; Fu, H.-J.; Yu, L.; Yu, C.-L.; Law, B.Y.-K. Protective effects of Radix Stellariae extract against Alzheimer’s disease via autophagy activation in Caenorhabditis elegans and cellular models. Biomed. Pharmacother. 2023, 165, 115261. [Google Scholar] [CrossRef]
- Zhu, F.-D.; Chen, X.; Yu, L.; Hu, M.-L.; Pan, Y.-R.; Qin, D.-L.; Wu, J.-M.; Li, L.; Law, B.Y.-K.; Wong, V.K.-W. Targeting autophagy to discover the Piper wallichii petroleum ether fraction exhibiting antiaging and anti-Alzheimer’s disease effects in Caenorhabditis elegans. Phytomedicine 2023, 117, 154916. [Google Scholar] [CrossRef]
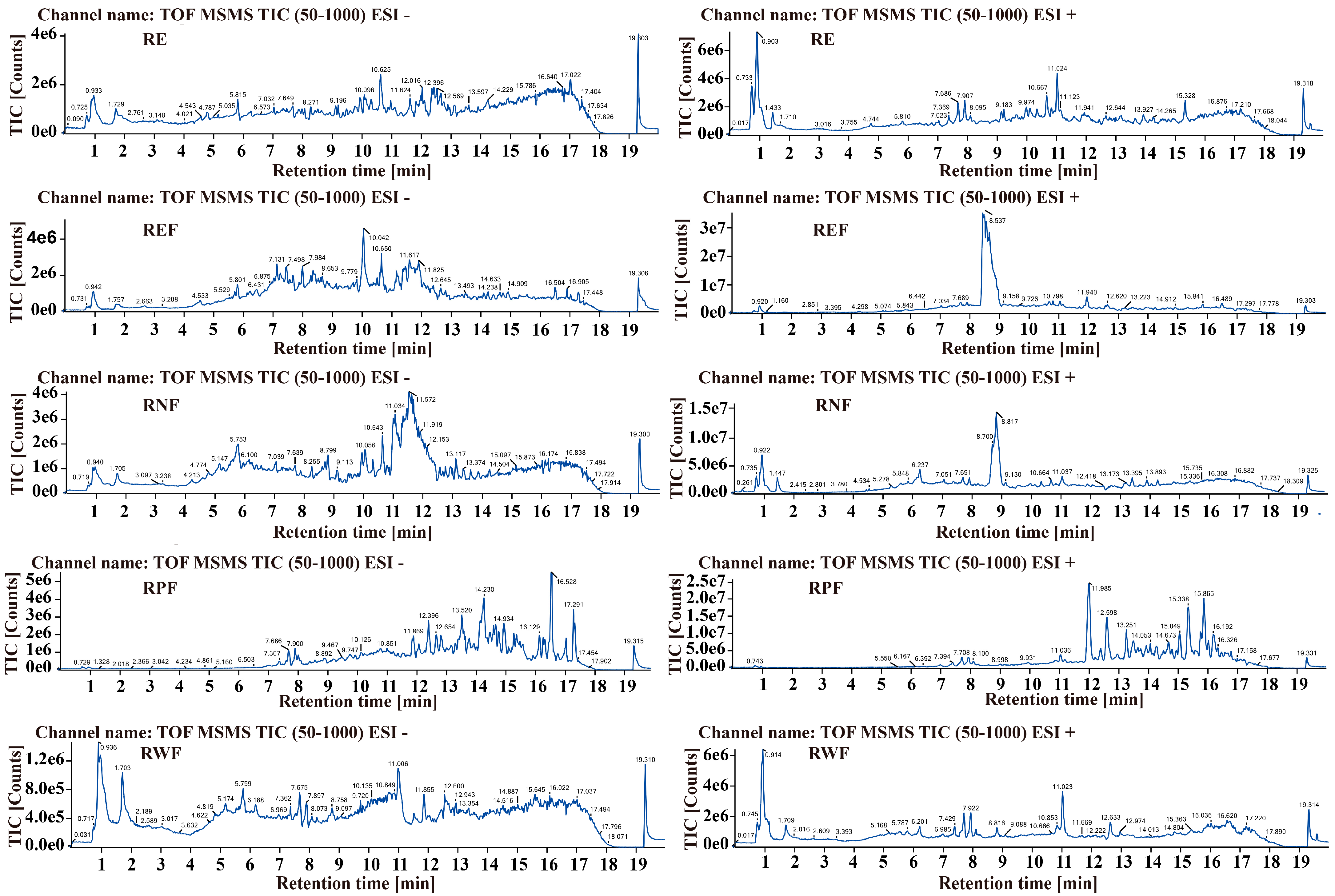

| Chemical Name | Retention Time (min) | Chemical Formula | Expected Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Distribution |
|---|---|---|---|---|---|---|
| Coumarin | 5.23 | C9H6O2 | 181.0121 | 181.0162 | 0.0041 | REF |
| L(+)-Arginine | 0.84 | C6H14N4O2 | 175.1159 | 175.1167 | 0.0008 | |
| Vitamin C | 1.86 | C6H8O6 | 175.021 | 175.0262 | 0.0052 | |
| L-Cystathionine | 4.18 | C7H14N2O4S | 257.0379 | 257.0311 | −0.0068 | |
| Palmitoleic acid | 11.16 | C16H30O2 | 277.2131 | 277.2162 | 0.0031 | |
| Genistein | 7.98 | C15H10O5 | 269.0423 | 269.0479 | 0.0056 | |
| Dammaradienol | 17.67 | C30H50O | 427.3741 | 427.3784 | 0.0043 | |
| Cianidanol | 1.74 | C15H14O6 | 289.0651 | 289.0682 | 0.0031 | RNF |
| Lauric acid | 10.77 | C12H24O2 | 223.1639 | 223.1691 | 0.0052 | RPF |
| Palmitic acid | 13.42 | C16H32O2 | 279.229 | 279.2282 | −0.0008 | |
| Thymidine | 1.78 | C10H14N2O5 | 277.0576 | 277.0587 | 0.0011 | RWF |
| D-Erythro-Sphingosine | 12.93 | C18H37NO2 | 300.2855 | 300.2897 | 0.0042 | |
| Gibberellin A38 | 9.57 | C20H26O6 | 397.1512 | 397.1512 | 0 | |
| D-Xylulose | 1.06 | C5H10O5 | 149.0427 | 149.0473 | 0.0046 | RE |
| Guanine | 1.78 | C5H5N5O | 152.0598 | 152.0571 | −0.0027 | |
| Guanosine | 1.78 | C10H13N5O5 | 284.1027 | 284.1005 | −0.0022 | |
| Gibberellin A64 | 8.97 | C20H26O5 | 345.1794 | 345.1732 | −0.0062 | |
| D-beta-Tocotrienol | 9.64 | C28H42O2 | 433.3259 | 433.3241 | −0.0018 | |
| [12]-Gingerol | 7.59 | C23H38O4 | 401.2617 | 401.2614 | −0.0003 | |
| Torvoside A | 8.46 | C45H76O18 | 903.4658 | 903.4636 | −0.0022 | |
| DL-2-Aminoadipic aicd | 1.63 | C6H11NO4 | 184.0653 | 184.0609 | −0.0044 | REF, RE |
| D-erythro-C18-Dihydro-D-sphingosine | 15.7 | C18H39NO2 | 324.2835 | 324.2899 | 0.0064 | |
| 2″-O-alpha-L-Rhamnopyranoside-Orientin | 6.23 | C27H30O15 | 595.1647 | 595.1658 | 0.0011 | RWF, RE |
| Adenosine | 1.55 | C10H13N5O4 | 268.1022 | 268.1044 | 0.0022 | |
| Ganoderenic acid E | 9.7 | C30H40O8 | 527.2629 | 527.2688 | 0.0059 | RNF, RE |
| Capsicoside C2 | 9.12 | C44H72O17 | 907.4485 | 907.4495 | 0.001 | |
| trans-Traumatic acid | 7.98 | C12H20O4 | 229.1411 | 229.1435 | 0.0024 | REF, RPF |
| (R)-(−)-Citramalic acid | 2.69 | C5H8O5 | 171.0253 | 171.0291 | 0.0038 | REF, RWF |
| Gallic acid | 4.65 | C7H6O5 | 169.0133 | 169.0161 | 0.0028 | |
| Homovanillic acid | 2.07 | C9H10O4 | 181.0514 | 181.0523 | 0.0009 | |
| Linolenic acid | 12.22 | C18H30O2 | 277.2133 | 277.2185 | 0.0052 | RWF, RPF |
| L-Valine | 0.95 | C5H11NO2 | 118.0857 | 118.0857 | 0 | RWF, RNF, RE |
| Solasodine | 17.32 | C27H43NO2 | 414.3526 | 414.3575 | 0.0049 | REF, RPF, RE |
| Decanoic acid | 9.74 | C10H20O2 | 195.1356 | 195.1384 | 0.0028 | |
| Phytosphingosine | 10.94 | C18H39NO3 | 318.2969 | 318.2999 | 0.003 | REF, RPF, RWF |
| pyridoxal | 5.35 | C8H9NO3 | 190.0485 | 190.0499 | 0.0014 | REF, RNF, RE |
| L-Phenylalanine | 1.86 | C9H11NO2 | 200.0547 | 200.0582 | 0.0035 | |
| Tryptamine | 6.6 | C10H12N2 | 183.0917 | 183.0915 | −0.0002 | |
| L-Leucine | 0.96 | C6H13NO2 | 132.101 | 132.1016 | 0.0006 | RWF, RNF, REF |
| Allantoic acid | 5.2 | C4H8N4O4 | 177.0534 | 177.0533 | −0.0001 | |
| L-Malic acid | 0.94 | C4H6O5 | 133.013 | 133.015 | 0.002 | REF, RNF, RWF, RE |
| Salicylic acid | 3.16 | C7H6O3 | 137.0233 | 137.0261 | 0.0028 | |
| Inositol | 0.92 | C6H12O6 | 179.0533 | 179.0579 | 0.0046 | |
| L-Citrulline | 0.93 | C6H13N3O3 | 198.0913 | 198.0979 | 0.0066 | RPF, REF, RNF, RWF |
| Hexadecanamide | 16.83 | C16H33NO | 256.2602 | 256.2634 | 0.0032 | RWF, RPF, RNF, RE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, H.-J.; Zhou, X.-Y.; Li, Y.-P.; Chen, X.; He, Y.-N.; Qin, D.-L.; Yu, L.; Yu, C.-L.; Wu, J.-M.; Wu, A.-G.; et al. The Protective Effects of Reineckia carnea Ether Fraction against Alzheimer’s Disease Pathology: An Exploration in Caenorhabditis elegans Models. Int. J. Mol. Sci. 2023, 24, 16536. https://doi.org/10.3390/ijms242216536
Fu H-J, Zhou X-Y, Li Y-P, Chen X, He Y-N, Qin D-L, Yu L, Yu C-L, Wu J-M, Wu A-G, et al. The Protective Effects of Reineckia carnea Ether Fraction against Alzheimer’s Disease Pathology: An Exploration in Caenorhabditis elegans Models. International Journal of Molecular Sciences. 2023; 24(22):16536. https://doi.org/10.3390/ijms242216536
Chicago/Turabian StyleFu, Hai-Jun, Xing-Yue Zhou, Ya-Ping Li, Xue Chen, Yan-Ni He, Da-Lian Qin, Lu Yu, Chong-Lin Yu, Jian-Ming Wu, An-Guo Wu, and et al. 2023. "The Protective Effects of Reineckia carnea Ether Fraction against Alzheimer’s Disease Pathology: An Exploration in Caenorhabditis elegans Models" International Journal of Molecular Sciences 24, no. 22: 16536. https://doi.org/10.3390/ijms242216536
APA StyleFu, H.-J., Zhou, X.-Y., Li, Y.-P., Chen, X., He, Y.-N., Qin, D.-L., Yu, L., Yu, C.-L., Wu, J.-M., Wu, A.-G., & Zhou, X.-G. (2023). The Protective Effects of Reineckia carnea Ether Fraction against Alzheimer’s Disease Pathology: An Exploration in Caenorhabditis elegans Models. International Journal of Molecular Sciences, 24(22), 16536. https://doi.org/10.3390/ijms242216536





