Antigenic and Structural Properties of the Lipopolysaccharide of the Uropathogenic Proteus mirabilis Dm55 Strain Classified to a New O85 Proteus Serogroup
Abstract
:1. Introduction
2. Results
2.1. Serological Studies
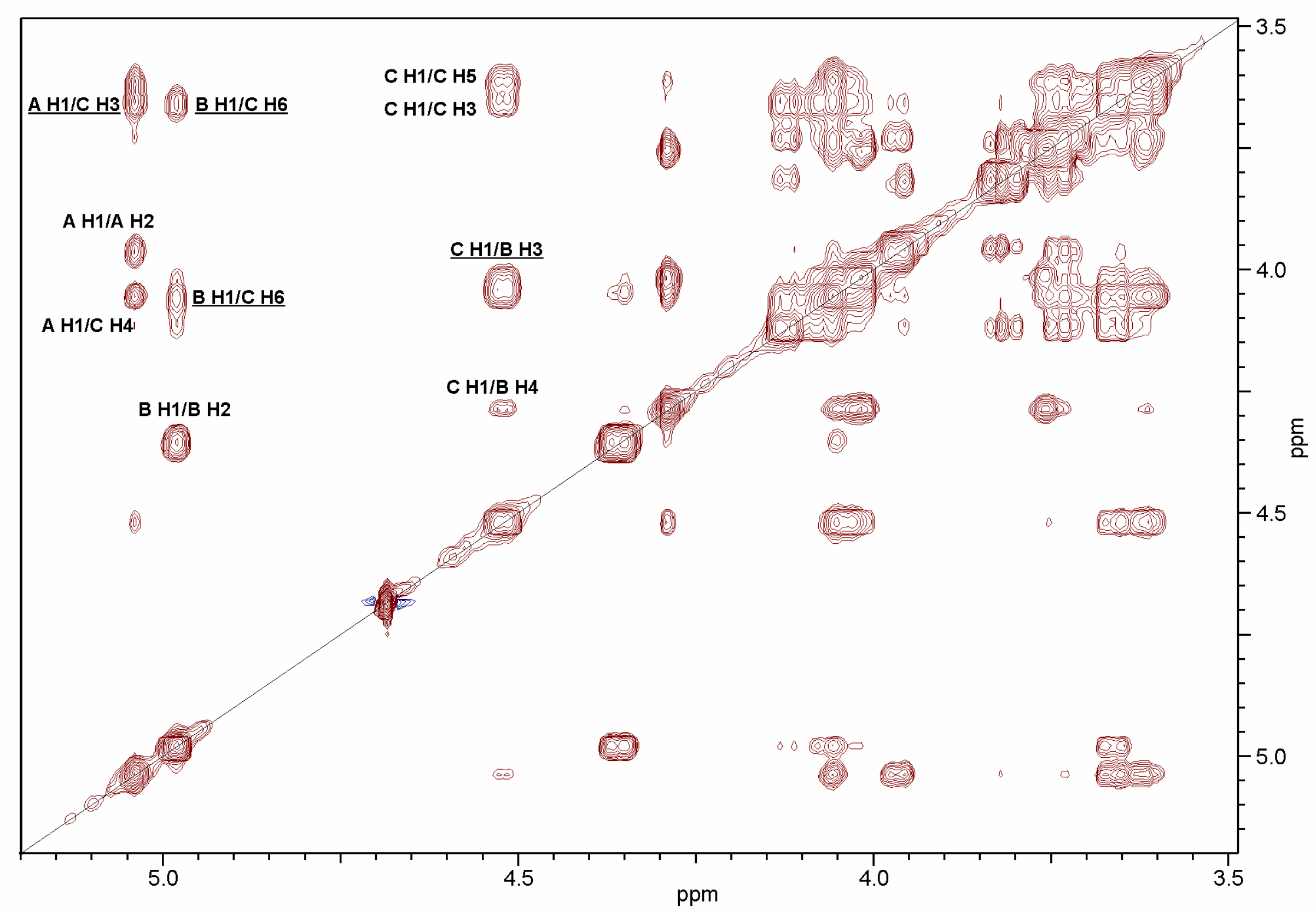
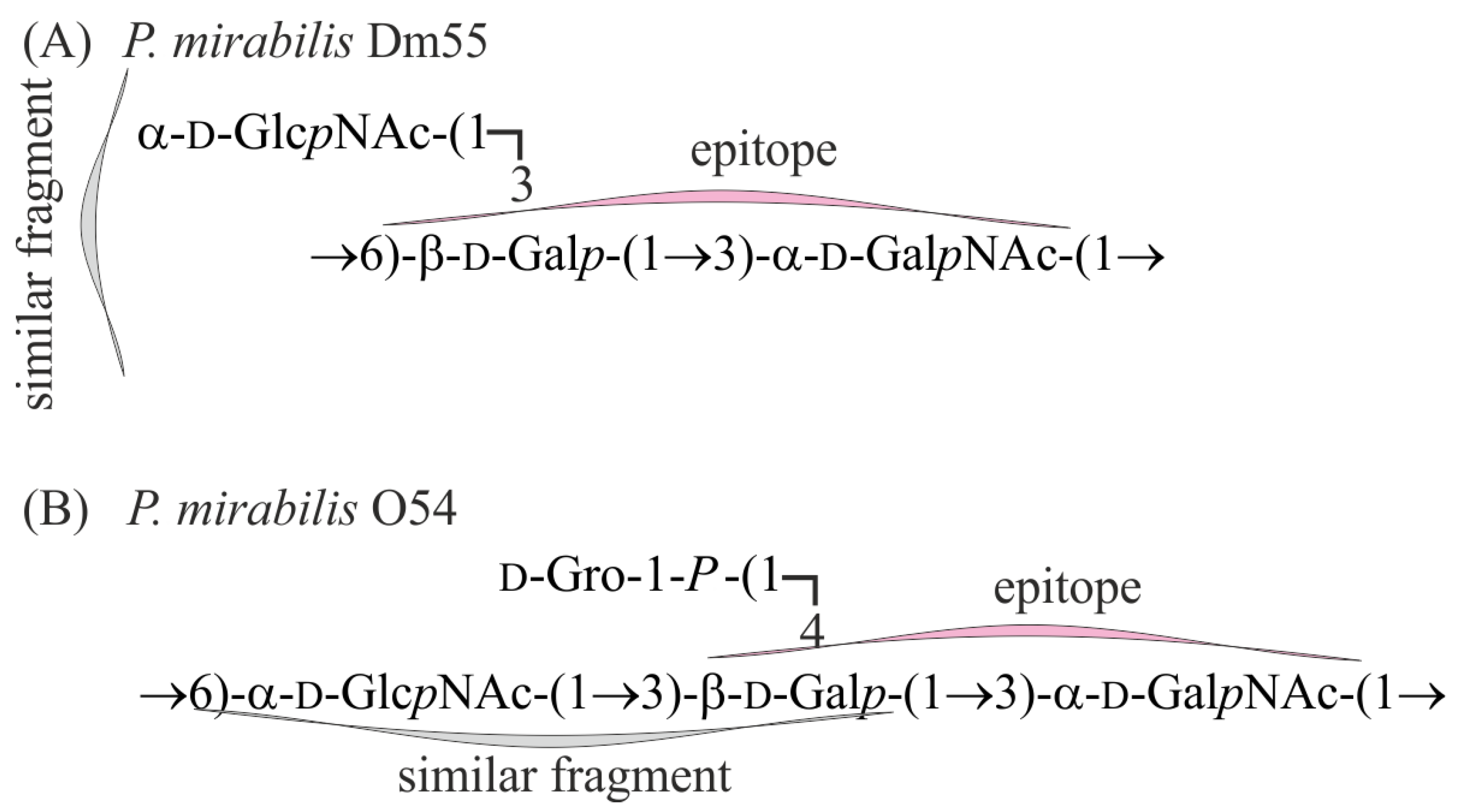
2.2. Structural Studies of O-Polysaccharide (OPS)
3. Discussion
4. Materials and Methods
4.1. The Studied Strains, Lipopolysaccharides (LPSs), and Polyclonal O-Specific Sera
4.2. Enzyme-Linked Immunosorbent Assay (ELISA)
4.3. Polyacrylamide Gel Electrophoresis (PAGE) of LPS and Western Blotting
4.4. Degradation of LPS and Isolation of O-Polysaccharide
4.5. Chemical Analyses
4.6. NMR Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka, D. Significance and roles of Proteus spp. bacteria in natural environments. Microb. Ecol. 2016, 72, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and urinary tract infections. Microbiol. Spectr. 2015, 3, 1–65. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.L.; Kamm, M.A.; Siew, C.N.; Morrisond, M. Proteus spp. as putative gastrointestinal pathogens. Clin. Microbiol. Rev. 2018, 31, e00085-17. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Mobley, H.L.T.; Pearson, M.M. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 2018, 8, 1–73. [Google Scholar] [CrossRef]
- Filipiak, A.; Chrapek, M.; Literacka, E.; Wawszczak, M.; Głuszek, S.; Majchrzak, M.; Wróbel, G.; Łysek-Gładysińska, M.; Gniadkowski, M.; Adamus-Białek, W. Pathogenic factors correlate with antimicrobial resistance among clinical Proteus mirabilis strains. Front. Microbiol. 2020, 11, 579389. [Google Scholar] [CrossRef]
- Flores-Mireles, A.; Hreha, T.N.; Hunstad, D.A. Pathophysiology, treatment, and prevention of catheter-associated urinary tract infection. Top. Spinal. Cord. Inj. Rehabil. 2019, 25, 228–240. [Google Scholar] [CrossRef]
- Ioannou, P.; Vougiouklakis, G. Infective endocarditis by Proteus species: A systematic review. GERMS 2020, 10, 229–239. [Google Scholar] [CrossRef]
- Rashid, T.; Ebringer, A. Rheumatoid arthritis is linked to Proteus—The evidence. Clin. Rheumatol. 2007, 26, 1036–1043. [Google Scholar] [CrossRef]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Perepelov, A.V.; Kondakova, A.N.; Senchenkova, S.N.; Sidorczyk, Z.; Rozalski, A.; Kaca, W. Structure and serology of O-antigens as the basis for classification of Proteus strains. Innate Immun. 2011, 17, 70–96. [Google Scholar] [CrossRef] [PubMed]
- Launay, O.; Lewis, D.J.M.; Anemona, A.; Loulergue, P.; Leahy, J.; Sciré, A.S.; Maugard, A.; Marchetti, E.; Zancan, S.; Huo, Z.; et al. Safety profile and immunologic responses of a novel vaccine against Shigella sonnei administered intramuscularly, intradermally and intranasally: Results from two parallel randomized phase 1 clinical studies in healthy adult volunteers in Europe. EBioMedicine 2017, 22, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka, D.; Palusiak, A.; Siwińska, M.; Zabłotni, A. The prevailing O serogroups among the serologically differentiated clinical Proteus spp. strains in central Poland. Sci. Rep. 2021, 11, 18982. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka, D.; Siwińska, M.; Senchenkova, S.N.; Levina, E.A.; Shashkov, A.S.; Knirel, Y.A. Structural and serological characterization of the O antigen of Proteus mirabilis clinical isolates classified into a new Proteus serogroup, O84. Int. J. Mol. Sci. 2023, 24, 4699. [Google Scholar] [CrossRef]
- Bock, K.; Pedersen, C. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 1983, 41, 27–66. [Google Scholar] [CrossRef]
- Jansson, P.-E.; Kenne, L.; Widmalm, G. Computer-assisted structural analysis of polysaccharides with an extended version of casper using 1H- and 13C-n.m.r. data. Carbohydr. Res. 1989, 188, 169–191. [Google Scholar] [CrossRef]
- Lipkind, G.M.; Shashkov, A.S.; Knirel, Y.A.; Vinogradov, E.V.; Kochetkov, N.K. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-N.M.R. data. Carbohydr. Res. 1988, 175, 59–75. [Google Scholar] [CrossRef]
- Lipkind, G.M.; Shashkov, A.S.; Mamyan, S.S.; Kochetkov, N.K. The Nuclear Overhauser effect and structural factors determining the conformations of disaccharide glycosides. Carbohydr. Res. 1988, 181, 522–531. [Google Scholar] [CrossRef]
- Shashkov, A.S.; Lipkind, G.M.; Knirel, Y.A.; Kochetkov, N.K. Stereochemical factors determining the effects of glycosylation on the 13C chemical shifts in carbohydrates. Magn. Reson. Chem. 1988, 26, 735–747. [Google Scholar] [CrossRef]
- Drzewiecka, D.; Arbatsky, N.P.; Shashkov, A.S.; Staczek, P.; Knirel, Y.A.; Sidorczyk, Z. Structure and serological properties of the O-antigen of two clinical Proteus mirabilis strains classified into a new Proteus O77 serogroup. FEMS Immunol. Med. Microbiol. 2008, 54, 185–194. [Google Scholar] [CrossRef]
- Schmitt, P.; Splettstösser, W.; Porsch-Ozcürümez, M.; Finke, E.J.; Grunow, R. A novel screening ELISA and a confirmatory Western blot useful for diagnosis and epidemiological studies of tularemia. Epidemiol. Infect. 2005, 133, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Møller, H.J.; Poulsen, J.H. Improved method for silver staining of glycoproteins in thin sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 1995, 226, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Senior, B.W. Media and tests to simplify the recognition and identification of members of the Proteeae. J. Med. Microbiol. 1997, 46, 39–44. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 1965, 5, 83–89. [Google Scholar]
- Sidorczyk, Z.; Zych, K.; Toukach, F.V.; Arbatsky, N.P.; Zabłotni, A.; Shashkov, A.S.; Knirel, Y.A. Structure of the O-polysaccharide and classification of Proteus mirabilis G1 in Proteus serogroup O3. Eur. J. Biochem. 2002, 269, 1406–1412. [Google Scholar] [CrossRef]
- Tsai, C.M.; Frasch, C.E. A Sensitive silver-stain for detecting lipopolysaccharide in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Leontein, K.; Lindberg, B.; Lönngren, J. Assignment of absolute configuration of sugars by g.l.c. of their acetylated glycosides formed from chiral alcohols. Carbohydr. Res. 1978, 62, 359–362. [Google Scholar] [CrossRef]
- Ciucanu, I.; Kerek, F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984, 131, 209–217. [Google Scholar] [CrossRef]
- Matsuura, M. Structural modifications of bacterial lipopolysaccharide that facilitate Gram-negative bacteria evasion of host innate immunity. Front. Immunol. 2013, 4, 109. [Google Scholar] [CrossRef]
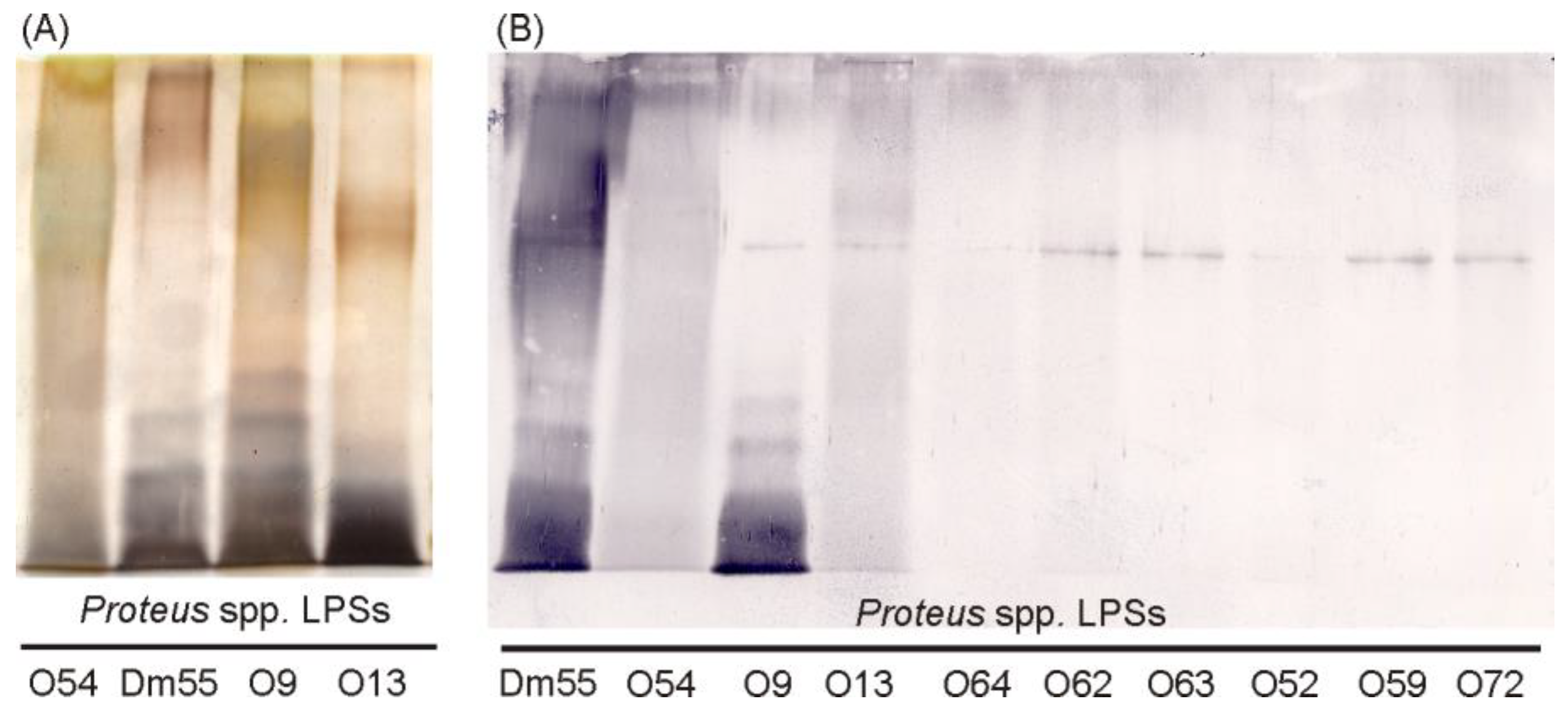
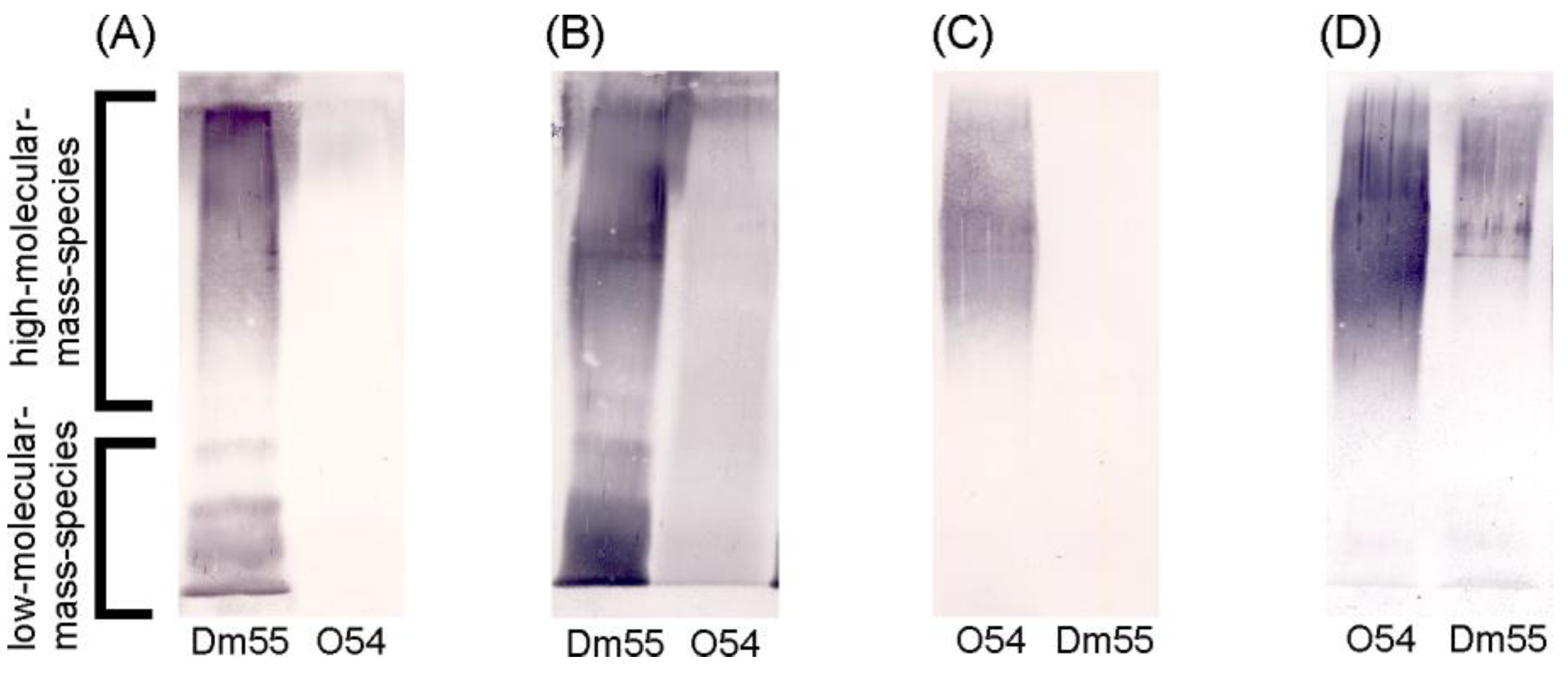




| Proteus spp. LPSs | Reciprocal Titre of: | |
|---|---|---|
| Dm55 Antiserum with the Cross-Reacting LPSs | Homologous Antisera | |
| Dm55 | - | 4,096,000 |
| O54 | 64,000 | 256,000 |
| O9 | 128,000 | 256,000 |
| O13 | 512,000 | 512,000 |
| O64 | 32,000 | 2,048,000 |
| O62 | 64,000 | >4,096,000 |
| O63 | 16,000 | 512,000 |
| O52 | 16,000 | 2,048,000 |
| O59 | 8000/16,000 | 128,000 |
| O72 | 64,000 | 128,000 |
| Reciprocal Titer in the Reaction with | ||
|---|---|---|
| P. mirabilis Dm55 Antiserum | Dm55 LPS | O54 LPS |
| not adsorbed | 4,096,000 | 64,000 |
| adsorbed with P. mirabilis O54 biomass | 512,000 | <2000 |
| P. mirabilis O54 Antiserum | ||
| not adsorbed | 64,000 | 256,000 |
| adsorbed with P. mirabilis Dm55 biomass | <2000 | 8000 |
| Sugar Residue | H-1 C-1 | H-2 C-2 | H-3 C-3 | H-4 C-4 | H-5 C-5 | H-6 C-6 | NAc | |
|---|---|---|---|---|---|---|---|---|
| α-d-GlcpNAc | A | 5.04 | 3.97 | 3.82 | 3.74 | 4.13 | 3.76 | 2.04 |
| 95.6 | 54.7 | 72.2 | 70.6 | 71.9 | 62.2 | 23.3; 175.6 | ||
| →3)-α-d-GalpNAc | B | 4.99 | 4.36 | 4.04 | 4.29 | 4.03 | 3.76 | 2.04 |
| 98.4 | 49.7 | 79.2 | 69.8 | 71.8 | 62.2 | 23.3; 175.6 | ||
| →3,6)-β-d-Galp | C | 4.53 | 3.62 | 3.66 | 4.06 | 3.62 | 3.66; 4.06 | |
| 105.9 | 70.3 | 78.7 | 66.4 | 76.0 | 65.8 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palusiak, A.; Turska-Szewczuk, A.; Drzewiecka, D. Antigenic and Structural Properties of the Lipopolysaccharide of the Uropathogenic Proteus mirabilis Dm55 Strain Classified to a New O85 Proteus Serogroup. Int. J. Mol. Sci. 2023, 24, 16424. https://doi.org/10.3390/ijms242216424
Palusiak A, Turska-Szewczuk A, Drzewiecka D. Antigenic and Structural Properties of the Lipopolysaccharide of the Uropathogenic Proteus mirabilis Dm55 Strain Classified to a New O85 Proteus Serogroup. International Journal of Molecular Sciences. 2023; 24(22):16424. https://doi.org/10.3390/ijms242216424
Chicago/Turabian StylePalusiak, Agata, Anna Turska-Szewczuk, and Dominika Drzewiecka. 2023. "Antigenic and Structural Properties of the Lipopolysaccharide of the Uropathogenic Proteus mirabilis Dm55 Strain Classified to a New O85 Proteus Serogroup" International Journal of Molecular Sciences 24, no. 22: 16424. https://doi.org/10.3390/ijms242216424
APA StylePalusiak, A., Turska-Szewczuk, A., & Drzewiecka, D. (2023). Antigenic and Structural Properties of the Lipopolysaccharide of the Uropathogenic Proteus mirabilis Dm55 Strain Classified to a New O85 Proteus Serogroup. International Journal of Molecular Sciences, 24(22), 16424. https://doi.org/10.3390/ijms242216424






