Neural Marker Expression in Adipose-Derived Stem Cells Grown in PEG-Based 3D Matrix Is Enhanced in the Presence of B27 and CultureOne Supplements
Abstract
:1. Introduction
2. Results
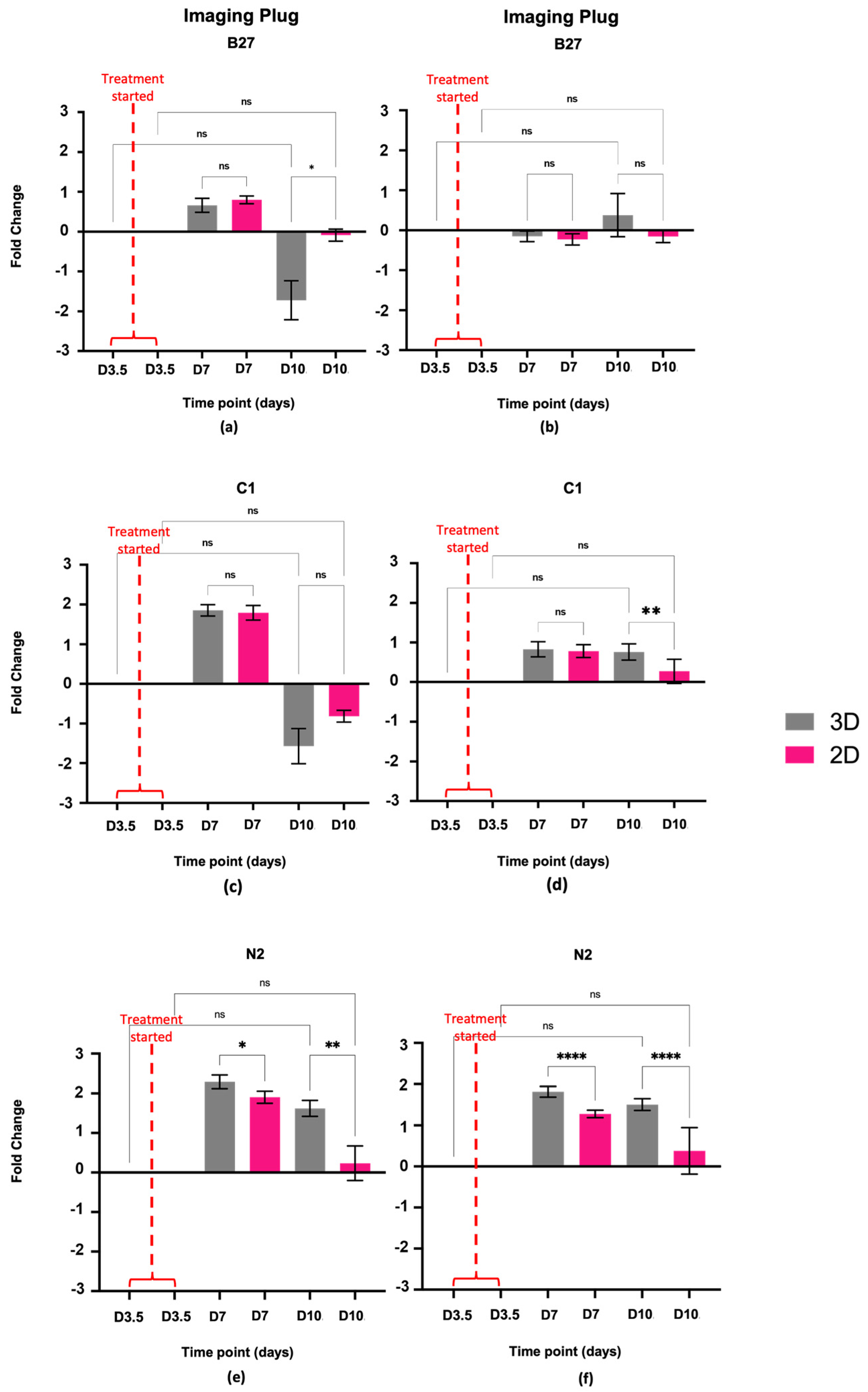
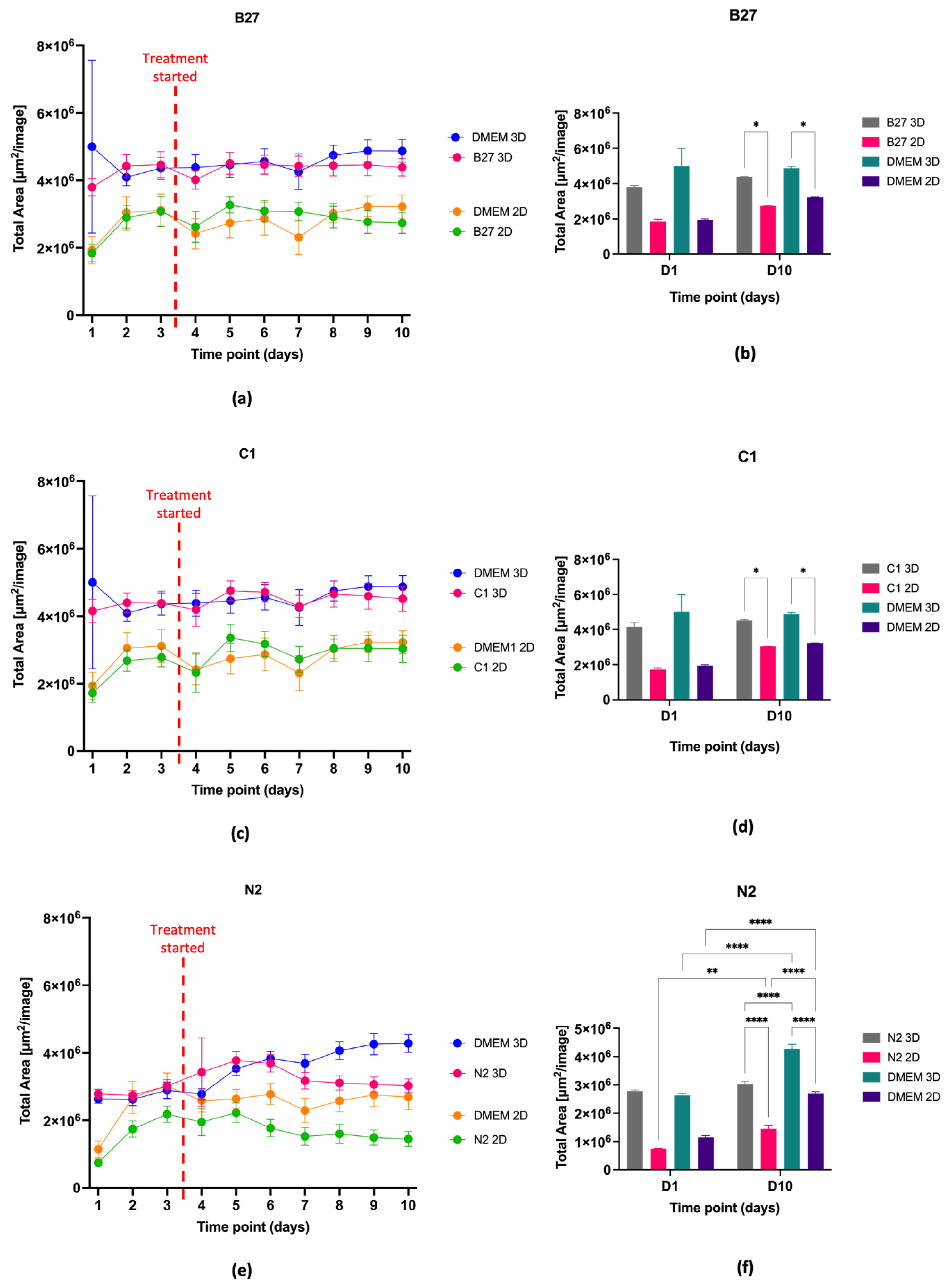
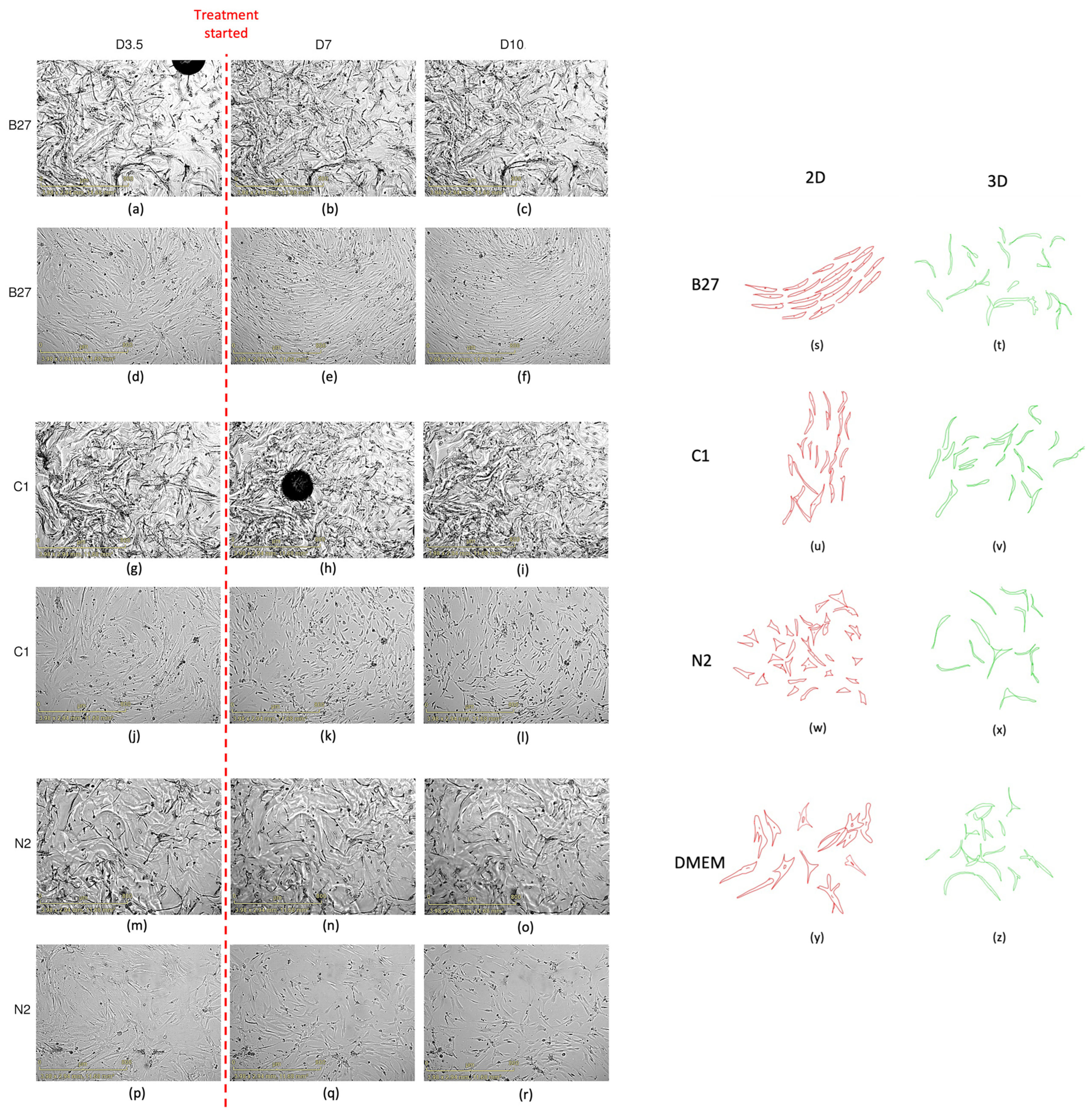
2.1. Cell Proliferation, Viability, and Morphology
2.1.1. Cell Proliferation and Viability
2.1.2. Cell Morphology
2.2. Cell Characterisation
2.2.1. Immunocytochemistry
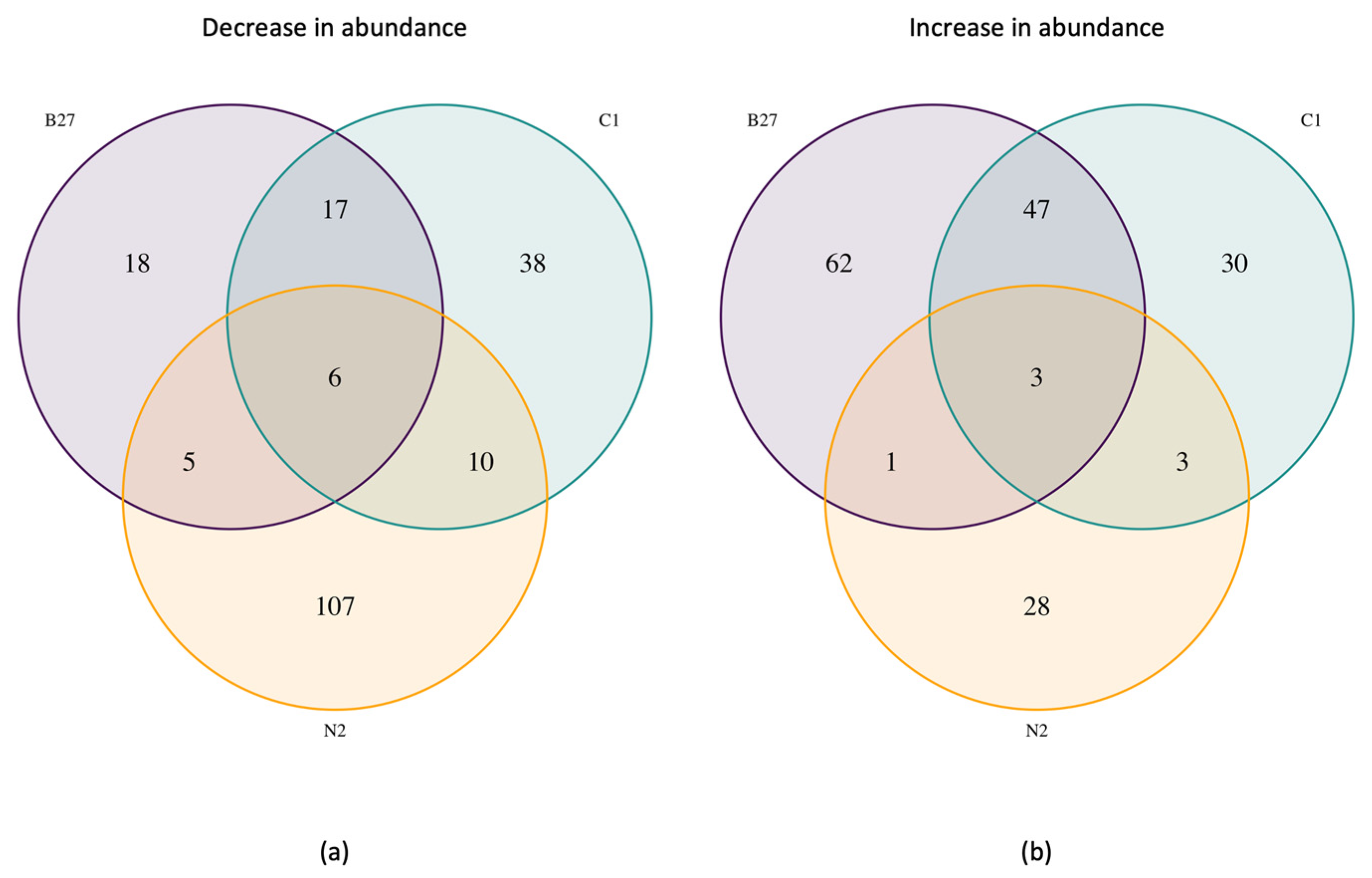
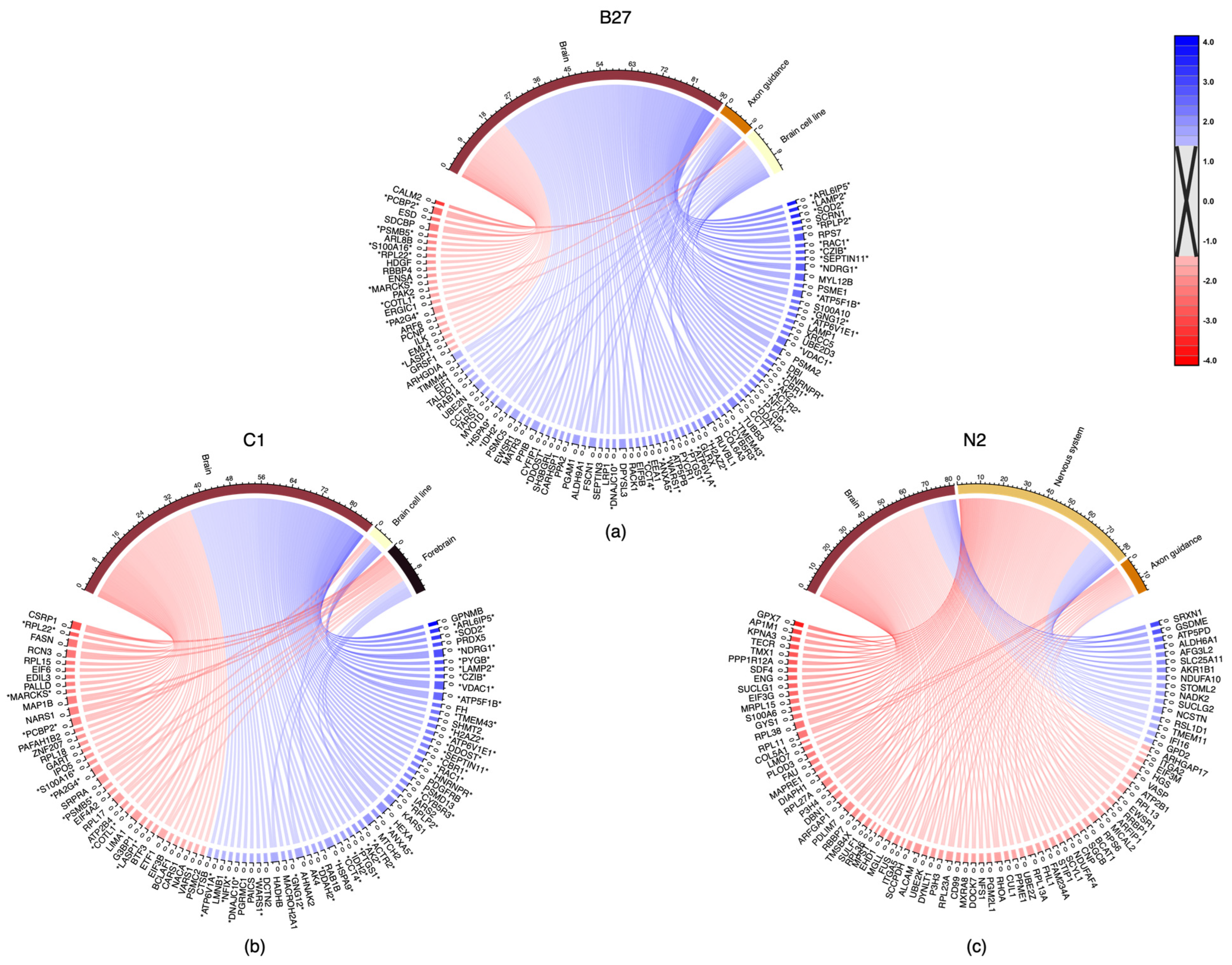
2.2.2. Proteomics
3. Discussion
3.1. Cell Viability and Confluence during Supplement Treatment
3.2. Morphological Changes during Supplement Treatment
3.3. Neural Surface Marker Expression Changes
3.4. Analysis of Proteome Changes across Supplement Cells
3.4.1. Proteome Changes Common in B27 and C1
3.4.2. Proteome Changes Unique to B27 Treatment
3.4.3. Proteome Changes Unique to C1 Treatment
3.4.4. Proteome Changes Unique to N2
4. Materials and Methods
4.1. The 3D Bioprinting of hADSCs in PEG-Based Hydrogels
4.2. Cell Morphology: Incucyte Imaging
4.3. Cell Viability and Proliferation: Alamar Blue
4.4. Cell Characterisation
4.4.1. Immunocytochemistry
4.4.2. Proteomics
Protein Extraction
LC-MS/MS Analysis
Data Processing and Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gimble, J.; Guilak, F. Adipose-derived adult stem cells: Isolation, characterization, and differentiation potential. Cytotherapy 2003, 5, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Cho, H.H.; Cho, Y.B.; Park, J.S.; Jeong, H.S. Functional neural differentiation of human adipose tissue-derived stem cells using bFGF and forskolin. BMC Cell Biol. 2010, 11, 25. [Google Scholar] [CrossRef]
- Park, J.; Lee, N.; Lee, J.; Choe, E.K.; Kim, M.K.; Lee, J.; Byun, M.S.; Chon, M.W.; Kim, S.W.; Lee, C.J.; et al. Small molecule-based lineage switch of human adipose-derived stem cells into neural stem cells and functional GABAergic neurons. Sci. Rep. 2017, 7, 10166. [Google Scholar] [CrossRef] [PubMed]
- Wislet-Gendebien, S.; Wautier, F.; Leprince, P.; Rogister, B. Astrocytic and neuronal fate of mesenchymal stem cells expressing nestin. Brain Res. Bull. 2005, 68, 95–102. [Google Scholar] [CrossRef]
- Tsuji, W.; Rubin, J.P.; Marra, K.G. Adipose-derived stem cells: Implications in tissue regeneration. World J. Stem Cells 2014, 6, 312–321. [Google Scholar] [CrossRef]
- Wankhade, U.D.; Shen, M.; Kolhe, R.; Fulzele, S. Advances in Adipose-Derived Stem Cells Isolation, Characterization, and Application in Regenerative Tissue Engineering. Stem Cells Int. 2016, 2016, 3206807. [Google Scholar] [CrossRef] [PubMed]
- Strioga, M.; Viswanathan, S.; Darinskas, A.; Slaby, O.; Michalek, J. Same or not the same? Comparison of adipose tissue-derived versus bone marrow-derived mesenchymal stem and stromal cells. Stem Cells Dev. 2012, 21, 2724–2752. [Google Scholar] [CrossRef]
- Rad, A.A.; Heidari, M.H.; Aliaghaei, A.; Broujeni, M.E.; Shojaei, A.; Abbaszadeh, H.-A. In vitro differentiation of adipose derived stem cells into functional dopaminergic neurons. Biomed. Pharmacol. J. 2017, 10, 595–605. [Google Scholar]
- Ahmadi, N.; Razavi, S.; Kazemi, M.; Oryan, S. Stability of neural differentiation in human adipose derived stem cells by two induction protocols. Tissue Cell 2012, 44, 87–94. [Google Scholar] [CrossRef]
- Soheilifar, M.H.; Javeri, A.; Amini, H.; Taha, M.F. Generation of Dopamine-Secreting Cells from Human Adipose Tissue-Derived Stem Cells In Vitro. Rejuvenation Res. 2018, 21, 360–368. [Google Scholar] [CrossRef]
- Faghih, H.; Javeri, A.; Amini, H.; Taha, M.F. Directed differentiation of human adipose tissue-derived stem cells to dopaminergic neurons in low-serum and serum-free conditions. Neurosci. Lett. 2019, 708, 134353. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Milthorpe, B.K.; Herbert, B.R.; Padula, M.P. Proteomic Analysis of Human Adipose Derived Stem Cells during Small Molecule Chemical Stimulated Pre-neuronal Differentiation. Int. J. Stem Cells 2017, 10, 193–217. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Hubert, T.; Milthorpe, B.K. Valproic Acid Promotes Early Neural Differentiation in Adult Mesenchymal Stem Cells Through Protein Signalling Pathways. Cells 2020, 9, 619. [Google Scholar] [CrossRef]
- Santos, J.; Dalla, P.V.; Milthorpe, B.K. Molecular Dynamics of Cytokine Interactions and Signalling of Mesenchymal Stem Cells Undergoing Directed Neural-like Differentiation. Life 2022, 12, 392. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.; Fu, R.H.; Huang, Y.C.; Chen, S.Y.; Hsu, C.J.; Lin, S.Z.; Tu, C.T.; Chang, L.H.; Wu, P.A.; Liu, S.P. Adipose-derived Stem Cells Stimulated with n-Butylidenephthalide Exhibit Therapeutic Effects in a Mouse Model of Parkinson’s Disease. Cell Transplant. 2018, 27, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.B.; Singh, P.; Feaster, M.M.; Ramnarain, A.; Pavlides, C.; Chen, Z.L.; Yu, W.M.; Feltri, M.L.; Strickland, S. Mesenchymal stem cells facilitate axon sorting, myelination, and functional recovery in paralyzed mice deficient in Schwann cell-derived laminin. Glia 2011, 59, 267–277. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Pan, X.; Zhang, Y.; Lin, L.; Wu, Y.; Huang, Y.; He, H. Adipose-derived stem cell transplantation improves learning and memory via releasing neurotrophins in rat model of temporal lobe epilepsy. Brain Res. 2021, 1750, 147121. [Google Scholar] [CrossRef]
- Zhou, F.; Gao, S.; Wang, L.; Sun, C.; Chen, L.; Yuan, P.; Zhao, H.; Yi, Y.; Qin, Y.; Dong, Z.; et al. Human adipose-derived stem cells partially rescue the stroke syndromes by promoting spatial learning and memory in mouse middle cerebral artery occlusion model. Stem Cell Res. Ther. 2015, 6, 92. [Google Scholar] [CrossRef]
- Ning, H.; Lin, G.; Lue, T.F.; Lin, C.S. Neuron-like differentiation of adipose tissue-derived stromal cells and vascular smooth muscle cells. Differentiation 2006, 74, 510–518. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Benya, P.D.; Shaffer, J.D. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 1982, 30, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Hongisto, V.; Jernstrom, S.; Fey, V.; Mpindi, J.P.; Kleivi Sahlberg, K.; Kallioniemi, O.; Perala, M. High-throughput 3D screening reveals differences in drug sensitivities between culture models of JIMT1 breast cancer cells. PLoS ONE 2013, 8, e77232. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Hopkins, A.M.; De Laporte, L.; Tortelli, F.; Spedden, E.; Staii, C.; Atherton, T.J.; Hubbell, J.A.; Kaplan, D.L.J. Silk hydrogels as soft substrates for neural tissue engineering. Adv. Funct. Mater. 2013, 23, 5140–5149. [Google Scholar] [CrossRef]
- Her, G.J.; Wu, H.C.; Chen, M.H.; Chen, M.Y.; Chang, S.C.; Wang, T.W. Control of three-dimensional substrate stiffness to manipulate mesenchymal stem cell fate toward neuronal or glial lineages. Acta Biomater. 2013, 9, 5170–5180. [Google Scholar] [CrossRef]
- Li, Y.; Huang, G.; Li, M.; Wang, L.; Elson, E.L.; Lu, T.J.; Genin, G.M.; Xu, F. An approach to quantifying 3D responses of cells to extreme strain. Sci. Rep. 2016, 6, 19550. [Google Scholar] [CrossRef]
- Gomila Pelegri, N.; Stanczak, A.M.; Bottomley, A.L.; Milthorpe, B.K.; Gorrie, C.A.; Padula, M.P.; Santos, J. Adipose-Derived Stem Cells Spontaneously Express Neural Markers When Grown in a PEG-Based 3D Matrix. Int. J. Mol. Sci. 2023, 24, 12139. [Google Scholar] [CrossRef]
- Pelegri, N.G.; Milthorpe, B.K.; Gorrie, C.A.; Santos, J. Neurogenic marker expression in differentiating human adipose derived adult mesenchymal stem cells. Stem Cell Investig. 2023, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G. Cell proliferation in development and differentiation. In The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- De Biasio, A.; Sánchez, R.; Prieto, J.; Villate, M.; Campos-Olivas, R.; Blanco, F.J. Reduced stability and increased dynamics in the human proliferating cell nuclear antigen (PCNA) relative to the yeast homolog. PLoS ONE 2011, 6, e16600. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J.; Moscona, A. Role of cell shape in growth control. Nature 1978, 273, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Kim, J.B. Three-dimensional tissue culture models in cancer biology. Semin. Cancer Biol. 2005, 15, 365–377. [Google Scholar] [CrossRef]
- Gulati, A. Understanding neurogenesis in the adult human brain. Indian J. Pharmacol. 2015, 47, 583. [Google Scholar] [CrossRef]
- UniProt. O75915·PRAF3_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/O75915/entry (accessed on 5 October 2023).
- Jia, F.; Yue, M.; Chandra, D.; Keramidas, A.; Goldstein, P.A.; Homanics, G.E.; Harrison, N.L. Taurine is a potent activator of extrasynaptic GABAA receptors in the thalamus. J. Neurosci. 2008, 28, 106–115. [Google Scholar] [CrossRef]
- Sturman, J.A. Taurine in development. J. Nutr. 1988, 118, 1169–1176. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, K.-Q.; Huang, Y.-N.; Chen, L.; Xu, T.-L. Taurine activates strychnine-sensitive glycine receptors in neurons of the rat inferior colliculus. Brain Res. 2004, 1021, 232–240. [Google Scholar] [CrossRef]
- Zhou, Y.; Danbolt, N.C. Glutamate as a neurotransmitter in the healthy brain. J. Neural Transm. 2014, 121, 799–817. [Google Scholar] [CrossRef]
- Pal, M.M. Glutamate: The master neurotransmitter and its implications in chronic stress and mood disorders. Front. Hum. Neurosci. 2021, 15, 722323. [Google Scholar] [CrossRef] [PubMed]
- Crupi, R.; Impellizzeri, D.; Cuzzocrea, S. Role of metabotropic glutamate receptors in neurological disorders. Front. Mol. Neurosci. 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Siddique, I.; Kamble, K.; Gupta, S.; Solanki, K.; Bhola, S.; Ahsan, N.; Gupta, S. ARL6IP5 Ameliorates α-Synuclein Burden by Inducing Autophagy via Preventing Ubiquitination and Degradation of ATG12. Int. J. Mol. Sci. 2023, 24, 499. [Google Scholar] [CrossRef] [PubMed]
- Vadodaria, K.C.; Brakebusch, C.; Suter, U.; Jessberger, S. Stage-specific functions of the small Rho GTPases Cdc42 and Rac1 for adult hippocampal neurogenesis. J. Neurosci. 2013, 33, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- UniProt. P63000·RAC1_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/P63000/entry (accessed on 5 October 2023).
- Gualdoni, S.; Albertinazzi, C.; Corbetta, S.; Valtorta, F.; de Curtis, I. Normal levels of Rac1 are important for dendritic but not axonal development in hippocampal neurons. Biol. Cell 2007, 99, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Serwanski, D.R.; Miralles, C.P.; Nagata, K.-i.; De Blas, A.L. Septin 11 is present in GABAergic synapses and plays a functional role in the cytoarchitecture of neurons and GABAergic synaptic connectivity. J. Biol. Chem. 2009, 284, 17253–17265. [Google Scholar] [CrossRef] [PubMed]
- UniProt. Q9NVA2·SEP11_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/Q9NVA2/entry (accessed on 5 October 2023).
- UniProt. P38606·VATA_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/P38606/entry (accessed on 6 October 2023).
- UniProt. Q92597·NDRG1_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/Q92597/entry (accessed on 6 October 2023).
- Duly, A.M.; Kao, F.C.; Teo, W.S.; Kavallaris, M. βIII-Tubulin Gene Regulation in Health and Disease. Front. Cell Dev. Biol. 2022, 10, 851542. [Google Scholar] [CrossRef]
- Caccamo, D.; Katsetos, C.; Herman, M.; Frankfurter, A.; Collins, V.; Rubinstein, L. Immunohistochemistry of a spontaneous murine ovarian teratoma with neuroepithelial differentiation. Neuron-associated beta-tubulin as a marker for primitive neuroepithelium. Lab. Investig. 1989, 60, 390–398. [Google Scholar]
- Jiang, Y.Q.; Oblinger, M.M. Differential regulation of beta III and other tubulin genes during peripheral and central neuron development. J. Cell Sci. 1992, 103, 643–651. [Google Scholar] [CrossRef]
- Linhartová, I.; Dráber, P.; Dráberová, E.; Viklický, V. Immunological discrimination of β-tubulin isoforms in developing mouse brain. Post-translational modification of non-class-III β-tubulins. Biochem. J. 1992, 288, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Hausrat, T.J.; Radwitz, J.; Lombino, F.L.; Breiden, P.; Kneussel, M. Alpha-and beta-tubulin isotypes are differentially expressed during brain development. Dev. Neurobiol. 2021, 81, 333–350. [Google Scholar] [CrossRef] [PubMed]
- UniProt. Q14195·DPYL3_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/Q14195/entry (accessed on 6 October 2023).
- Deinhardt, K.; Kim, T.; Spellman, D.S.; Mains, R.E.; Eipper, B.A.; Neubert, T.A.; Chao, M.V.; Hempstead, B.L. Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Sci. Signal 2011, 4, ra82. [Google Scholar] [CrossRef]
- UniProt. Q16658·FSCN1_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/Q16658/entry (accessed on 7 October 2023).
- Zhang, F.-R.; Tao, L.-H.; Shen, Z.-Y.; Lv, Z.; Xu, L.-Y.; Li, E.-M. Fascin expression in human embryonic, fetal, and normal adult tissue. J. Histochem. Cytochem. 2008, 56, 193–199. [Google Scholar] [CrossRef] [PubMed]
- UniProt. Q7L576·CYFP1_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/Q7L576/entry (accessed on 7 October 2023).
- Werner, B.; Yadav, S. Phosphoregulation of the septin cytoskeleton in neuronal development and disease. Cytoskeleton 2022, 80, 275–289. [Google Scholar] [CrossRef]
- Xue, J.; Tsang, C.W.; Gai, W.P.; Malladi, C.S.; Trimble, W.S.; Rostas, J.A.; Robinson, P.J. Septin 3 (G-septin) is a developmentally regulated phosphoprotein enriched in presynaptic nerve terminals. J. Neurochem. 2004, 91, 579–590. [Google Scholar] [CrossRef]
- Tsang, C.W.; Fedchyshyn, M.; Harrison, J.; Xie, H.; Xue, J.; Robinson, P.J.; Wang, L.-Y.; Trimble, W.S. Superfluous role of mammalian septins 3 and 5 in neuronal development and synaptic transmission. Mol. Cell. Biol. 2008, 28, 7012–7029. [Google Scholar] [CrossRef]
- Kinoshita, A.; Shah, T.; Tangredi, M.M.; Strickland, D.K.; Hyman, B.T. The intracellular domain of the low density lipoprotein receptor-related protein modulates transactivation mediated by amyloid precursor protein and Fe65. J. Biol. Chem. 2003, 278, 41182–41188. [Google Scholar] [CrossRef]
- May, P.; Herz, J. LDL receptor-related proteins in neurodevelopment. Traffic 2003, 4, 291–301. [Google Scholar] [CrossRef]
- Gregorio, I.; Braghetta, P.; Bonaldo, P.; Cescon, M. Collagen VI in healthy and diseased nervous system. Dis. Models Mech. 2018, 11, dmm032946. [Google Scholar] [CrossRef]
- Raasakka, A.; Kursula, P. The myelin membrane-associated enzyme 2’,3’-cyclic nucleotide 3’-phosphodiesterase: On a highway to structure and function. Neurosci. Bull. 2014, 30, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Funa, K.; Sasahara, M. The roles of PDGF in development and during neurogenesis in the normal and diseased nervous system. J. Neuroimmune Pharmacol. 2014, 9, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Sil, S.; Periyasamy, P.; Thangaraj, A.; Chivero, E.T.; Buch, S. PDGF/PDGFR axis in the neural systems. Mol. Aspects Med. 2018, 62, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.K.; Tanaka, H.; Iwanaga, T.; Odani, S.; Kuwano, R. The presence of the 50-kDa subunit of dynactin complex in the nerve growth cone. Biochem. Biophys. Res. Commun. 1997, 233, 295–299. [Google Scholar] [CrossRef]
- UniProt. Q13561·DCTN2_HUMAN. 2023. Available online: https://www.uniprot.org/uniprotkb/Q13561/entry (accessed on 7 October 2023).
- Neel, D.V.; Basu, H.; Gunner, G.; Bergstresser, M.D.; Giadone, R.M.; Chung, H.; Miao, R.; Chou, V.; Brody, E.; Jiang, X.; et al. Gasdermin-E mediates mitochondrial damage in axons and neurodegeneration. Neuron 2023, 111, 1222–1240.e1229. [Google Scholar] [CrossRef]
- Patron, M.; Sprenger, H.-G.; Langer, T. m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration. Cell Res. 2018, 28, 296–306. [Google Scholar] [CrossRef]
- Li, Z.; Qiu, D.-h.; Liu, H.-Y.; Lei, Q.-F. Analysis of the nicastrin promoter rs10752637 polymorphism and its association with Alzheimer’s disease. Eur. J. Neurosci. 2009, 30, 1831–1836. [Google Scholar]
- Rao, S.S.; Winter, J.O. Adhesion molecule-modified biomaterials for neural tissue engineering. Front. Neuroeng. 2009, 2, 6. [Google Scholar] [CrossRef]
- Krishnan, U.M. Biomaterials in the treatment of Parkinson’s disease. Neurochem. Int. 2021, 145, 105003. [Google Scholar] [CrossRef]
- Shah, A.D.; Goode, R.J.; Huang, C.; Powell, D.R.; Schittenhelm, R.B. LFQ-analyst: An easy-to-use interactive web platform to analyze and visualize label-free proteomics data preprocessed with MaxQuant. J. Proteome Res. 2019, 19, 204–211. [Google Scholar] [CrossRef]



| Treatment | Cell Type | Base Media | Supplement |
|---|---|---|---|
| B27 | hADSC | Neurobasal media | 1% B27 supplement |
| CultureOne | hADSC | Neurobasal media | 1% CultureOne (C1) |
| N2 | hADSC | Neurobasal media | 1% N2 supplement |
| DMEM (undifferentiated control) | hADSC | DMEM/F12 + Glutamax—control media | 10% FBS |
| Positive Staining controls | SHSY-5Y or U87MG | DMEM/F12 + Glutamax—control media | 10% FBS |
| Negative staining controls | hADSC | DMEM/F12 + Glutamax—control media | 10% FBS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomila Pelegri, N.; Stanczak, A.M.; Bottomley, A.L.; Cummins, M.L.; Milthorpe, B.K.; Gorrie, C.A.; Padula, M.P.; Santos, J. Neural Marker Expression in Adipose-Derived Stem Cells Grown in PEG-Based 3D Matrix Is Enhanced in the Presence of B27 and CultureOne Supplements. Int. J. Mol. Sci. 2023, 24, 16269. https://doi.org/10.3390/ijms242216269
Gomila Pelegri N, Stanczak AM, Bottomley AL, Cummins ML, Milthorpe BK, Gorrie CA, Padula MP, Santos J. Neural Marker Expression in Adipose-Derived Stem Cells Grown in PEG-Based 3D Matrix Is Enhanced in the Presence of B27 and CultureOne Supplements. International Journal of Molecular Sciences. 2023; 24(22):16269. https://doi.org/10.3390/ijms242216269
Chicago/Turabian StyleGomila Pelegri, Neus, Aleksandra M. Stanczak, Amy L. Bottomley, Max L. Cummins, Bruce K. Milthorpe, Catherine A. Gorrie, Matthew P. Padula, and Jerran Santos. 2023. "Neural Marker Expression in Adipose-Derived Stem Cells Grown in PEG-Based 3D Matrix Is Enhanced in the Presence of B27 and CultureOne Supplements" International Journal of Molecular Sciences 24, no. 22: 16269. https://doi.org/10.3390/ijms242216269
APA StyleGomila Pelegri, N., Stanczak, A. M., Bottomley, A. L., Cummins, M. L., Milthorpe, B. K., Gorrie, C. A., Padula, M. P., & Santos, J. (2023). Neural Marker Expression in Adipose-Derived Stem Cells Grown in PEG-Based 3D Matrix Is Enhanced in the Presence of B27 and CultureOne Supplements. International Journal of Molecular Sciences, 24(22), 16269. https://doi.org/10.3390/ijms242216269









