Membrane-Bound Transcription Factor ZmNAC074 Positively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
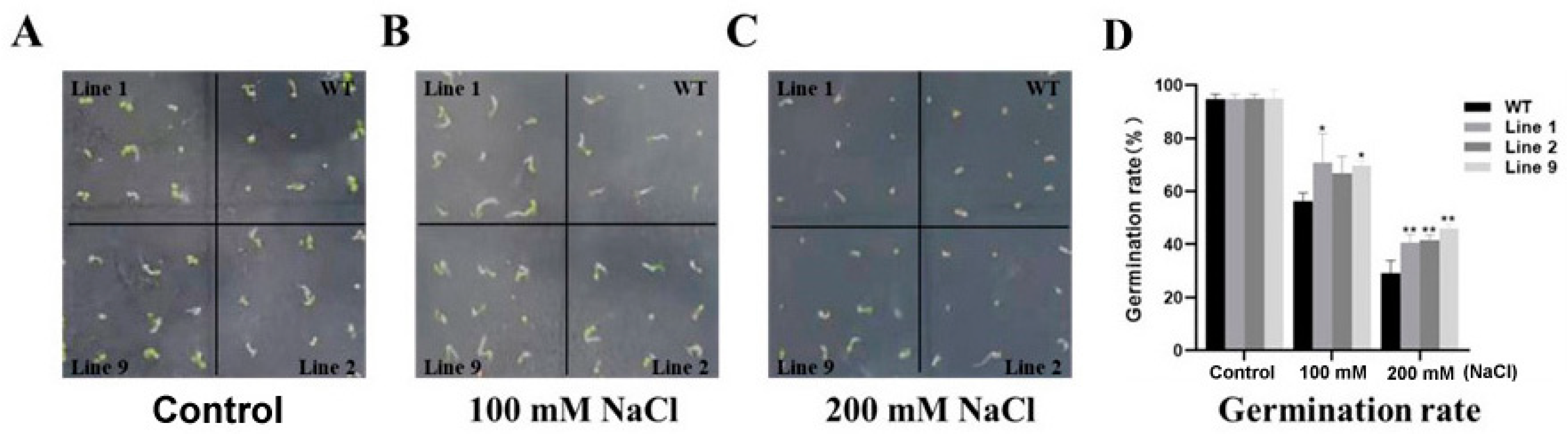
2.1. Germination of ZmNAC074-Overexpressing Transgenic Arabidopsis Seeds under Salt Stress
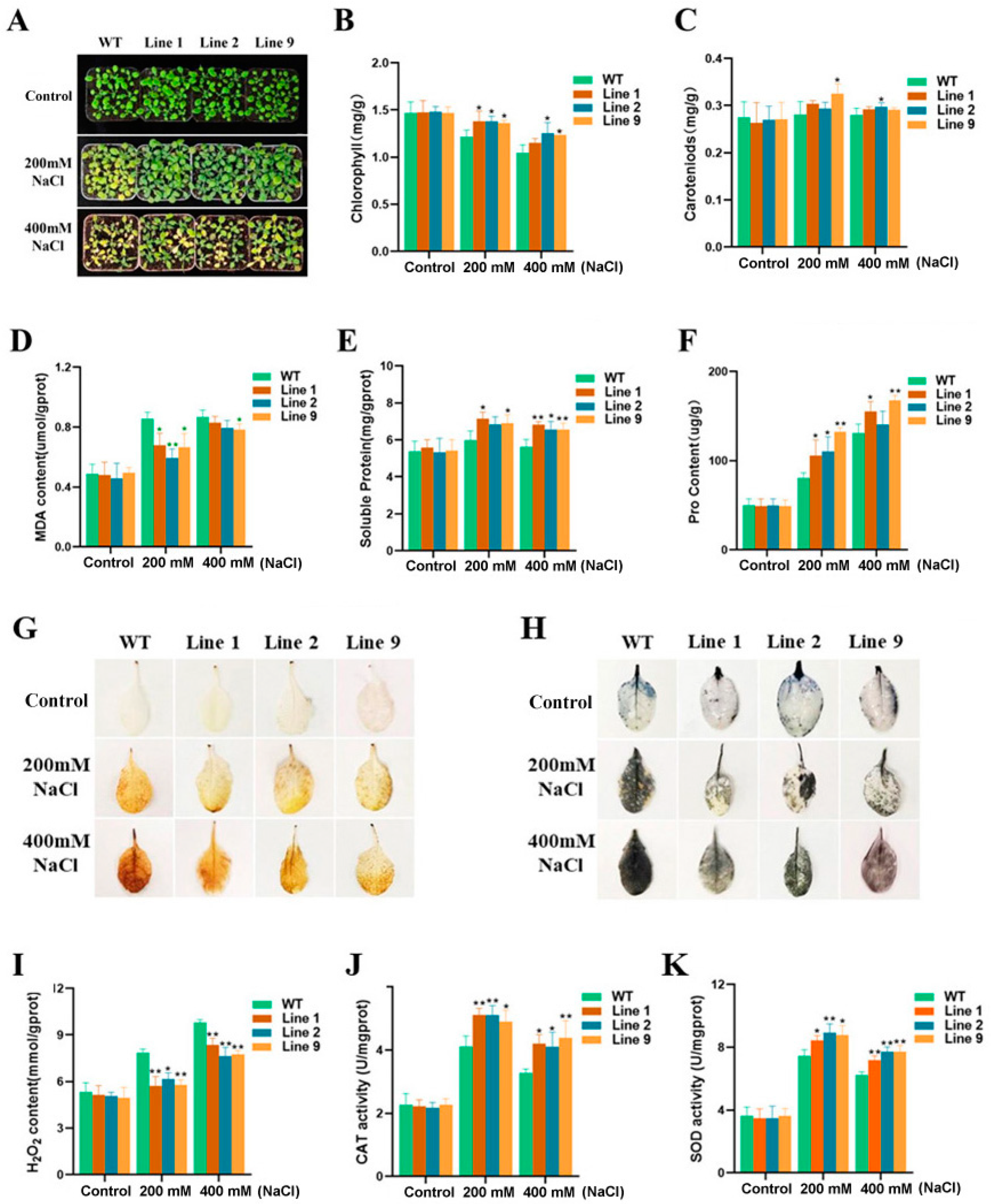
2.2. The Phenotype of ZmNAC074-Overexpressing Transgenic Arabidopsis Seedlings under Salt Stress and the Determination of Physiological Indexes
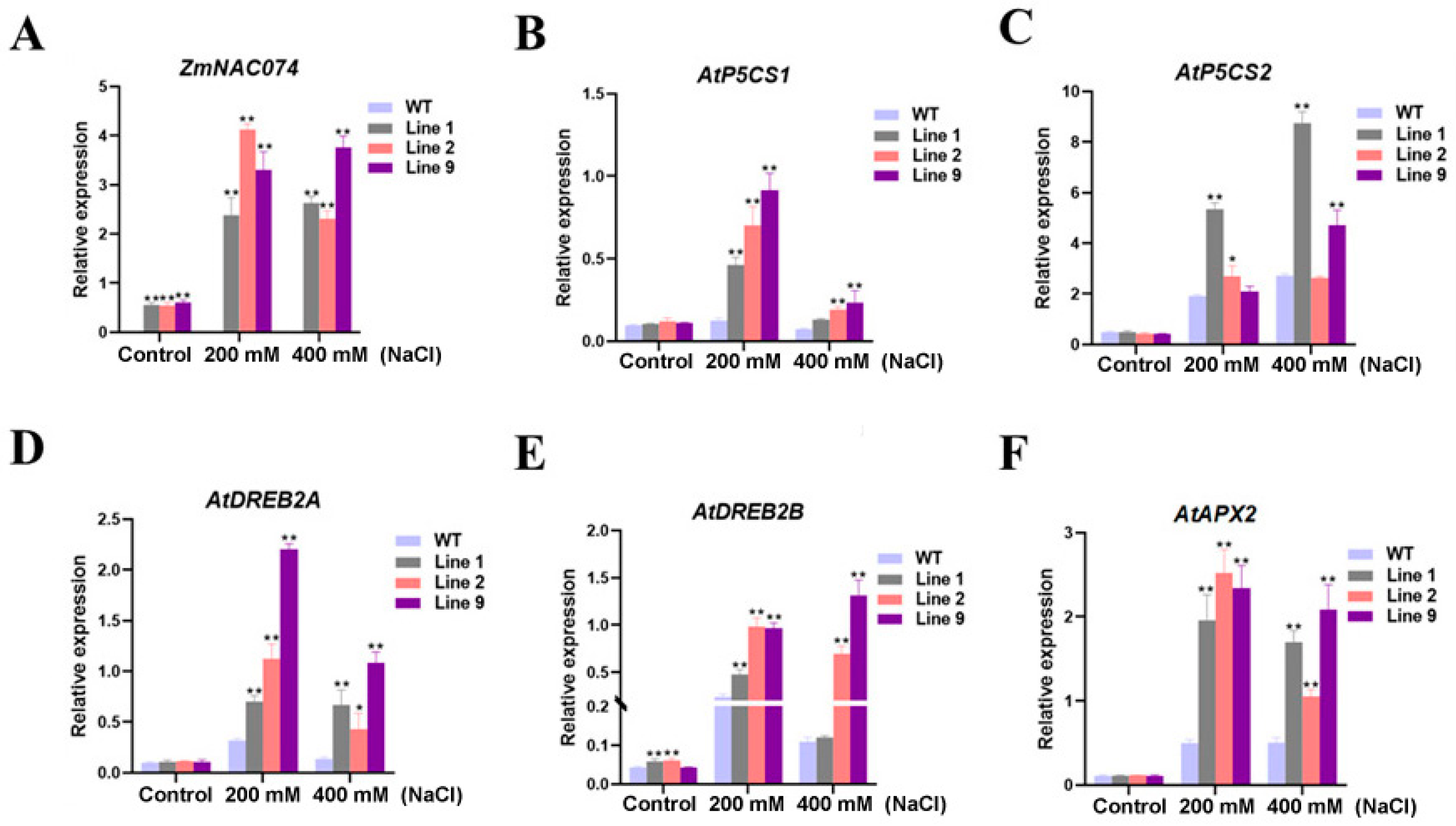
2.3. Overexpression of ZmNAC074 Regulates the Expression of Stress-Responsive Genes under Salt Stress
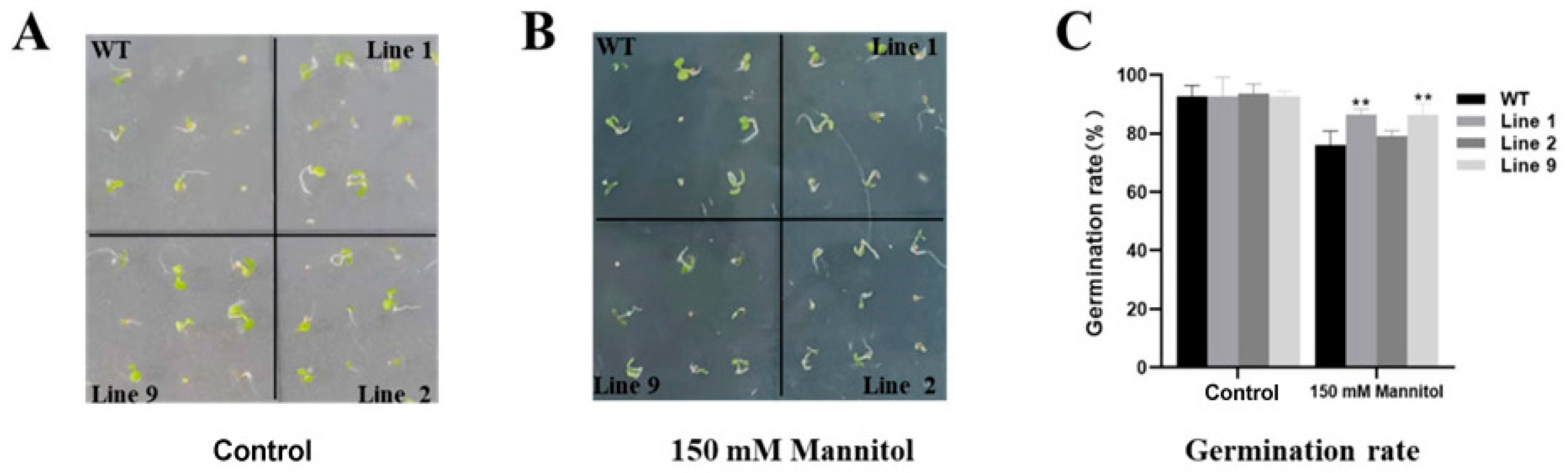
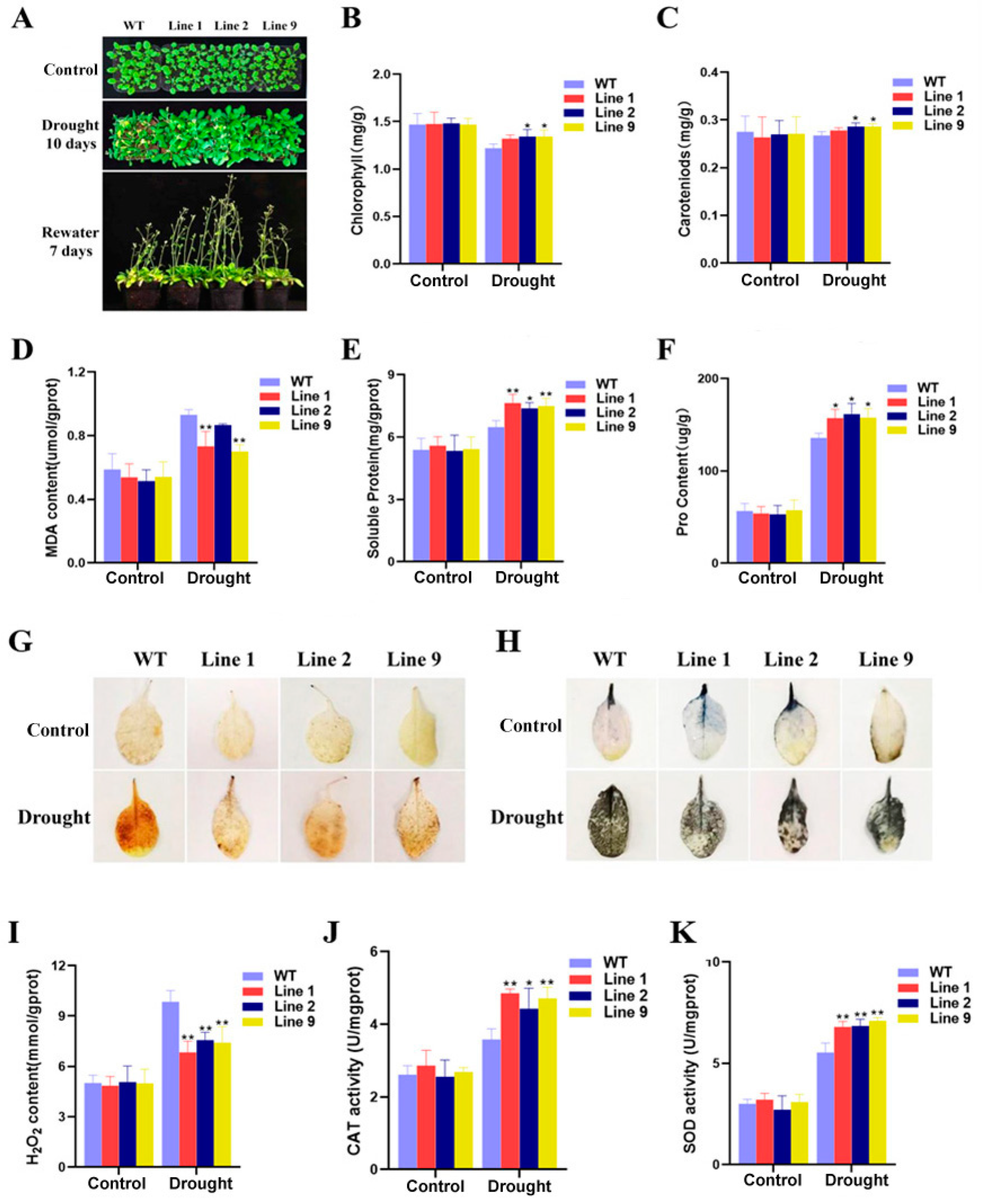
2.4. Germination of ZmNAC074-Overexpressing Transgenic Arabidopsis Seeds under Drought Stress
2.5. Phenotypic of ZmNAC074-Overexpressing Transgenic Arabidopsis Seedlings under Drought Stress and Determination of Physiological Indexes
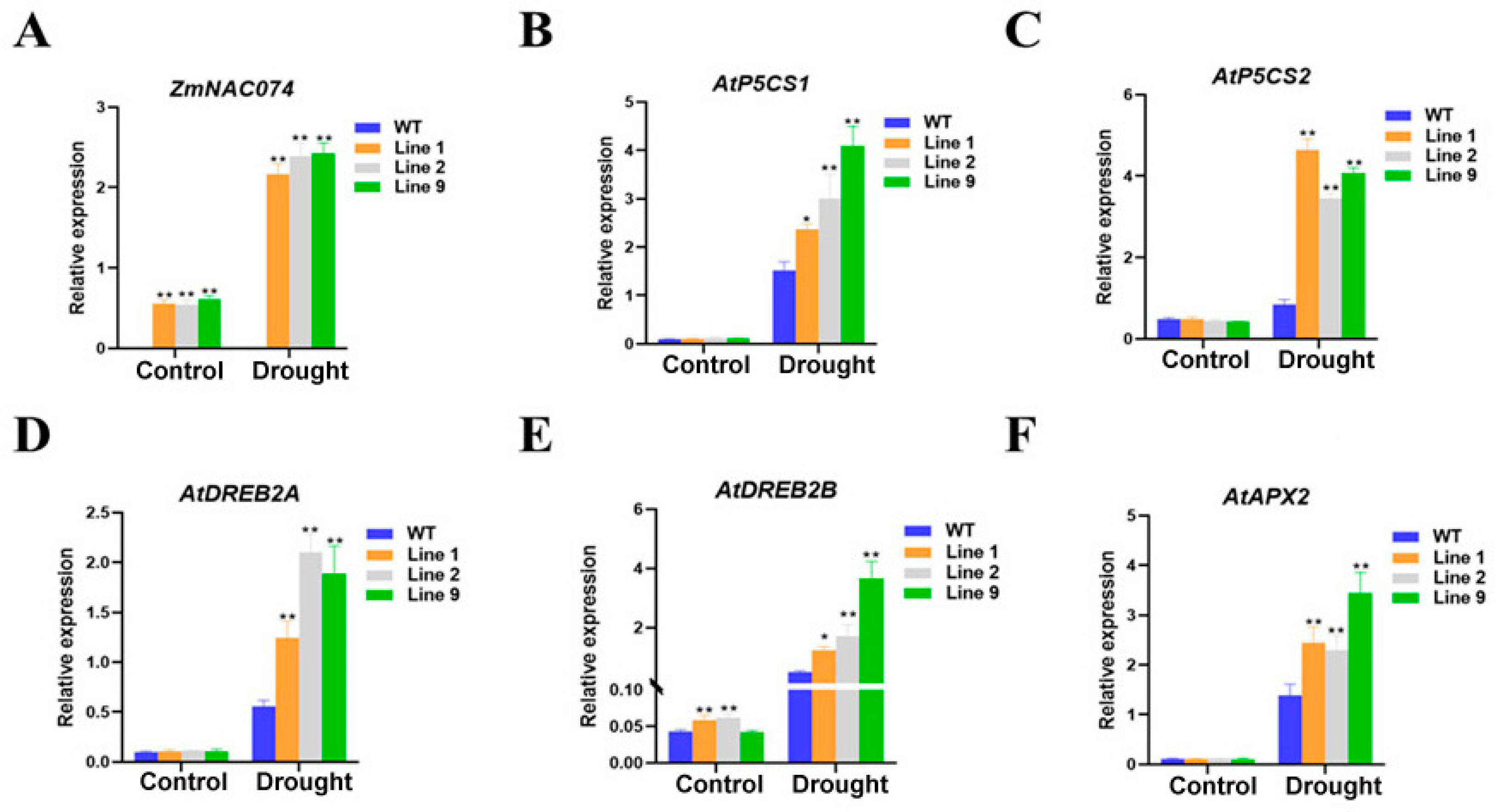
2.6. Overexpression of ZmNAC074 Regulates the Expression of Stress-Responsive Genes under Drought Stress
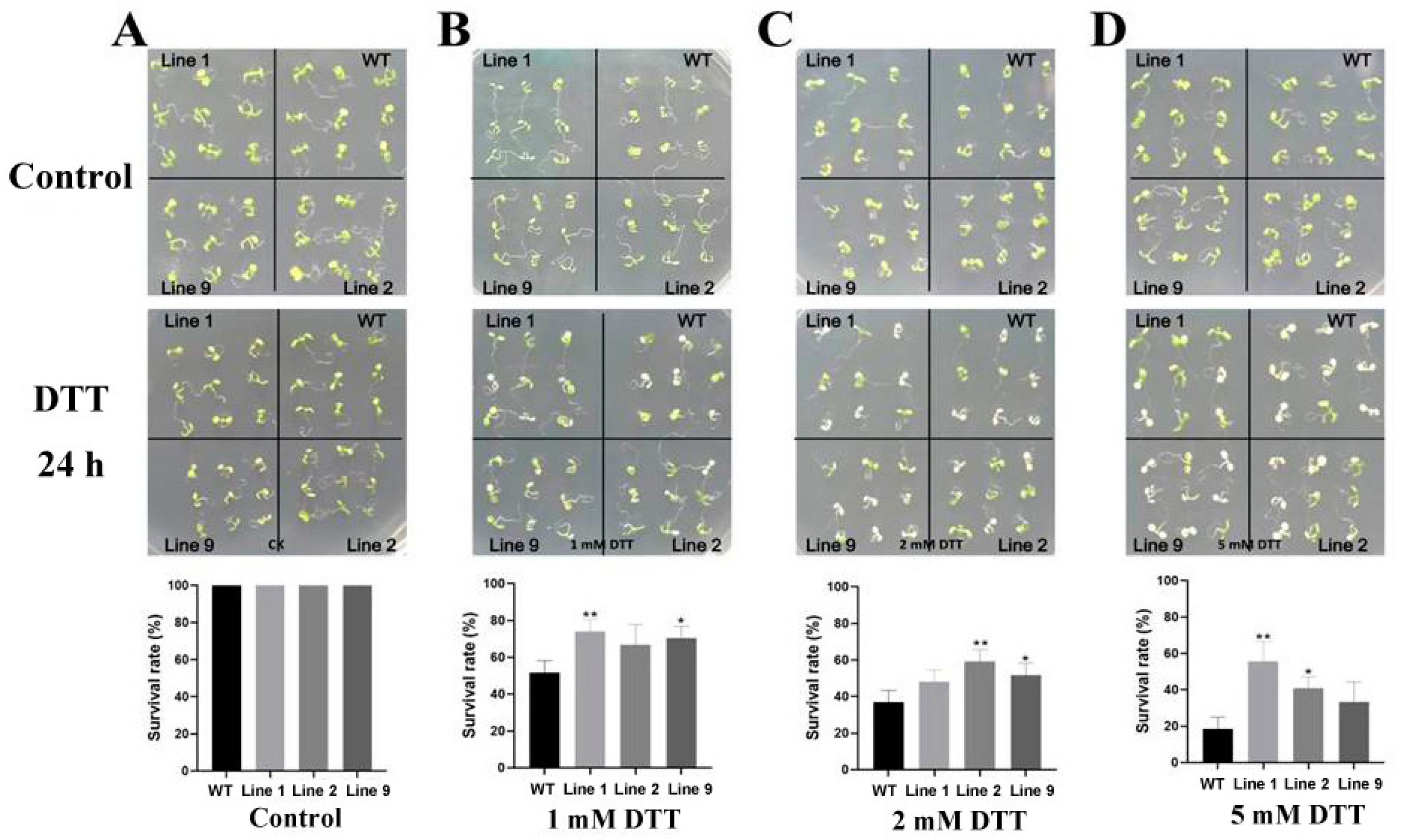
2.7. Survival Rate of Transgenic Arabidopsis under ER Stress
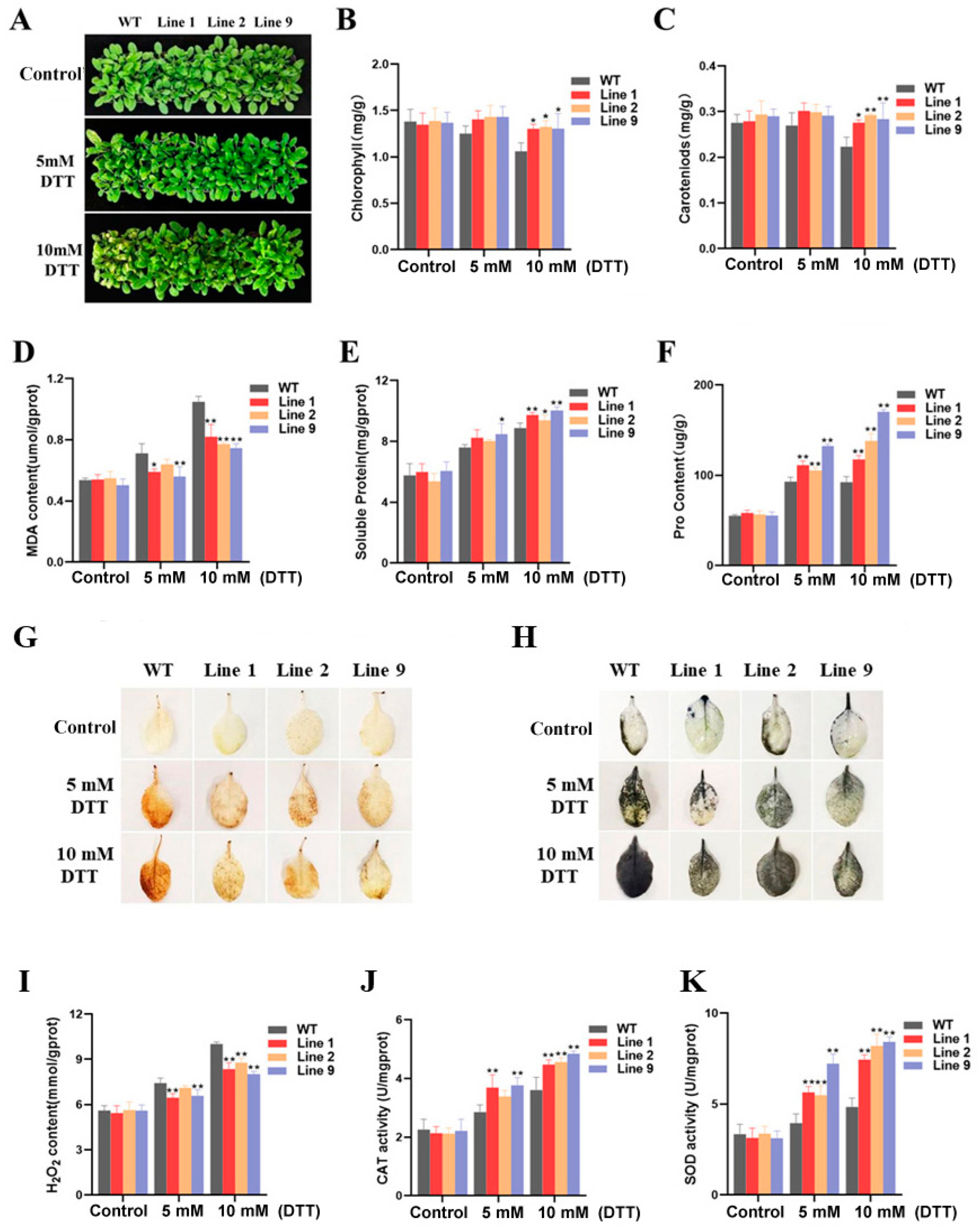
2.8. Phenotypic and Physiological Indexes of Transgenic Arabidopsis under ER Stress
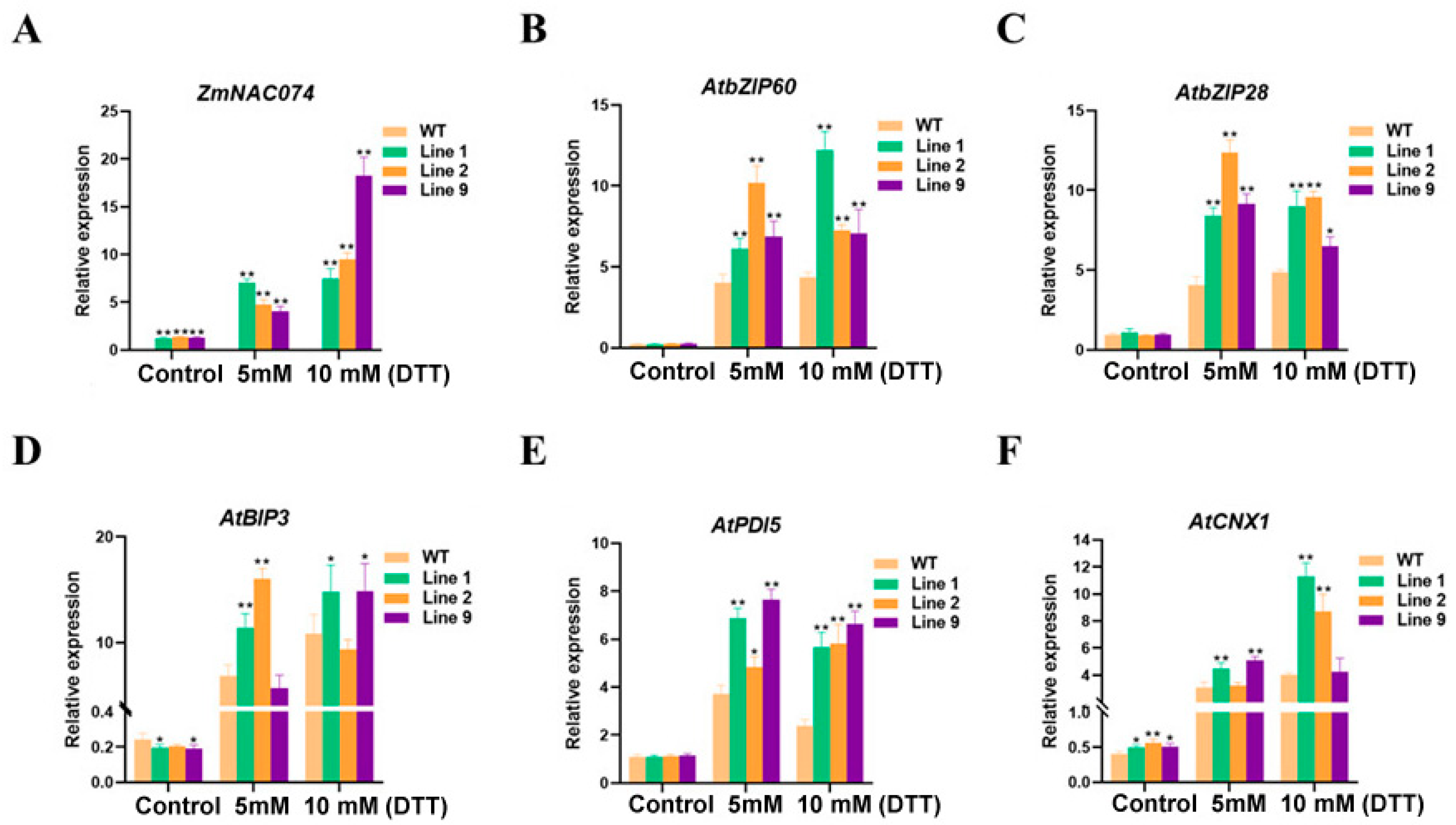
2.9. Expression Analysis of Stress-Responsive Genes in Transgenic Arabidopsis under ER Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. RNA Extraction and Quantitative Real-Time PCR
4.3. Germination Assay
4.4. Salt, Drought and ER Stress Treatments
4.5. Measurements of Physiological Indexes
4.6. Statistical Analysis
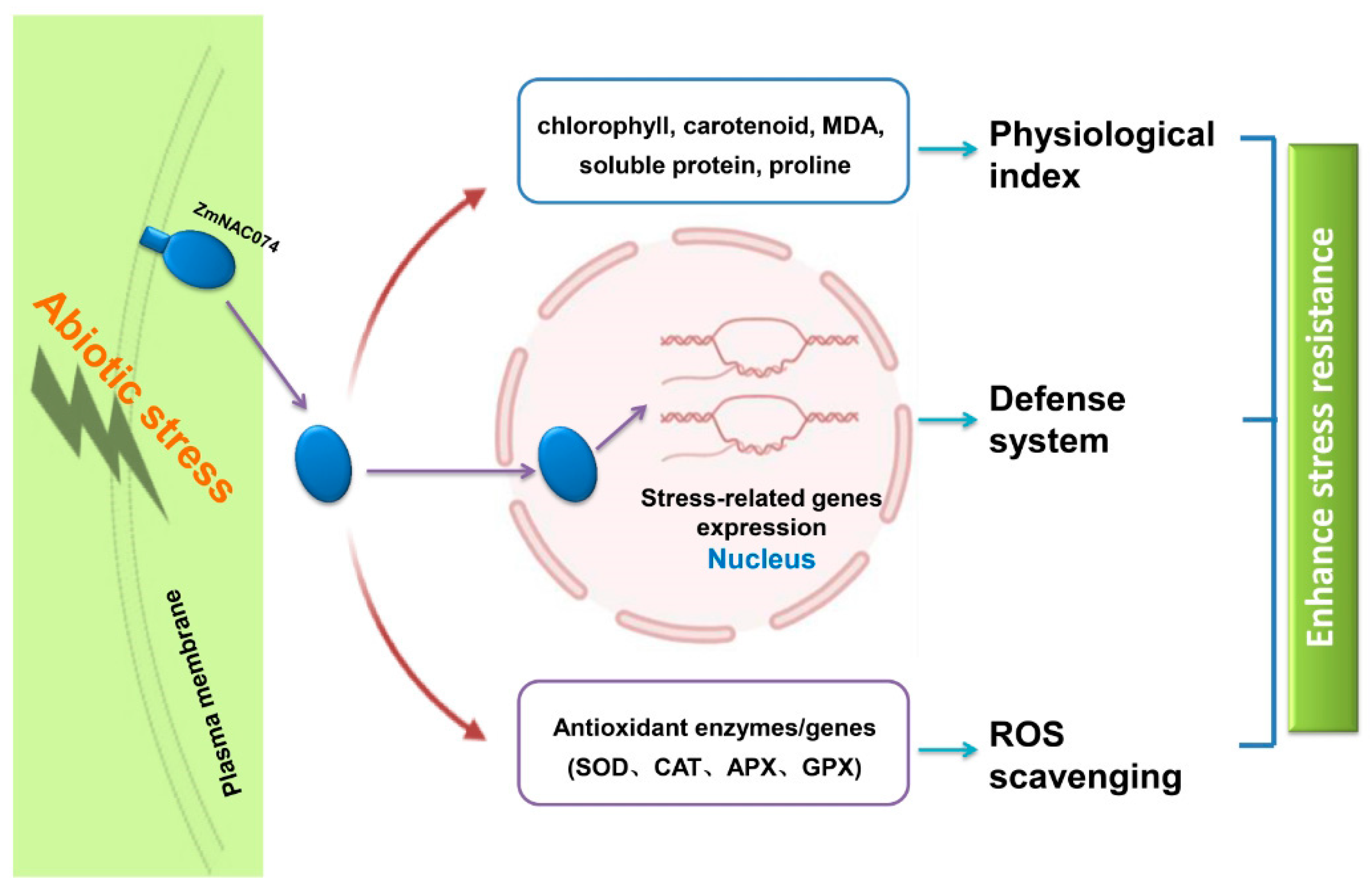
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sekhar, K.M.; Kota, V.R.; Reddy, T.P.; Rao, K.V.; Reddy, A.R. Amelioration of plant responses to drought under elevated CO2 by rejuvenating photosynthesis and nitrogen use efficiency: Implications for future climate-resilient crops. Photosynth. Res. 2021, 150, 21–40. [Google Scholar]
- Felemban, A.; Braguy, J.; Zurbriggen, M.D.; Al-Babili, S. Apocarotenoids Involved in Plant Development and Stress Response. Front. Plant Sci. 2019, 10, 1168. [Google Scholar]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic Stress and Reactive Oxygen Species: Generation, Signaling, and Defense Mechanisms. Antioxidants 2021, 10, 277. [Google Scholar]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [PubMed]
- Muhammad Aslam, M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar]
- Hai, N.N.; Chuong, N.N.; Tu, N.H.C.; Kisiala, A.; Hoang, X.L.T.; Thao, N.P. Role and Regulation of Cytokinins in Plant Response to Drought Stress. Plants 2020, 9, 422. [Google Scholar]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.J. The Role of the Plant Antioxidant System in Drought Tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [PubMed]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Singh, M.B.; Lohani, N.; Bhalla, P.L. The Role of Endoplasmic Reticulum Stress Response in Pollen Development and Heat Stress Tolerance. Front. Plant Sci. 2021, 12, 661062. [Google Scholar]
- Wan, S.; Jiang, L. Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 2016, 253, 753–764. [Google Scholar]
- Wang, J.; Shi, H.; Mao, X.; Li, R.Z. Transcription factors networks and their roles in plant responses to environmental stress. Ying Yong Sheng Tai Xue Bao 2006, 17, 1740–1746. [Google Scholar] [PubMed]
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Yang, H.; Hu, R.; Wei, D.; Tang, Q.; Wang, Z. The role of NAC transcription factors in flower development in plants. Sheng Wu Gong Cheng Xue Bao 2022, 38, 2687–2699. [Google Scholar] [PubMed]
- Lip, S.; Kurowska, M.; Szarejko, I. NAC transcription factors family and increased tolerance to water deficiency in plants. Postep. Biochem. 2013, 59, 295–304. [Google Scholar]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Deng, Z.; Wen, L.; Li, X.; Guo, Y. Potato NAC Transcription Factor StNAC053 Enhances Salt and Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 2568. [Google Scholar] [CrossRef]
- Yang, Z.T.; Lu, S.J.; Wang, M.J.; Bi, D.L.; Sun, L.; Zhou, S.F.; Song, Z.T.; Liu, J.X. A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 2014, 79, 1033–1043. [Google Scholar] [CrossRef]
- Sun, L.; Yang, Z.T.; Song, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Liu, J.X. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 2013, 76, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Poole, N.; Donovan, J.; Erenstein, O. Viewpoint: Agri-nutrition research: Revisiting the contribution of maize and wheat to human nutrition and health. Food Policy 2021, 100, 101976. [Google Scholar] [CrossRef] [PubMed]
- Sheoran, S.; Kaur, Y.; Kumar, S.; Shukla, S.; Rakshit, S.; Kumar, R. Recent Advances for Drought Stress Tolerance in Maize (Zea mays L.): Present Status and Future Prospects. Front. Plant Sci. 2022, 13, 872566. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nishiyama, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 2010, 1, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Xi, Y.; Ling, Q.; Zhou, Y.; Liu, X.; Qian, Y. ZmNAC074, a maize stress-responsive NAC transcription factor, confers heat stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 986628. [Google Scholar] [CrossRef] [PubMed]
- Lephatsi, M.M.; Meyer, V.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Plant Responses to Abiotic Stresses and Rhizobacterial Biostimulants: Metabolomics and Epigenetics Perspectives. Metabolites 2021, 11, 457. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Yue, X.; Zeng, H.; Zhu, J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 2014, 26, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.H.; Melencion, S.M.B.; Alinapon, C.V.; Kim, M.J.; Lee, E.S.; Paeng, S.K.; Park, J.H.; Nawkar, G.M.; Jung, Y.J.; Chae, H.B.; et al. The membrane-tethered NAC transcription factor, AtNTL7, contributes to ER-stress resistance in Arabidopsis. Biochem Biophys. Res. Commun. 2017, 488, 641–647. [Google Scholar] [CrossRef]
- Vargas-Hernández, B.Y.; Núñez-Muñoz, L.; Calderón-Pérez, B.; Xoconostle-Cázares, B.; Ruiz-Medrano, R. The NAC Transcription Factor ANAC087 Induces Aerial Rosette Development and Leaf Senescence in Arabidopsis. Front. Plant Sci. 2022, 13, 818107. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, J.; Zhang, N.; Xin, M.; Peng, H.; Hu, Z.; Ni, Z.; Du, J. The Wheat NAC Transcription Factor TaNAC2L Is Regulated at the Transcriptional and Post-Translational Levels and Promotes Heat Stress Tolerance in Transgenic Arabidopsis. PLoS ONE 2015, 10, e0135667. [Google Scholar] [CrossRef]
- Muhammad, I.; Shalmani, A.; Ali, M.; Yang, Q.H.; Ahmad, H.; Li, F.B. Mechanisms Regulating the Dynamics of Photosynthesis Under Abiotic Stresses. Front. Plant Sci. 2021, 11, 615942. [Google Scholar] [CrossRef]
- Davey, M.W.; Stals, E.; Panis, B.; Keulemans, J.; Swennen, R.L. High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 2005, 347, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Cao, X.; Khan, M.A.R. Proline, a multifaceted signaling molecule in plant responses to abiotic stress: Understanding the physiological mechanisms. Plant Biol. 2022, 24, 227–239. [Google Scholar] [CrossRef]
- Qiu, J.R.; Huang, Z.; Xiang, X.Y.; Xu, W.X.; Wang, J.T.; Chen, J.; Song, L.; Xiao, Y.; Li, X.; Ma, J.; et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 2020, 20, 542. [Google Scholar] [CrossRef]
- Székely, G.; Abrahám, E.; Cséplo, A.; Rigó, G.; Zsigmond, L.; Csiszár, J.; Ayaydin, F.; Strizhov, N.; Jásik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ding, Y.; Zhu, C. Sensitivity and Responses of Chloroplasts to Heat Stress in Plants. Front. Plant Sci. 2020, 11, 375. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A Sweetpotato Auxin Response Factor Gene (IbARF5) Is Involved in Carotenoid Biosynthesis and Salt and Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1307. [Google Scholar] [CrossRef]
- Guihur, A.; Rebeaud, M.E.; Goloubinoff, P. How do plants feel the heat and survive? Trends Biochem. Sci. 2022, 47, 824–838. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signaling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Huchzermeyer, B.; Menghani, E.; Khardia, P.; Shilu, A. Metabolic Pathway of Natural Antioxidants, Antioxidant Enzymes and ROS Providence. Antioxidants 2022, 11, 761. [Google Scholar] [CrossRef]
- Singh, A.; Mehta, S.; Yadav, S.; Nagar, G.; Ghosh, R.; Roy, A.; Chakraborty, A.; Singh, I.K. How to Cope with the Challenges of Environmental Stresses in the Era of Global Climate Change: An Update on ROS Stave off in Plants. Int. J. Mol. Sci. 2022, 23, 1995. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhao, X.; Chi, X.; Wang, Y.; Jia, C.; Zhang, H.; Zhou, G.; Hu, R. A novel Miscanthus NAC transcription factor MlNAC10 enhances drought and salinity tolerance in transgenic Arabidopsis. J. Plant Physiol. 2019, 233, 84–93. [Google Scholar] [CrossRef]
- Li, Q.; Wei, H.; Liu, L.; Yang, X.; Zhang, X.; Xie, Q. Unfolded protein response activation compensates endoplasmic reticulum-associated degradation deficiency in Arabidopsis. J. Integr. Plant Biol. 2017, 59, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Cho, Y.; Sharma, O.; Kanehara, K. What’s unique? The unfolded protein response in plants. J. Exp. Bot. 2022, 73, 1268–1276. [Google Scholar] [CrossRef]
- Yang, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Bi, D.L.; Sun, L.; Song, Z.T.; Zhang, S.S.; Zhou, S.F.; Liu, J.X. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014, 10, e1004243. [Google Scholar] [CrossRef]
- Wang, H.; Niu, H.; Zhai, Y.; Lu, M. Characterization of BiP Genes from Pepper (Capsicum annuum L.) and the Role of CaBiP1 in Response to Endoplasmic Reticulum and Multiple Abiotic Stresses. Front. Plant Sci. 2017, 8, 1122. [Google Scholar] [CrossRef]
- Liu, L.; Li, J. Communications Between the Endoplasmic Reticulum and Other Organelles During Abiotic Stress Response in Plants. Front. Plant Sci. 2019, 10, 749. [Google Scholar] [CrossRef] [PubMed]
- Ruberti, C.; Brandizzi, F. Conserved and plant-unique strategies for overcoming endoplasmic reticulum stress. Front. Plant Sci. 2014, 5, 69. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Xi, Y.; Xia, L.; Qiu, Z.; Liu, L.; Ma, H. Membrane-Bound Transcription Factor ZmNAC074 Positively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2023, 24, 16157. https://doi.org/10.3390/ijms242216157
Qian Y, Xi Y, Xia L, Qiu Z, Liu L, Ma H. Membrane-Bound Transcription Factor ZmNAC074 Positively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2023; 24(22):16157. https://doi.org/10.3390/ijms242216157
Chicago/Turabian StyleQian, Yexiong, Yan Xi, Lingxue Xia, Ziling Qiu, Li Liu, and Hui Ma. 2023. "Membrane-Bound Transcription Factor ZmNAC074 Positively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis" International Journal of Molecular Sciences 24, no. 22: 16157. https://doi.org/10.3390/ijms242216157
APA StyleQian, Y., Xi, Y., Xia, L., Qiu, Z., Liu, L., & Ma, H. (2023). Membrane-Bound Transcription Factor ZmNAC074 Positively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences, 24(22), 16157. https://doi.org/10.3390/ijms242216157





