Dependency of B-Cell Acute Lymphoblastic Leukemia and Multiple Myeloma Cell Lines on MEN1 Extends beyond MEN1–KMT2A Interaction
Abstract
1. Introduction
2. Results
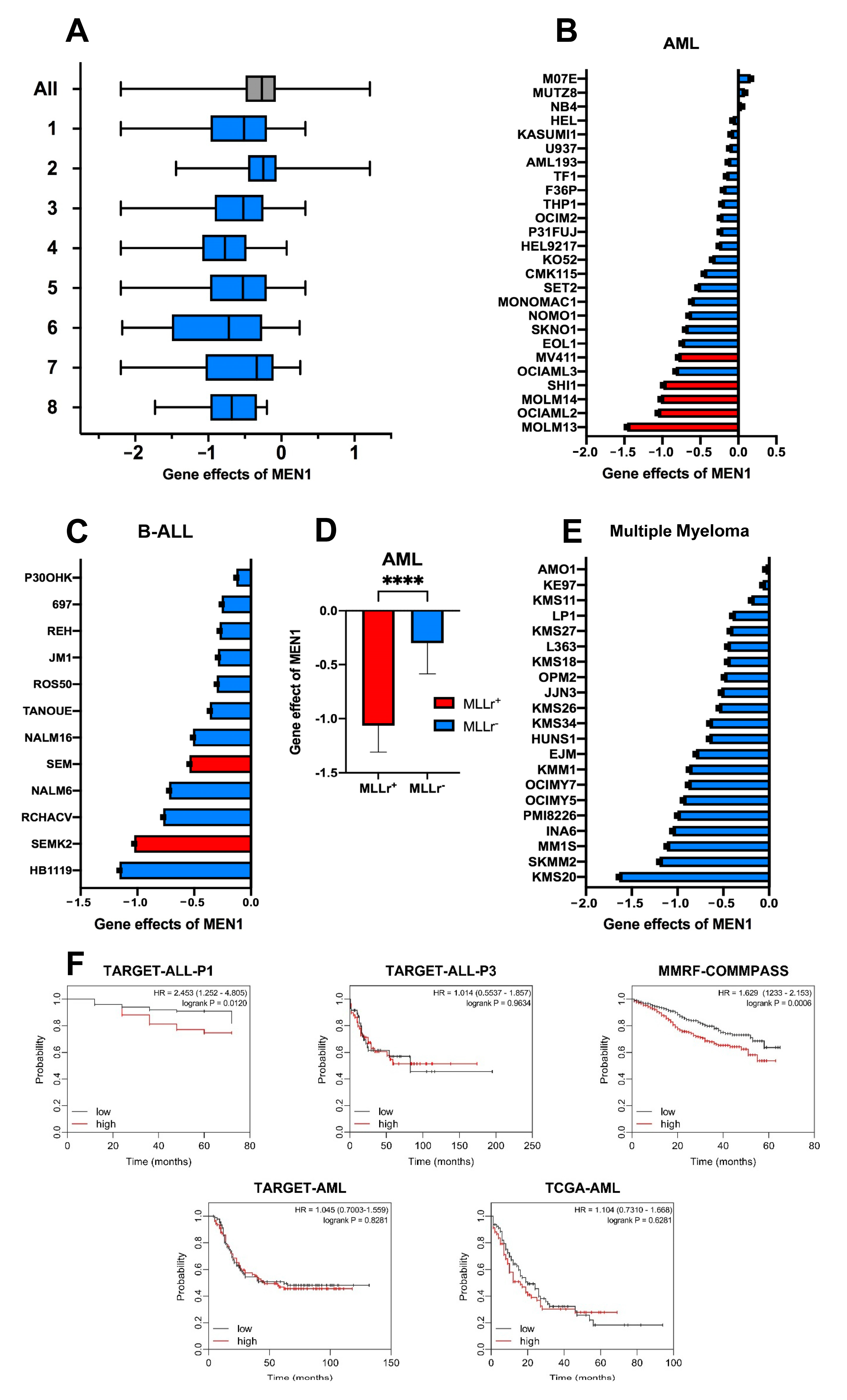
2.1. MEN1 Dependency Is not Unique to Neoplasms Harboring MLLr, NPM1m, or MN1m
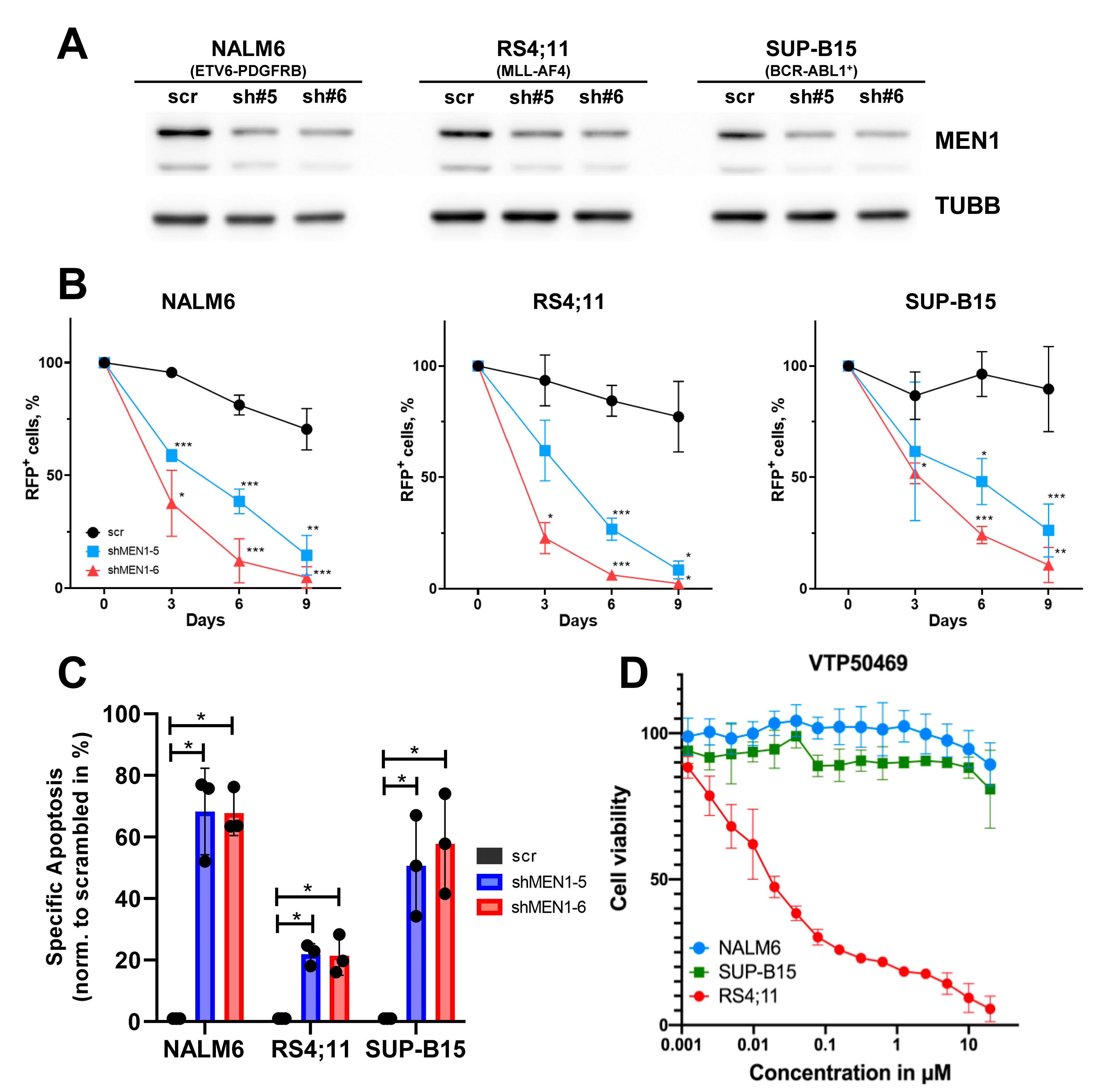
2.2. MEN1 Knockdown Decreases Fitness and Induces Apoptosis in B-ALL Cell Lines Independently of Mutational Background
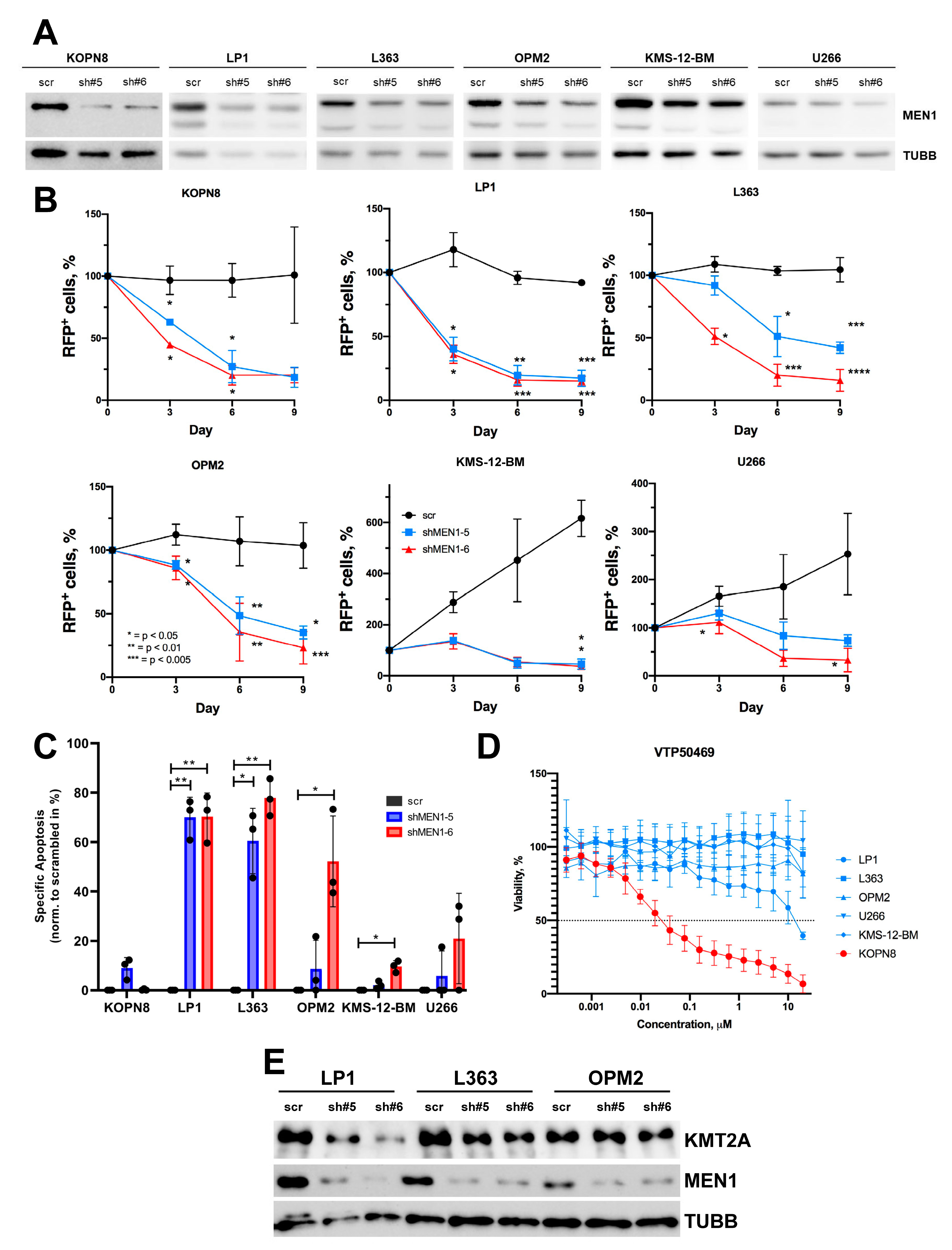
2.3. MEN1 Knockdown Decreases Fitness and Induces Apoptosis in MM Cell Lines
2.4. CRISPR/Cas9-Dependent Inactivation of MEN1 Decreases the Fitness of MM and B-ALL Cells Harboring Edited Alleles
3. Discussion
4. Materials and Methods
4.1. Cell lines and Culture Conditions
4.2. shRNA-Mediated Knockdown of MEN1
4.3. CRISPR/Cas9 Gene Editing and Analysis of the Dynamics of CRISPR-Induced InDels
4.4. Immunoblotting
4.5. Assessment of Apoptosis, Cell Viability, and Competitive Co-Culture Assay
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matkar, S.; Thiel, A.; Hua, X. Menin: A scaffold protein that controls gene expression and cell signaling. Trends Biochem. Sci. 2013, 38, 394–402. [Google Scholar] [CrossRef]
- Winters, A.C.; Bernt, K.M. MLL-Rearranged Leukemias—An Update on Science and Clinical Approaches. Front. Pediatr. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Evans, K.; Gadrey, J.Y.; Eschle, B.K.; Hatton, C.; Uckelmann, H.J.; Ross, K.N.; Perner, F.; Olsen, S.N.; Pritchard, T.; et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 2019, 36, 660–673.e11. [Google Scholar] [CrossRef]
- Cierpicki, T.; Grembecka, J. Challenges and opportunities in targeting the menin-MLL interaction. Future Med. Chem. 2014, 6, 447–462. [Google Scholar] [CrossRef]
- Ozyerli-Goknar, E.; Nizamuddin, S.; Timmers, H.T.M. A Box of Chemistry to Inhibit the MEN1 Tumor Suppressor Gene Promoting Leukemia. ChemMedChem 2021, 16, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Issa, G.C.; Ravandi, F.; DiNardo, C.D.; Jabbour, E.; Kantarjian, H.M.; Andreeff, M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia 2021, 35, 2482–2495. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, C.; Xie, H.M.; Kingsley, M.C.; Haladyna, J.N.; Riedel, S.S.; Alikarami, F.; Lenard, A.; McGeehan, G.M.; Ernst, P.; Bernt, K.M. Menin is necessary for long term maintenance of meningioma-1 driven leukemia. Leukemia 2021, 35, 1405–1417. [Google Scholar] [CrossRef]
- Uckelmann, H.J.; Armstrong, S.A. Chromatin Complexes Maintain Self-Renewal of Myeloid Progenitors in AML: Opportunities for Therapeutic Intervention. Stem Cell Rep. 2020, 15, 6–12. [Google Scholar] [CrossRef]
- Grieselhuber, N.R.; Mims, A.S. Novel Targeted Therapeutics in Acute Myeloid Leukemia: An Embarrassment of Riches. Curr. Hematol. Malig. Rep. 2021, 16, 192–206. [Google Scholar] [CrossRef]
- Swaminathan, M.; Bourgeois, W.; Armstrong, S.A.; Wang, E.S. Menin Inhibitors in Acute Myeloid Leukemia—What Does the Future Hold? Cancer J. 2022, 28, 62–66. [Google Scholar] [CrossRef]
- Stein, E.M.; Aldoss, I.; DiPersio, J.F.; Stone, R.M.; Arellano, M.L.; Rosen, G.; Meyers, M.L.; Huang, Y.; Smith, S.; Bagley, R.G.; et al. Safety and Efficacy of Menin Inhibition in Patients (Pts) with MLL-Rearranged and NPM1 Mutant Acute Leukemia: A Phase (Ph) 1, First-in-Human Study of SNDX-5613 (AUGMENT 101). Blood 2021, 138, 699. [Google Scholar] [CrossRef]
- AML Prognoses Better with Menin–MLL Inhibitor? Cancer Discov. 2021, 11, 216–217. [CrossRef]
- Svoboda, L.K.; Bailey, N.; Van Noord, R.A.; Krook, M.A.; Harris, A.; Cramer, C.; Jasman, B.; Patel, R.M.; Thomas, D.; Borkin, D.; et al. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget 2017, 8, 458–471. [Google Scholar] [CrossRef]
- Dreijerink, K.M.A.; Groner, A.C.; Vos, E.S.M.; Font-Tello, A.; Gu, L.; Chi, D.; Reyes, J.; Cook, J.; Lim, E.; Lin, C.Y.; et al. Enhancer-Mediated Oncogenic Function of the Menin Tumor Suppressor in Breast Cancer. Cell Rep. 2017, 18, 2359–2372. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Khan, A.P.; Asangani, I.A.; Cieślik, M.; Prensner, J.R.; Wang, X.; Iyer, M.K.; Jiang, X.; Borkin, D.; Escara-Wilke, J.; et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 2015, 21, 344–352. [Google Scholar] [CrossRef]
- Brzezinka, K.; Nevedomskaya, E.; Lesche, R.; Haegebarth, A.; Ter Laak, A.; Fernández-Montalván, A.E.; Eberspaecher, U.; Werbeck, N.D.; Moenning, U.; Siegel, S.; et al. Characterization of the Menin-MLL Interaction as Therapeutic Cancer Target. Cancers 2020, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Heuck, C.J.; Qu, P.; van Rhee, F.; Waheed, S.; Usmani, S.Z.; Epstein, J.; Zhang, Q.; Edmondson, R.; Hoering, A.; Crowley, J.; et al. Five gene probes carry most of the discriminatory power of the 70-gene risk model in multiple myeloma. Leukemia 2014, 28, 2410–2413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, C.; Wendlandt, E.B.; Darbro, B.; Xu, H.; Thomas, G.S.; Tricot, G.; Chen, F.; Shaughnessy, J.D.; Zhan, F. Genetic Analysis of Multiple Myeloma Identifies Cytogenetic Alterations Implicated in Disease Complexity and Progression. Cancers 2021, 13, 517. [Google Scholar] [CrossRef]
- Heikamp, E.B.; Henrich, J.A.; Perner, F.; Wong, E.M.; Hatton, C.; Wen, Y.; Barwe, S.P.; Gopalakrishnapillai, A.; Xu, H.; Uckelmann, H.J.; et al. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood 2022, 139, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Fiskus, W.; Boettcher, S.; Daver, N.; Mill, C.P.; Sasaki, K.; Birdwell, C.E.; Davis, J.A.; Takahashi, K.; Kadia, T.M.; DiNardo, C.D.; et al. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J. 2022, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Fiskus, W.; Mill, C.P.; Birdwell, C.; Davis, J.A.; Das, K.; Boettcher, S.; Kadia, T.M.; DiNardo, C.D.; Takahashi, K.; Loghavi, S.; et al. Targeting of epigenetic co-dependencies enhances anti-AML efficacy of Menin inhibitor in AML with MLL1-r or mutant NPM1. Blood Cancer J. 2023, 13, 53. [Google Scholar] [CrossRef]
- Singh, S.; Narang, A.S.; Mahato, R.I. Subcellular Fate and Off-Target Effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011, 28, 2996–3015. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, M.M.; Biayna, J.; Supek, F. TP53-dependent toxicity of CRISPR/Cas9 cuts is differential across genomic loci and can confound genetic screening. Nat. Commun. 2022, 13, 4520. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, C.; Wang, B.; Wang, F.; Pei, B.; Cheng, C.; Yang, W.; Zhao, Z. Transduction with Lentiviral Vectors Altered the Expression Profile of Host MicroRNAs. J. Virol. 2018, 92, e00503-18. [Google Scholar] [CrossRef]
- Lyu, P.; Javidi-Parsijani, P.; Atala, A.; Lu, B. Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’ genome editing. Nucleic Acids Res. 2019, 47, e99. [Google Scholar] [CrossRef]
- Peretz, L.; Besser, E.; Hajbi, R.; Casden, N.; Ziv, D.; Kronenberg, N.; Gigi, L.B.; Sweetat, S.; Khawaled, S.; Aqeilan, R.; et al. Combined shRNA over CRISPR/cas9 as a methodology to detect off-target effects and a potential compensatory mechanism. Sci. Rep. 2018, 8, 93. [Google Scholar] [CrossRef]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Joberty, G.; Fälth-Savitski, M.; Paulmann, M.; Bösche, M.; Doce, C.; Cheng, A.T.; Drewes, G.; Grandi, P. A Tandem Guide RNA-Based Strategy for Efficient CRISPR Gene Editing of Cell Populations with Low Heterogeneity of Edited Alleles. Crispr J. 2020, 3, 123–134. [Google Scholar] [CrossRef]
- Klossowski, S.; Miao, H.; Kempinska, K.; Wu, T.; Purohit, T.; Kim, E.; Linhares, B.M.; Chen, D.; Jih, G.; Perkey, E.; et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J. Clin. Investig. 2020, 130, 981–997. [Google Scholar] [CrossRef]
- Sukhodolets, K.E.; Hickman, A.B.; Agarwal, S.K.; Sukhodolets, M.V.; Obungu, V.H.; Novotny, E.A.; Crabtree, J.S.; Chandrasekharappa, S.C.; Collins, F.S.; Spiegel, A.M.; et al. The 32-kilodalton subunit of replication protein A interacts with menin, the product of the MEN1 tumor suppressor gene. Mol. Cell Biol. 2003, 23, 493–509. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Mao, H.; Schnepp, R.W.; Sykes, S.M.; Silva, A.C.; D’Andrea, A.D.; Hua, X. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res. 2003, 63, 4204–4210. [Google Scholar] [PubMed]
- Nelakurti, D.D.; Pappula, A.L.; Rajasekaran, S.; Miles, W.O.; Petreaca, R.C. Comprehensive Analysis of MEN1 Mutations and Their Role in Cancer. Cancers 2020, 12, 2616. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, J.-E.; Cho, E.-J.; Liu, J.O.; Youn, H.-D. Menin, a Tumor Suppressor, Represses JunD-Mediated Transcriptional Activity by Association with an mSin3A-Histone Deacetylase Complex. Cancer Res. 2003, 63, 6135. [Google Scholar] [PubMed]
- Leng, L.; Zhuang, K.; Liu, Z.; Huang, C.; Gao, Y.; Chen, G.; Lin, H.; Hu, Y.; Wu, D.; Shi, M.; et al. Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation. Neuron 2018, 100, 551–563.e7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xing, B.; Feng, Z.; Ma, J.; Ma, X.; Hua, X. Menin Upregulates FOXO1 Protein Stability by Repressing Skp2-Mediated Degradation in β Cells. Pancreas 2019, 48, 267–274. [Google Scholar] [CrossRef]
- Gehringer, F.; Weissinger, S.E.; Swier, L.J.; Möller, P.; Wirth, T.; Ushmorov, A. FOXO1 Confers Maintenance of the Dark Zone Proliferation and Survival Program and Can Be Pharmacologically Targeted in Burkitt Lymphoma. Cancers 2019, 11, 1427. [Google Scholar] [CrossRef]
- Wang, F.; Demir, S.; Gehringer, F.; Osswald, C.D.; Seyfried, F.; Enzenmüller, S.; Eckhoff, S.M.; Maier, T.; Holzmann, K.; Debatin, K.M.; et al. Tight regulation of FOXO1 is essential for maintenance of B-cell precursor acute lymphoblastic leukemia. Blood 2018, 131, 2929–2942. [Google Scholar] [CrossRef]
- Wang, Y.; Ozawa, A.; Zaman, S.; Prasad, N.B.; Chandrasekharappa, S.C.; Agarwal, S.K.; Marx, S.J. The tumor suppressor protein menin inhibits AKT activation by regulating its cellular localization. Cancer Res. 2011, 71, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Shojaee, S.; Chan, L.N.; Buchner, M.; Cazzaniga, V.; Cosgun, K.N.; Geng, H.; Qiu, Y.H.; von Minden, M.D.; Ernst, T.; Hochhaus, A.; et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat. Med. 2016, 22, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Gehringer, F.; Weissinger, S.E.; Möller, P.; Wirth, T.; Ushmorov, A. Physiological levels of the PTEN-PI3K-AKT axis activity are required for maintenance of Burkitt lymphoma. Leukemia 2020, 34, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Müschen, M. Autoimmunity checkpoints as therapeutic targets in B cell malignancies. Nat. Rev. Cancer 2018, 18, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Nishii, K.; Katayama, N.; Chen, F.; Usui, E.; Kadowaki, S.; Mitani, H.; Masuya, M.; Kageyama, S.I.; Kita, K.; Shiku, H. Aggressive neoplastic plasma cell growth with MLL gene rearrangement after high-dose therapy with autologous stem cell support for multiple myeloma. Bone Marrow Transpl. 2001, 27, 555–558. [Google Scholar] [CrossRef][Green Version]
- Sparbier, C.E.; Gillespie, A.; Gomez, J.; Kumari, N.; Motazedian, A.; Chan, K.L.; Bell, C.C.; Gilan, O.; Chan, Y.C.; Popp, S.; et al. Targeting Menin disrupts the KMT2A/B and polycomb balance to paradoxically activate bivalent genes. Nat. Cell Biol. 2023, 25, 258–272. [Google Scholar] [CrossRef]
- Kurmasheva, R.T.; Bandyopadhyay, A.; Favours, E.; Pozo, V.D.; Ghilu, S.; Phelps, D.A.; McGeehan, G.M.; Erickson, S.W.; Smith, M.A.; Houghton, P.J. Evaluation of VTP-50469, a menin-MLL1 inhibitor, against Ewing sarcoma xenograft models by the pediatric preclinical testing consortium. Pediatr. Blood Cancer 2020, 67, e28284. [Google Scholar] [CrossRef]
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.; Abraham, R.T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem 1995, 270, 815–822. [Google Scholar] [CrossRef]
- Bai, H.; Zhang, S.-Q.; Lei, H.; Wang, F.; Ma, M.; Xin, M. Menin-MLL protein-protein interaction inhibitors: A patent review (2014–2021). Expert Opin. Ther. Pat. 2022, 32, 507–522. [Google Scholar] [CrossRef]
- Somanath, P.; Lu, D.; Law, B.; Archer, T.; Rughwani, T.; Kumar, L.; Kinoshita, T.; Balakrishnan, M.; Butler, T. Preclinical activity of irreversible Menin inhibitor, BMF-219, in chronic lymphocytic leukemia. J. Clin. Oncol. 2022, 40, 7541. [Google Scholar] [CrossRef]
- Somanath, P.; Lu, D.; Law, B.; Archer, T.C.; Cacovean, A.; Palmer, J.T.; Kinoshita, T.; Butler, T. Novel Irreversible Menin Inhibitor, BMF-219, Shows Potent Single Agent Activity in Clinically Relevant DLBCL Cells. Blood 2021, 138, 4318. [Google Scholar] [CrossRef]
- Schreiber, S.L. A Chemical Biology View of Bioactive Small Molecules and a Binder-Based Approach to Connect Biology to Precision Medicines. Isr. J. Chem. 2019, 59, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolffhardt, T.M.; Ketzer, F.; Telese, S.; Wirth, T.; Ushmorov, A. Dependency of B-Cell Acute Lymphoblastic Leukemia and Multiple Myeloma Cell Lines on MEN1 Extends beyond MEN1–KMT2A Interaction. Int. J. Mol. Sci. 2023, 24, 16472. https://doi.org/10.3390/ijms242216472
Wolffhardt TM, Ketzer F, Telese S, Wirth T, Ushmorov A. Dependency of B-Cell Acute Lymphoblastic Leukemia and Multiple Myeloma Cell Lines on MEN1 Extends beyond MEN1–KMT2A Interaction. International Journal of Molecular Sciences. 2023; 24(22):16472. https://doi.org/10.3390/ijms242216472
Chicago/Turabian StyleWolffhardt, Tatjana Magdalena, Franz Ketzer, Stefano Telese, Thomas Wirth, and Alexey Ushmorov. 2023. "Dependency of B-Cell Acute Lymphoblastic Leukemia and Multiple Myeloma Cell Lines on MEN1 Extends beyond MEN1–KMT2A Interaction" International Journal of Molecular Sciences 24, no. 22: 16472. https://doi.org/10.3390/ijms242216472
APA StyleWolffhardt, T. M., Ketzer, F., Telese, S., Wirth, T., & Ushmorov, A. (2023). Dependency of B-Cell Acute Lymphoblastic Leukemia and Multiple Myeloma Cell Lines on MEN1 Extends beyond MEN1–KMT2A Interaction. International Journal of Molecular Sciences, 24(22), 16472. https://doi.org/10.3390/ijms242216472




