Transcriptome and miRNAs Profiles Reveal Regulatory Network and Key Regulators of Secondary Xylem Formation in “84K” Poplar
Abstract
:1. Introduction
2. Results
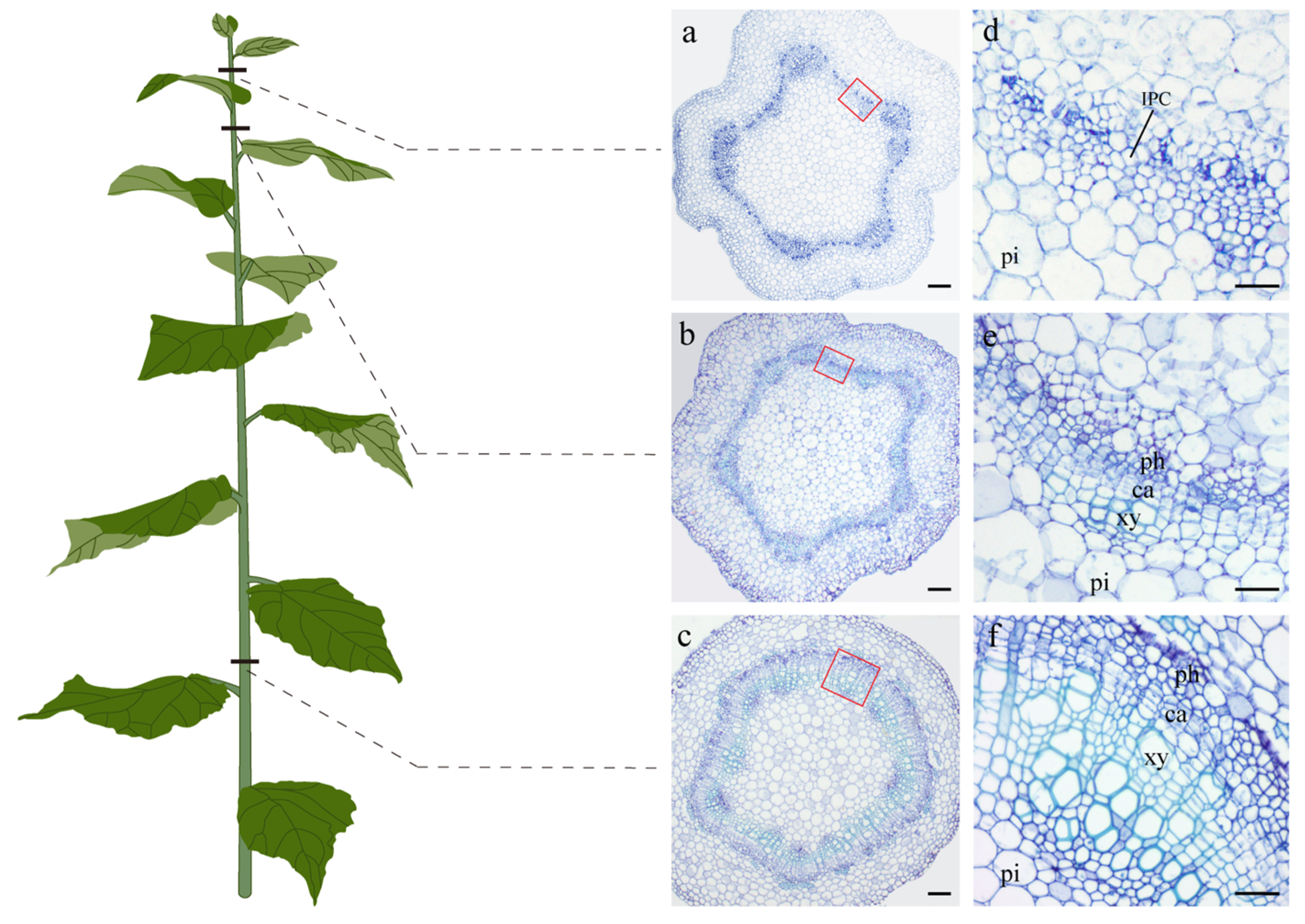
2.1. Anatomical Observations of Three Representative Developmental Stages of “84K” Poplar Stems
2.2. Transcriptome Analysis during Secondary Growth of “84K” Poplar Stems
2.2.1. RNA Sequencing
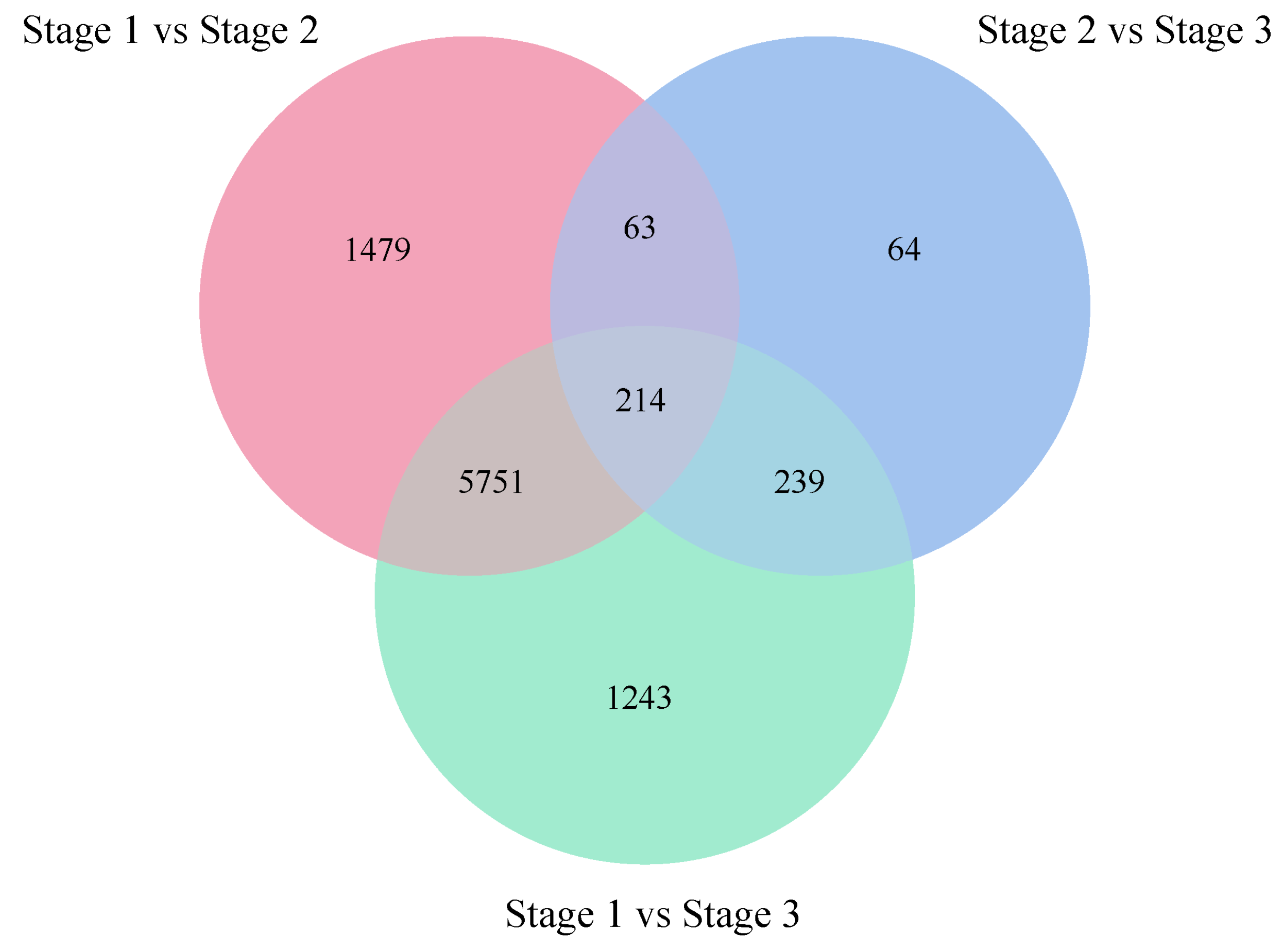
2.2.2. Identification and Functional Annotation of Differentially Expression Genes
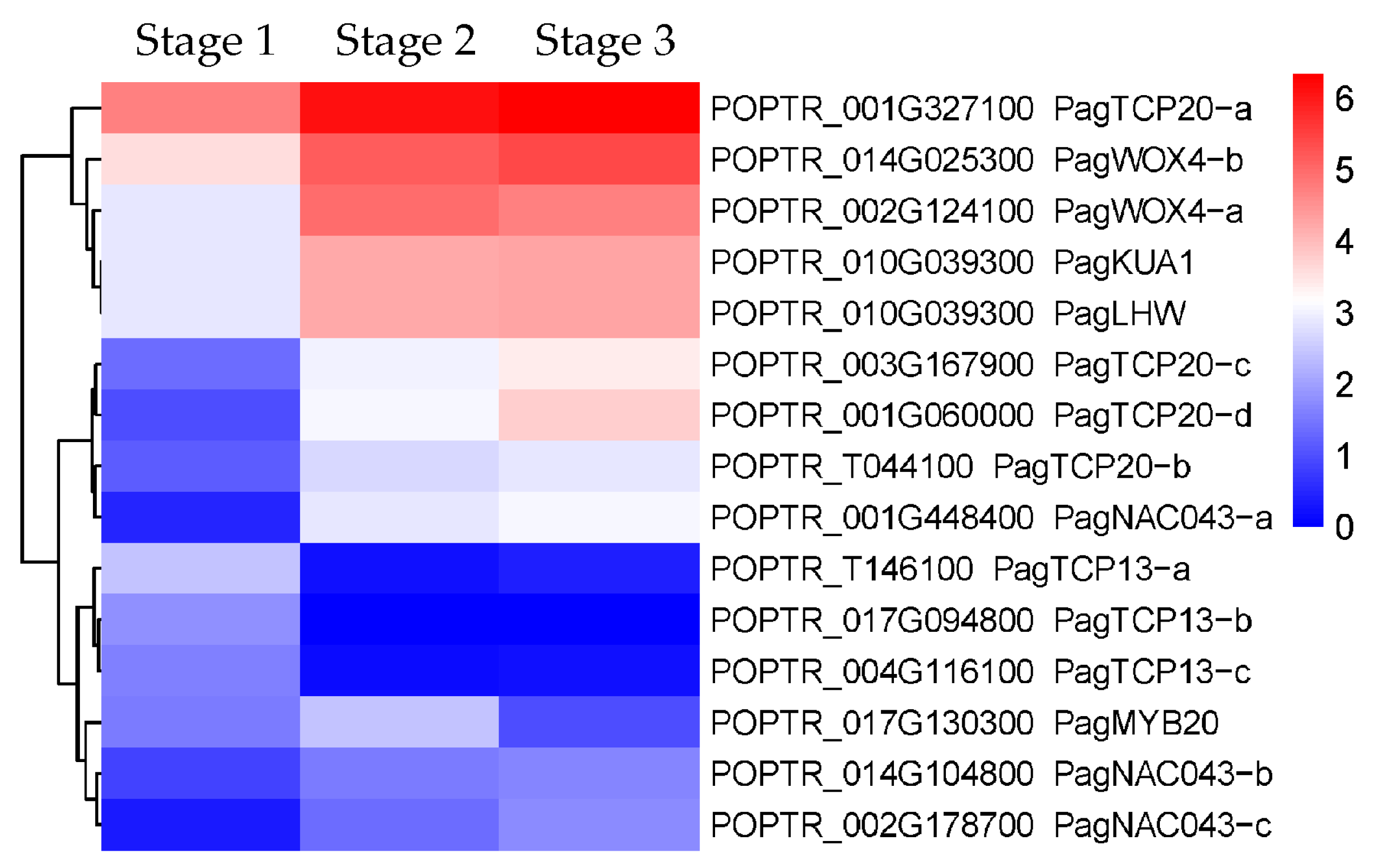
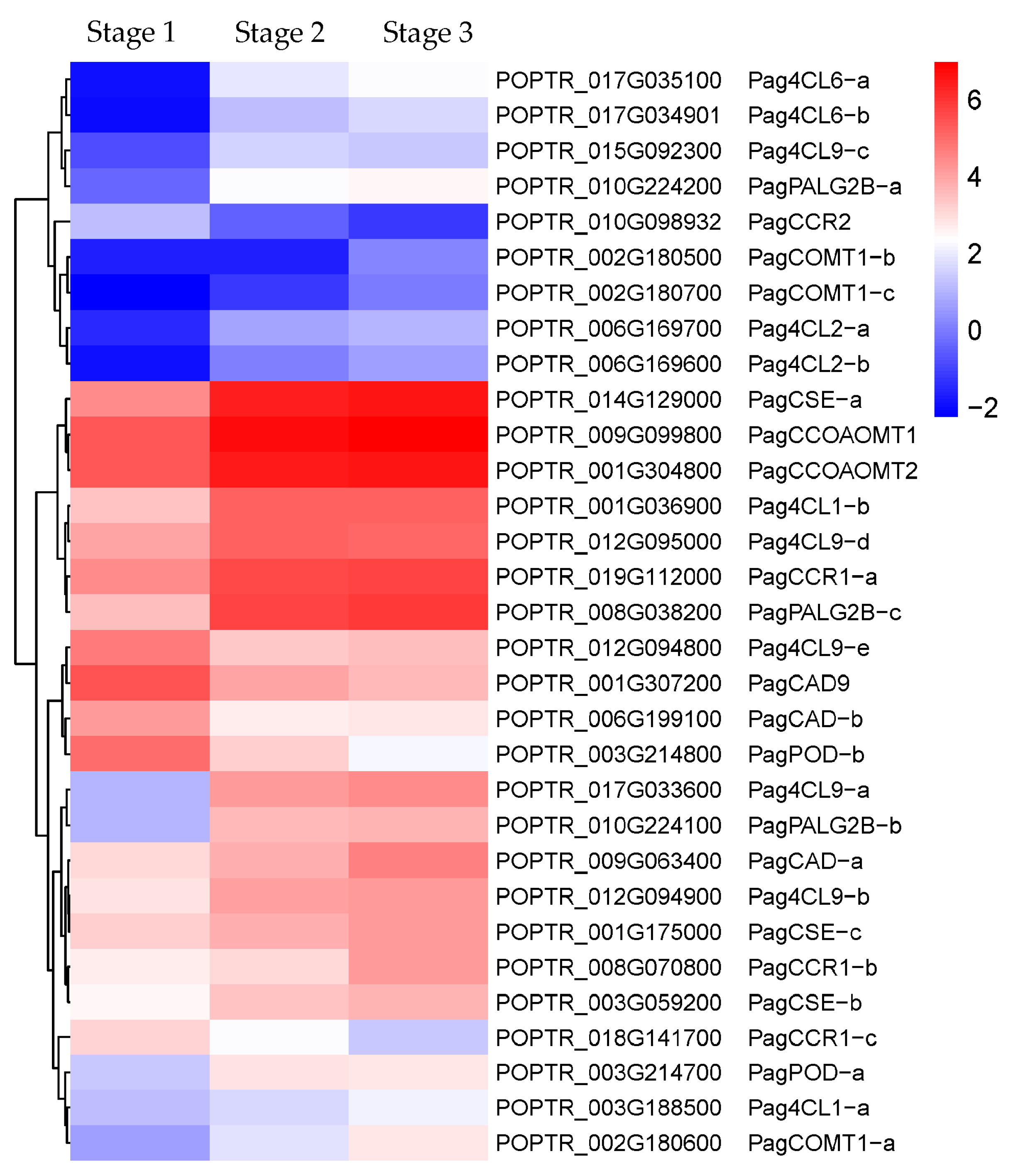
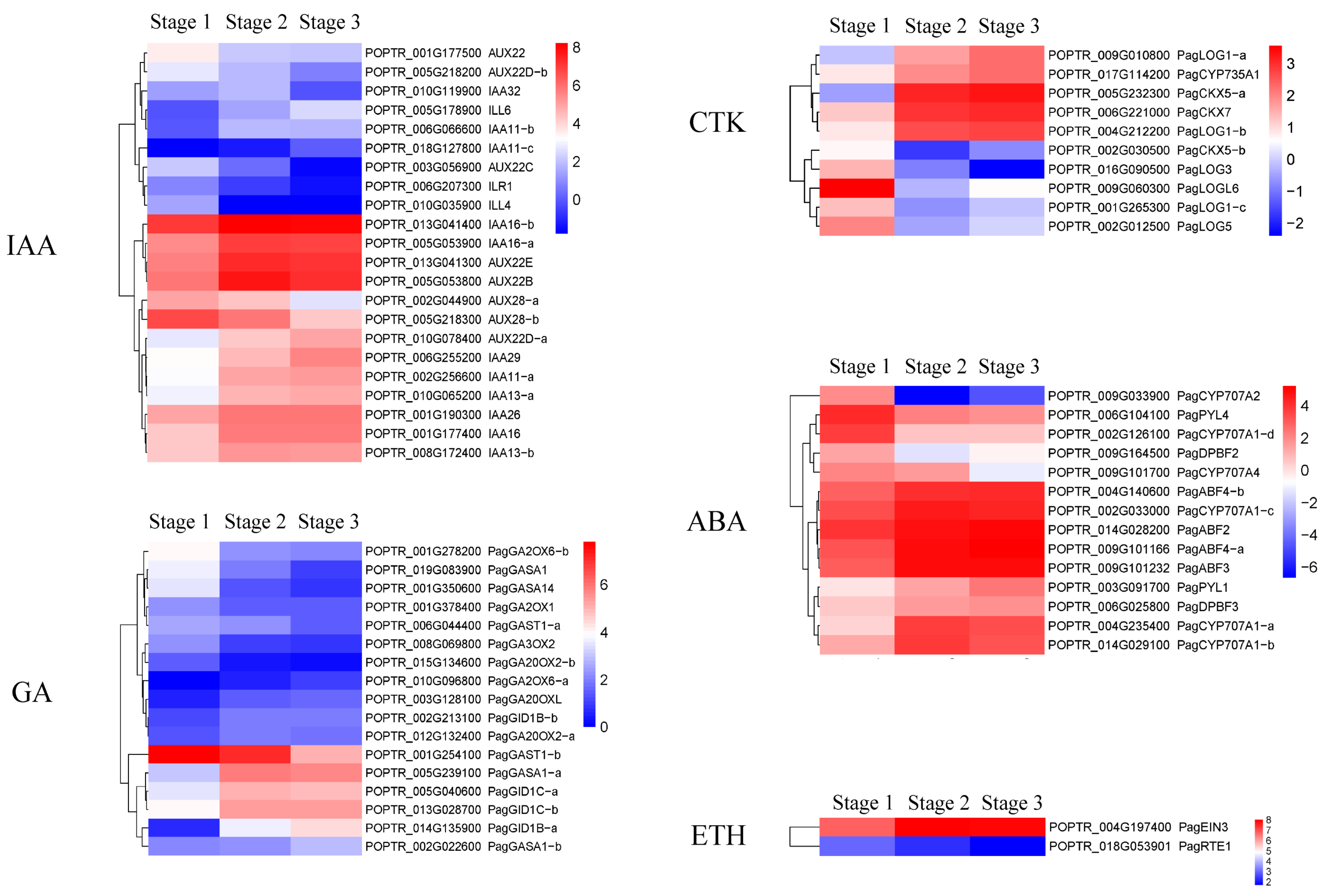
2.2.3. DEGs Related to the Secondary Xylem Formation
2.3. Analysis of miRNAs and Their Targets during Secondary Growth of “84K” Poplar Stems
2.3.1. Sequencing of Small RNAs and Identification of miRNAs
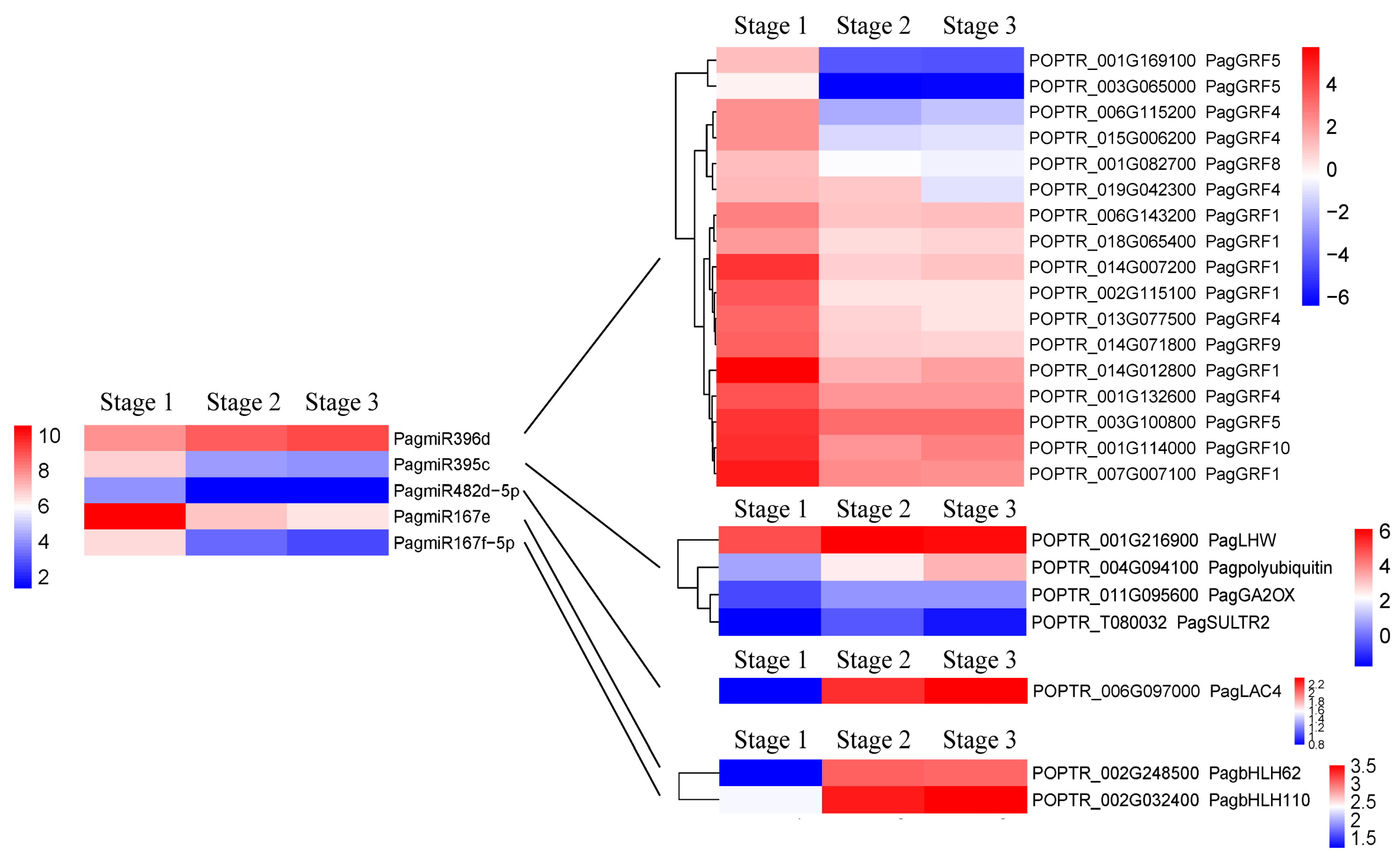
2.3.2. DEMs and Their Targets Related to Secondary Xylem Formation
2.4. RT-qPCR Validation of DEMs and Their Targets
3. Discussion
3.1. Transcription Factors Regulate Secondary Xylem Formation
3.1.1. Regulation of Cambial Activity and Its Differentiation into Xylem
3.1.2. Regulation of Xylem Cell Expansion and Secondary Cell Wall Formation
3.1.3. Lignin Biosynthesis-Related Enzymes Genes Regulate Secondary Xylem Formation
3.1.4. Plant Hormones Regulate Secondary Xylem Formation
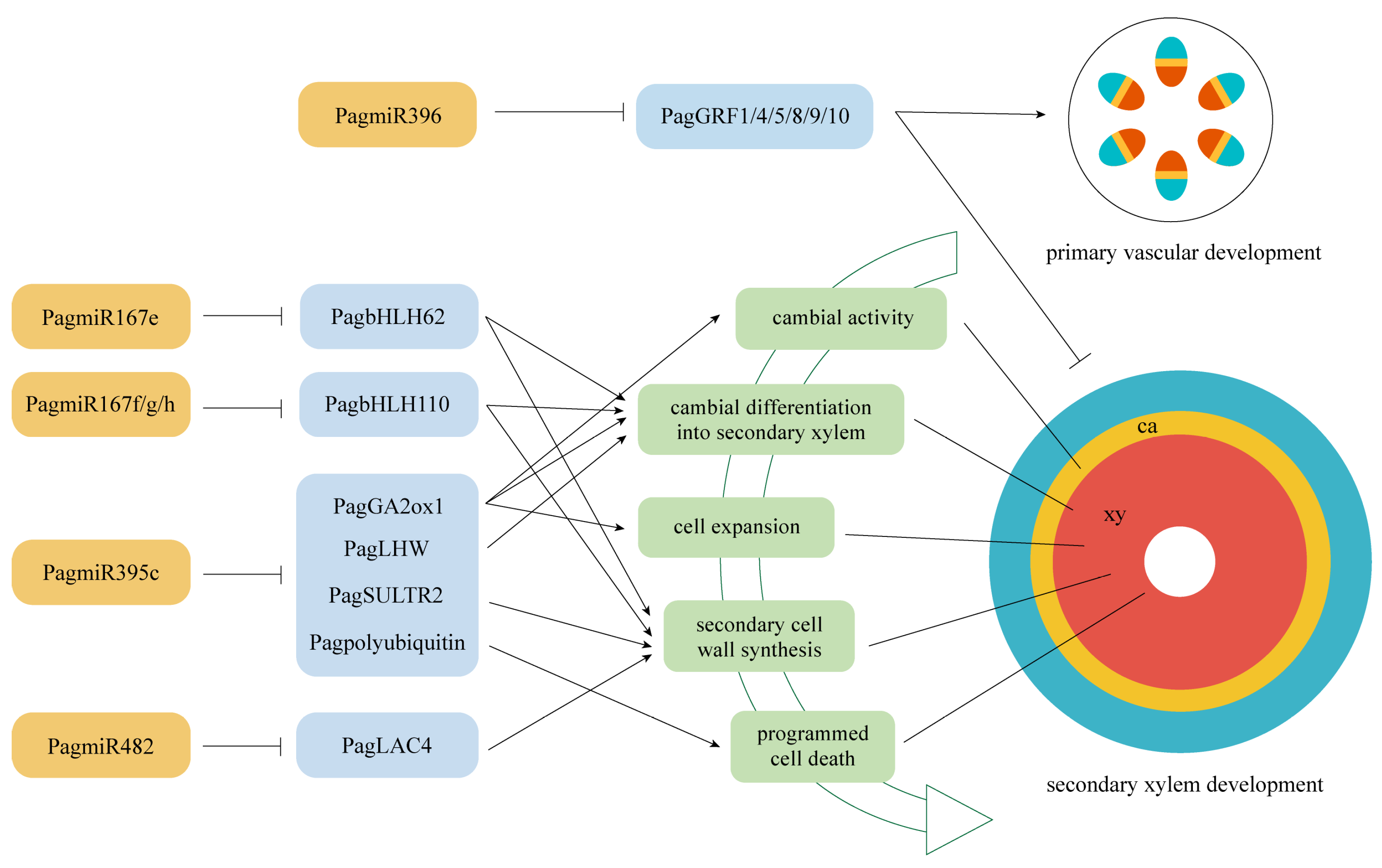
3.2. MiRNAs and Their Targets Regulate Secondary Xylem Formation
3.2.1. PagmiR396d-PagGRFs Modules
3.2.2. PagmiR395c-PagGA2ox1/PagLHW/PagSULTR2/PagPolyubiquitin 1 Modules
3.2.3. PagmiR482d-PagLAC4 Module
3.2.4. PagmiR167e-PagbHLH62 and PagmiR167f/g/h-PagbHLH110 Modules
4. Materials and Methods
4.1. Plant Materials and Sample Collection
4.2. Histochemistry and Microscopic Observation
4.3. RNA Extraction
4.4. Transcriptome Sequencing and Analysis
4.5. Small RNA Sequencing and Analysis
4.6. MiRNA Target Identification and Annotation
4.7. Real-Time Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fischer, U.; Kucukoglu, M.; Helariutta, Y.; Bhalerao, R.P. The dynamics of cambial stem cell activity. Annu. Rev. Plant Biol. 2019, 70, 293–319. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Wang, H.; Zhao, P.; Yu, Q.; He, Y.; Deng, W.; Guo, H. The regulation of xylem development by transcription factors and their upstream microRNAs. Int. J. Mol. Sci. 2022, 23, 10134. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.H.; Zhong, R. Molecular control of wood formation in trees. J. Exp. Bot. 2015, 66, 4119–4131. [Google Scholar] [CrossRef]
- Camargo, E.L.O.; Ployet, R.; Cassan-Wang, H.; Mounet, F.; Grima-Pettenati, J. Digging in wood: New insights in the regulation of wood formation in tree species. Adv. Bot. Res. 2019, 89, 201–233. [Google Scholar]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 2022, 13, 1030890. [Google Scholar] [CrossRef]
- Růžička, K.; Ursache, R.; Hejátko, J.; Helariutta, Y. Xylem development—From the cradle to the grave. New Phytol. 2015, 207, 519–535. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H. Signaling, transcriptional regulation, and asynchronous pattern formation governing plant xylem development. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2016, 92, 98–107. [Google Scholar] [CrossRef]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef]
- Chao, Q.; Gao, Z.F.; Zhang, D.; Zhao, B.G.; Dong, F.Q.; Fu, C.X.; Liu, L.J.; Wang, B.C. The developmental dynamics of the Populus stem transcriptome. Plant Biotechnol. J. 2019, 17, 206–219. [Google Scholar] [CrossRef]
- Kim, M.H.; Cho, J.S.; Jeon, H.W.; Sangsawang, K.; Shim, D.; Choi, Y.I.; Park, E.J.; Lee, H.; Ko, J.H. Wood transcriptome profiling identifies critical pathway genes of secondary wall biosynthesis and novel regulators for vascular cambium development in Populus. Genes 2019, 10, 690. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, B.; Zhang, J.; Hu, J. Insights of molecular mechanism of xylem development in five black poplar cultivars. Front. Plant Sci. 2020, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, J.; Liu, Y.; Liu, X.; Guo, X.; Li, H.; Liu, D.; Lu, H. Integrated transcriptomic and proteomic analysis in the roadmap of the xylem development stage in Populus tomentosa. Front. Plant Sci. 2021, 12, 724559. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhao, P.; Liu, S.; Yang, Q.; Guo, H. The control of developmental phase transitions by microRNAs and their targets in seed plants. Int. J. Mol. Sci. 2020, 21, 1971. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Zheng, Y.; Kudapa, H.; Jagadeeswaran, G.; Hivrale, V.; Varshney, R.K.; Sunkar, R. High throughput sequencing of small RNA component of leaves and inflorescence revealed conserved and novel miRNAs as well as phasiRNA loci in chickpea. Plant Sci. 2015, 235, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, X.; Li, W.; Lu, Y.; Dai, X.; Zhou, Z.; Li, Q. MicroRNA comparison between poplar and larch provides insight into the different mechanism of wood formation. Plant Cell Rep. 2020, 39, 1199–1217. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Tian, D.; Wang, J.; Chen, A.; Miao, Y.; Chen, Y.; Li, J.; Wu, X.; Zheng, B.; Guo, W.; et al. Overexpression of miR390b promotes stem elongation and height growth in Populus. Hortic. Res. 2023, 10, uhac258. [Google Scholar] [CrossRef]
- Kondo, Y.; Tamaki, T.; Fukuda, H. Regulation of xylem cell fate. Front. Plant Sci. 2014, 5, 315. [Google Scholar] [CrossRef]
- Lu, D.; Wang, T.; Persson, S.; Mueller-Roeber, B.; Schippers, J.H.M. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat. Commun. 2014, 5, 3767. [Google Scholar] [CrossRef]
- Hou, J.; Xu, H.; Fan, D.; Ran, L.; Li, J.; Wu, S.; Luo, K.; He, X.Q. MiR319a-targeted PtoTCP20 regulates secondary growth via interactions with PtoWOX4 and PtoWND6 in Populus tomentosa. New Phytol. 2020, 228, 1354–1368. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, J.; Li, H.; Chiang, V.L.; Fu, Y. Cooperative regulation of flavonoid and lignin biosynthesis in plants. Crit. Rev. Plant Sci. 2021, 40, 109–126. [Google Scholar] [CrossRef]
- Fan, F.; Zhou, Z.; Qin, H.; Tan, J.; Ding, G. Exogenous brassinosteroid facilitates xylem development in Pinus massoniana seedlings. Int. J. Mol. Sci. 2021, 22, 7615. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hou, J.; Xu, H.; Zhang, Y.; Huang, R.; Wang, D.; He, X.Q. The PtoTCP20-miR396d-PtoGRF15 module regulates secondary vascular development in Populus. Plant Commun. 2023, 4, 100494. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, D.; Wang, Z.; Chen, N.; Ma, X.; He, X.Q. MiR395c regulates secondary xylem development through sulfate metabolism in poplar. Front. Plant Sci. 2022, 13, 897376. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Bergmann, D.C. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 2007, 134, 2959–2968. [Google Scholar] [CrossRef]
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657. [Google Scholar] [CrossRef]
- Hur, Y.S.; Kim, J.; Kim, S.; Son, O.; Kim, W.Y.; Kim, G.T.; Ohme-Takagi, M.; Cheon, C.I. Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes 2019, 10, 644. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal. Behav. 2015, 10, e989746. [Google Scholar] [CrossRef]
- Xiao, R.; Zhang, C.; Guo, X.; Li, H.; Lu, H. MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 2021, 22, 3560. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef]
- Tribot, A.; Amer, G.; Abdou Alio, M.; de Baynast, H.; Delattre, C.; Pons, A.; Mathias, J.D.; Callois, J.M.; Vial, C.; Michaud, P.; et al. Wood-lignin: Supply, extraction processes and use as bio-based material. Eur. Polym. J. 2019, 112, 228–240. [Google Scholar] [CrossRef]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Zanardo, D.I.L.; Lima, R.B.; Ferrarese, M.d.L.L.; Bubna, G.A.; Ferrarese-Filho, O. Soybean root growth inhibition and lignification induced by p-coumaric acid. Environ. Exp. Bot. 2009, 66, 25–30. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Cheng, X.; Su, X.; Zhao, Y.; Jiang, T.; Jin, Q.; Lin, Y.; Cai, Y. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. Peerj 2019, 7, e8064. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hua, H.; Xu, J.; Mo, J.; Zhao, H.; Yang, J. Cloning and functional analysis of lignin biosynthesis genes Cf4CL and CfCCoAOMT in cryptomeria fortunei. Genes 2019, 10, 619. [Google Scholar] [CrossRef]
- Yao, T.; Feng, K.; Xie, M.; Barros, J.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Chen, J.G. Phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants. Front. Plant Sci. 2021, 12, 704697. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl Shikimate Esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Jang, H.A.; Bae, E.K.; Kim, M.H.; Park, S.J.; Choi, N.Y.; Pyo, S.W.; Lee, C.; Jeong, H.Y.; Lee, H.; Choi, Y.I.; et al. CRISPR-Knockout of CSE gene improves saccharification efficiency by reducing lignin content in hybrid poplar. Int. J. Mol. Sci. 2021, 22, 9750. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, N.; Hisano, H.; Cao, Y.; Wu, F.; Liu, W.; Bao, Y.; Wang, Z.; Fu, C. Simultaneous regulation of F5H in COMT-RNAi transgenic switchgrass alters effects of COMT suppression on syringyl lignin biosynthesis. Plant Biotechnol. J. 2019, 17, 836–845. [Google Scholar] [CrossRef]
- Kim, M.H.; Bae, E.K.; Lee, H.; Ko, J.H. Current understanding of the genetics and molecular mechanisms regulating wood formation in plants. Genes 2022, 13, 1181. [Google Scholar] [CrossRef]
- Agusti, J.; Lichtenberger, R.; Schwarz, M.; Nehlin, L.; Greb, T. Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genet. 2011, 7, e1001312. [Google Scholar] [CrossRef]
- Bhalerao, R.P.; Fischer, U. Environmental and hormonal control of cambial stem cell dynamics. J. Exp. Bot. 2017, 68, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Savidge, R.A. The role of plant hormones in higher plant cellular differentiation. II. Experiments with the vascular cambium, and sclereid and tracheid differentiation in the pine, Pinus contorta. Histochem. J. 1983, 15, 447–466. [Google Scholar] [CrossRef]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Alonso Serra, J.; Koskinen, P.; Zhang, J.; Elo, A.; Mähönen, A.P.; Street, N.; et al. Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, B.; Zhao, Y.; Guo, Y.; Liu, G.; Li, R.; Zeisler-Diehl, V.V.; Chen, Y.; He, X.; Schreiber, L.; et al. Article non-coding RNA analyses of seasonal cambium activity in Populus tomentosa. Cells 2022, 11, 640. [Google Scholar] [CrossRef] [PubMed]
- Love, J.; Björklund, S.; Vahala, J.; Hertzberg, M.; Kangasjärvi, J.; Sundberg, B. Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. USA 2009, 106, 5984–5989. [Google Scholar] [CrossRef]
- Arend, M.; Fromm, J. Concomitant analysis of cambial abscisic acid and cambial growth activity in poplar. Trees 2013, 27, 1271–1276. [Google Scholar] [CrossRef]
- Liu, X.; Huang, S.; Xie, H. Advances in the regulation of plant development and stress response by miR167. FBL 2021, 26, 655–665. [Google Scholar]
- Liu, Y.; Yan, J.; Wang, K.; Li, D.; Yang, R.; Luo, H.; Zhang, W. MiR396-GRF module associates with switchgrass biomass yield and feedstock quality. Plant Biotechnol. J. 2021, 19, 1523–1536. [Google Scholar] [CrossRef]
- Busov, V.B.; Meilan, R.; Pearce, D.W.; Ma, C.; Rood, S.B.; Strauss, S.H. Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 2003, 132, 1283–1291. [Google Scholar] [CrossRef]
- Dayan, J.; Schwarzkopf, M.; Avni, A.; Aloni, R. Enhancing plant growth and fiber production by silencing GA 2-oxidase. Plant Biotechnol. J. 2010, 8, 425–435. [Google Scholar] [CrossRef]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784. [Google Scholar] [CrossRef] [PubMed]
- Ai, Q.; Liang, G.; Zhang, H.; Yu, D. Control of sulfate concentration by miR395-targeted APS genes in Arabidopsis thaliana. Plant Divers. 2016, 38, 92–100. [Google Scholar] [CrossRef]
- Malcheska, F.; Honsel, A.; Wildhagen, H.; DÜRr, J.; Larisch, C.; Rennenberg, H.; Herschbach, C. Differential expression of specific sulphate transporters underlies seasonal and spatial patterns of sulphate allocation in trees. Plant Cell Environ. 2013, 36, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Escamez, S.; Tuominen, H. Programmes of cell death and autolysis in tracheary elements: When a suicidal cell arranges its own corpse removal. J. Exp. Bot. 2014, 65, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Guo, M.; Cheng, J.; Cheng, H.; Liu, X.; Ji, H.; Liu, G.; Cheng, Y.; Yang, C.; Turner, S. Aspartic proteases modulate programmed cell death and secondary cell wall synthesis during wood formation in poplar. J. Exp. Bot. 2022, 73, 6876–6890. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ahn, J.W.; Jin, U.H.; Choi, D.; Paek, K.H.; Pai, H.S. Activation of the programmed cell death pathway by in-hibition of proteasome function in plants. J. Biol. Chem. 2003, 278, 19406–19415. [Google Scholar] [CrossRef]
- Chou, E.Y.; Schuetz, M.; Hoffmann, N.; Watanabe, Y.; Sibout, R.; Samuels, A.L. Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J. Exp. Bot. 2018, 69, 1849–1859. [Google Scholar] [CrossRef]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Le Bris, P.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.Y.; Dixon, R.A. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef]
- Qin, S.; Fan, C.; Li, X.; Li, Y.; Hu, J.; Li, C.; Luo, K. LACCASE14 is required for the deposition of guaiacyl lignin and affects cell wall digestibility in poplar. Biotechnol. Biofuels 2020, 13, 197. [Google Scholar] [CrossRef]
- Lu, S.; Li, Q.; Wei, H.; Chang, M.; Tunlaya-Anukit, S.; Kim, H.; Liu, J.; Song, J.; Sun, Y.; Yuan, L.; et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2013, 110, 26. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Mu, X.; Grabowski, P.; Schmutz, J.; Lu, C. Enhancing microRNA167A expression in seed decreases the α-linolenic acid content and increases seed size in Camelina sativa. Plant J. 2019, 98, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.A.; Choi, Y.I.; Cho, J.S.; Lee, H. The poplar basic helix-loop-helix transcription factor BEE3-Like gene affects biomass production by enhancing proliferation of xylem cells in poplar. Biochem. Biophys. Res. Commun. 2015, 462, 64–70. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, S.; Han, X.; Ma, J.; Deng, W.; Wang, X.; Guo, H.; Xia, X. Integrated transcriptome and miRNA analysis uncovers molecular regulators of aerial stem-to-rhizome transition in the medical herb Gynostemma pentaphyllum. BMC Genom. 2019, 20, 865. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhao, P.; He, Y.; Su, Y.; Zhou, X.; Guo, H. Transcriptome and miRNAs Profiles Reveal Regulatory Network and Key Regulators of Secondary Xylem Formation in “84K” Poplar. Int. J. Mol. Sci. 2023, 24, 16438. https://doi.org/10.3390/ijms242216438
Wang H, Zhao P, He Y, Su Y, Zhou X, Guo H. Transcriptome and miRNAs Profiles Reveal Regulatory Network and Key Regulators of Secondary Xylem Formation in “84K” Poplar. International Journal of Molecular Sciences. 2023; 24(22):16438. https://doi.org/10.3390/ijms242216438
Chicago/Turabian StyleWang, Huilin, Pan Zhao, Yumei He, Yuting Su, Xinyi Zhou, and Huihong Guo. 2023. "Transcriptome and miRNAs Profiles Reveal Regulatory Network and Key Regulators of Secondary Xylem Formation in “84K” Poplar" International Journal of Molecular Sciences 24, no. 22: 16438. https://doi.org/10.3390/ijms242216438
APA StyleWang, H., Zhao, P., He, Y., Su, Y., Zhou, X., & Guo, H. (2023). Transcriptome and miRNAs Profiles Reveal Regulatory Network and Key Regulators of Secondary Xylem Formation in “84K” Poplar. International Journal of Molecular Sciences, 24(22), 16438. https://doi.org/10.3390/ijms242216438






