Circulating Extracellular Vesicle-Derived microRNAs as Novel Diagnostic and Prognostic Biomarkers for Non-Viral-Related Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Participants
2.2. Characterization of EVs
2.3. Profiling of EV-Derived microRNAs
2.4. Functional Gene Annotation and Pathway Enrichment Analysis
2.5. Plasma EV-miRNA Expression in the Validation Set
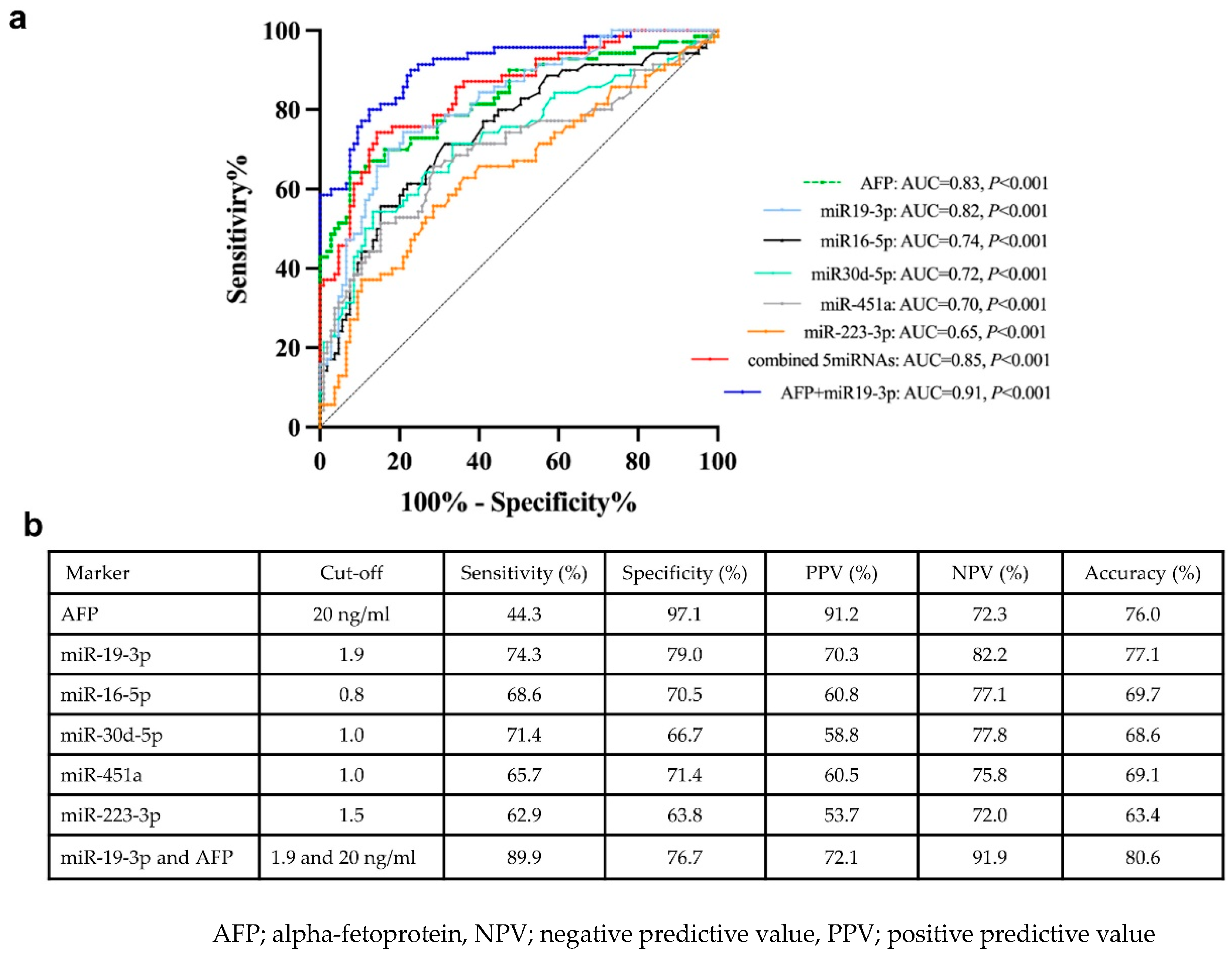
2.6. Diagnostic Role of Plasma EV-miRNAs
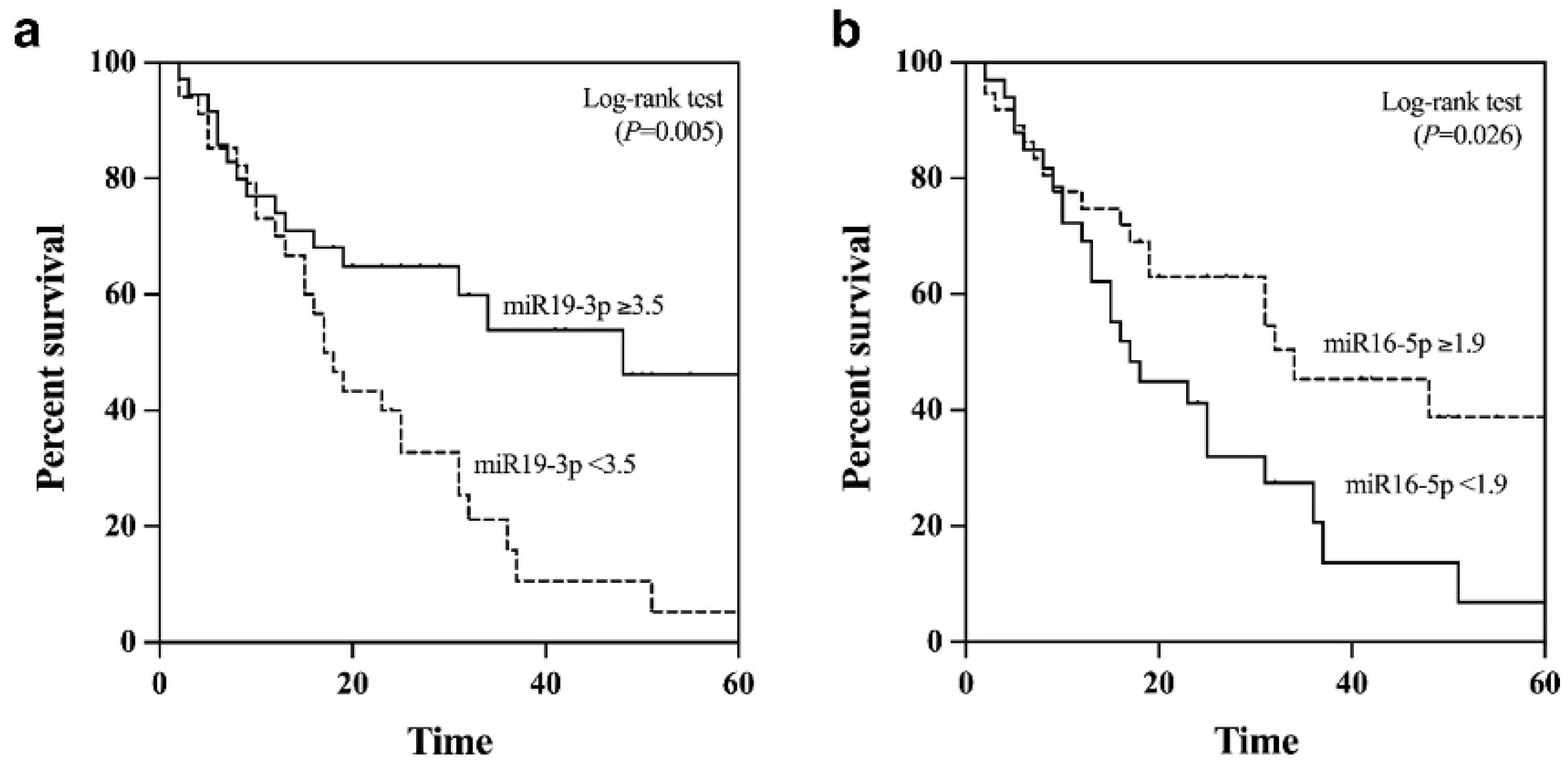
2.7. Prognostic Role of Plasma EV-miRNAs Regarding Overall Survival
3. Discussion
4. Materials and Methods
4.1. Research Subjects and Participant Consent
4.2. Blood Collection and Plasma Processing
4.3. EV Isolation
4.4. EV Characterization
4.4.1. NTA
4.4.2. TEM
4.4.3. Western Blotting
4.5. NanoString miRNA Expression Analysis
4.6. EV and Tissue miRNA Extraction
4.7. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, B.; Yeo, Y.H.; Feng, Y.; Xie, X.; Lee, D.H.; Fujii, H.; Wu, Y.; Kam, L.Y.; Ji, F.; et al. Prevalence, incidence, and outcome of non-alcoholic fatty liver disease in Asia, 1999–2019: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2019, 4, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e1. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Hsu, C.T.; Yeh, M.L.; Huang, C.F.; Huang, C.I.; Liang, P.C.; Lin, Y.H.; Hsieh, M.Y.; Wei, Y.J.; Hsieh, M.H.; et al. Early Fibrosis but Late Tumor Stage and Worse Outcomes in Hepatocellular Carcinoma Patients Without Hepatitis B or Hepatitis C. Dig. Dis. Sci. 2020, 65, 2120–2129. [Google Scholar] [CrossRef]
- Toyoda, H.; Kariyama, K.; Hiraoka, A.; Tsuji, K.; Ishikawa, T.; Hatanaka, T.; Naganuma, A.; Yasuda, S.; Nouso, K.; Kakizaki, S.; et al. Improved survival of viral hepatocellular carcinoma but not non-viral hepatocellular carcinoma from 2000 to 2020: A multi-centre cohort study of 6007 patients from high-volume academic centres in Japan. Aliment. Pharmacol. Ther. 2022, 56, 694–701. [Google Scholar] [CrossRef]
- von Felden, J.; Garcia-Lezana, T.; Schulze, K.; Losic, B.; Villanueva, A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut 2020, 69, 2025–2034. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef]
- Li, A.; Zhang, T.; Zheng, M.; Liu, Y.; Chen, Z. Exosomal proteins as potential markers of tumor diagnosis. J. Hematol. Oncol. 2017, 10, 175. [Google Scholar] [CrossRef]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef]
- Amorim, M.G.; Valieris, R.; Drummond, R.D.; Pizzi, M.P.; Freitas, V.M.; Sinigaglia-Coimbra, R.; Calin, G.A.; Pasqualini, R.; Arap, W.; Silva, I.T. A total transcriptome profiling method for plasma-derived extracellular vesicles: Applications for liquid biopsies. Sci. Rep. 2017, 7, 14395. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, G.M.; Liu, L.L.; Liu, C.; Liu, F.; Jiang, D.N.; Pu, X.Y. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol. Med. Rep. 2015, 12, 4683–4691. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis. Markers 2015, 2015, 893594. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. α-fetoprotein for hepatocellular carcinoma diagnosis: The demise of a brilliant star. Gastroenterology 2009, 137, 26–29. [Google Scholar] [CrossRef]
- Szabo, G.; Momen-Heravi, F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Vulf, M.; Shunkina, D.; Komar, A.; Bograya, M.; Zatolokin, P.; Kirienkova, E.; Gazatova, N.; Kozlov, I.; Litvinova, L. Analysis of miRNAs profiles in serum of patients with steatosis and steatohepatitis. Front. Cell Dev. Biol. 2021, 9, 736677. [Google Scholar] [CrossRef]
- Newman, L.A.; Useckaite, Z.; Johnson, J.; Sorich, M.J.; Hopkins, A.M.; Rowland, A. Selective isolation of liver-derived extracellular vesicles redefines performance of miRNA biomarkers for non-alcoholic fatty liver disease. Biomedicines 2022, 10, 195. [Google Scholar] [CrossRef]
- Sorop, A.; Iacob, R.; Iacob, S.; Constantinescu, D.; Chitoiu, L.; Fertig, T.E.; Dinischiotu, A.; Chivu-Economescu, M.; Bacalbasa, N.; Savu, L. Plasma small extracellular vesicles derived miR-21-5p and miR-92a-3p as potential biomarkers for hepatocellular carcinoma screening. Front. Genet. 2020, 11, 712. [Google Scholar] [CrossRef]
- Lin, J.; Lin, W.; Bai, Y.; Liao, Y.; Lin, Q.; Chen, L.; Wu, Y. Identification of exosomal hsa-miR-483-5p as a potential biomarker for hepatocellular carcinoma via microRNA expression profiling of tumor-derived exosomes. Exp. Cell Res. 2022, 417, 113232. [Google Scholar] [CrossRef]
- Zou, X.; Wei, J.; Huang, Z.; Zhou, X.; Lu, Z.; Zhu, W.; Miao, Y. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019, 8, 2810–2822. [Google Scholar] [CrossRef]
- Martens-Uzunova, E.S.; Kusuma, G.D.; Crucitta, S.; Lim, H.K.; Cooper, C.; Riches, J.E.; Azad, A.; Ochiya, T.; Boyle, G.M.; Southey, M.C. Androgens alter the heterogeneity of small extracellular vesicles and the small RNA cargo in prostate cancer. J. Extracell. Vesicles 2021, 10, e12136. [Google Scholar] [CrossRef] [PubMed]
- Ardizzone, A.; Calabrese, G.; Campolo, M.; Filippone, A.; Giuffrida, D.; Esposito, F.; Colarossi, C.; Cuzzocrea, S.; Esposito, E.; Paterniti, I. Role of miRNA-19a in cancer diagnosis and poor prognosis. Int. J. Mol. Sci. 2021, 22, 4697. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Teng, C.; Ma, J.; Fu, N.; Wang, L.; Wen, J.; Wang, T.-Y. miR-19 family: A promising biomarker and therapeutic target in heart, vessels and neurons. Life Sci. 2019, 232, 116651. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Du, L.; Yang, X.; Zhang, X.; Wang, L.; Yang, Y.; Li, J.; Wang, C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br. J. Cancer 2014, 111, 1985–1992. [Google Scholar] [CrossRef]
- Sochor, M.; Basova, P.; Pesta, M.; Dusilkova, N.; Bartos, J.; Burda, P.; Pospisil, V.; Stopka, T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014, 14, 448. [Google Scholar] [CrossRef]
- Han, Z.-B.; Zhong, L.; Teng, M.-J.; Fan, J.-W.; Tang, H.-M.; Wu, J.-Y.; Chen, H.-Y.; Wang, Z.-W.; Qiu, G.-Q.; Peng, Z.-H. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol. Oncol. 2012, 6, 445–457. [Google Scholar] [CrossRef]
- Hung, C.-L.; Yen, C.-S.; Tsai, H.-W.; Su, Y.-C.; Yen, C.-J. Upregulation of MicroRNA-19b predicts good prognosis in patients with hepatocellular carcinoma presenting with vascular invasion or multifocal disease. BMC Cancer 2015, 15, 665. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Li, Z.; Li, B.; Li, Z.; Li, R.; Guo, Q.; Xiong, L.; Yu, L.; Zhao, J. A systematic investigation based on microRNA-mediated gene regulatory network reveals that dysregulation of microRNA-19a/Cyclin D1 axis confers an oncogenic potential and a worse prognosis in human hepatocellular carcinoma. RNA Biol. 2015, 12, 643–657. [Google Scholar] [CrossRef][Green Version]
- Jiang, X.-M.; Yu, X.-N.; Liu, T.-T.; Zhu, H.-R.; Shi, X.; Bilegsaikhan, E.; Guo, H.-Y.; Song, G.-Q.; Weng, S.-Q.; Huang, X.-X. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed. Pharmacother. 2018, 105, 1147–1154. [Google Scholar] [CrossRef]
- Hu, B.; Tang, W.G.; Fan, J.; Xu, Y.; Sun, H.X. Differentially expressed miRNAs in hepatocellular carcinoma cells under hypoxic conditions are associated with transcription and phosphorylation. Oncol. Lett. 2018, 15, 467–474. [Google Scholar] [CrossRef]
- Sun, H.-X.; Yang, Z.-F.; Tang, W.-G.; Ke, A.-W.; Liu, W.-R.; Li, Y.; Gao, C.; Hu, B.; Fu, P.-Y.; Yu, M.-C. MicroRNA-19a-3p regulates cell growth through modulation of the PIK3IP1-AKT pathway in hepatocellular carcinoma. J. Cancer 2020, 11, 2476. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Chen, X.; Chen, S.; Ye, W.; Hou, K.; Liang, M. MiR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. J. Transl. Med. 2016, 14, 122. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, W.; Shen, P.; Bai, B.; Cao, L.-L. miR-19 is a potential clinical biomarker for gastrointestinal malignancy: A systematic review and meta-analysis. BioMed Res. Int. 2020, 2020, 2810150. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Makler, A.; Asghar, W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert Rev. Mol. Diagn. 2020, 20, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9, 1697028. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020. [Google Scholar] [CrossRef]
- Llovet, J.M.; Di Bisceglie, A.M.; Bruix, J.; Kramer, B.S.; Lencioni, R.; Zhu, A.X.; Sherman, M.; Schwartz, M.; Lotze, M.; Talwalkar, J. Design and endpoints of clinical trials in hepatocellular carcinoma. J. Natl. Cancer Inst. 2008, 100, 698–711. [Google Scholar] [CrossRef]
- Karlas, T.; Petroff, D.; Sasso, M.; Fan, J.-G.; Mi, Y.-Q.; de Lédinghen, V.; Kumar, M.; Lupsor-Platon, M.; Han, K.-H.; Cardoso, A.C. Individual patient data meta-analysis of controlled attenuation parameter (CAP) technology for assessing steatosis. J. Hepatol. 2017, 66, 1022–1030. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef] [PubMed]
- Povero, D.; Yamashita, H.; Ren, W.; Subramanian, M.G.; Myers, R.P.; Eguchi, A.; Simonetto, D.A.; Goodman, Z.D.; Harrison, S.A.; Sanyal, A.J. Characterization and proteome of circulating extracellular vesicles as potential biomarkers for NASH. Hepatol. Commun. 2020, 4, 1263–1278. [Google Scholar] [CrossRef] [PubMed]
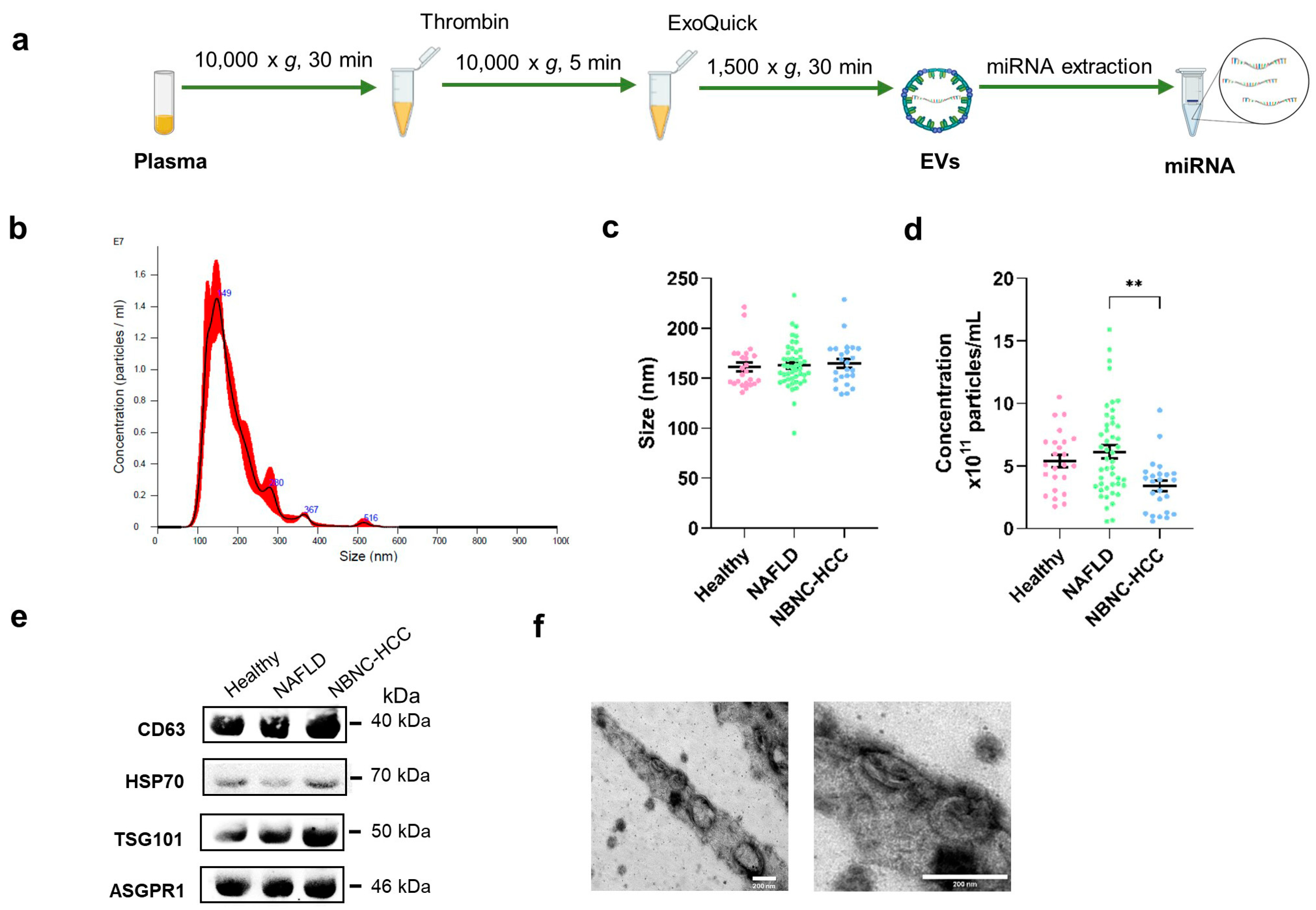
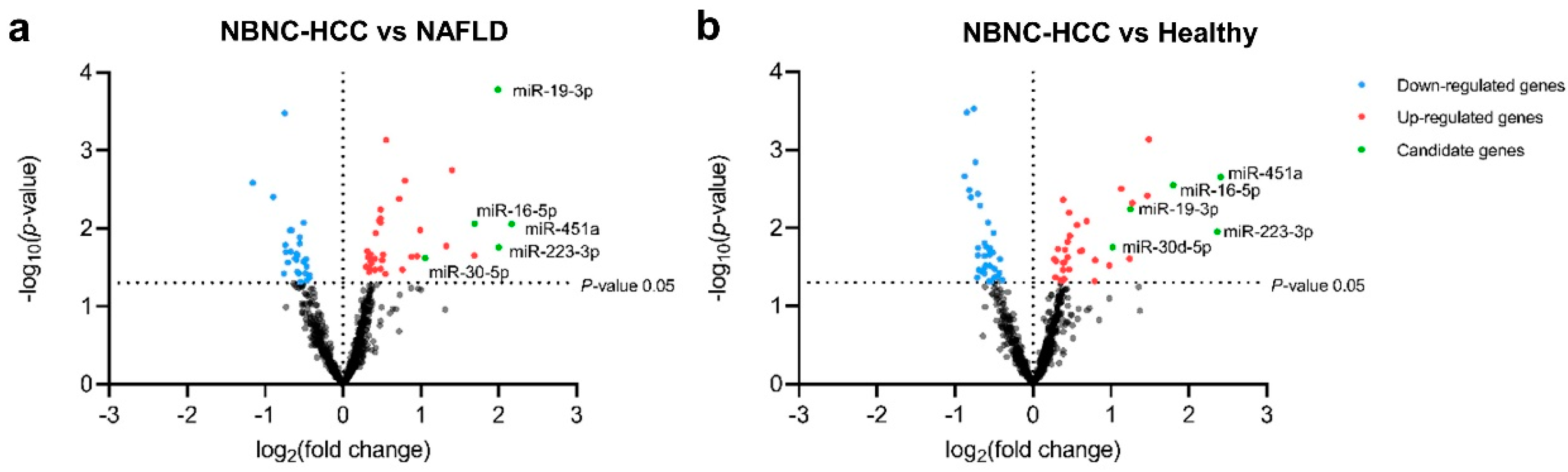
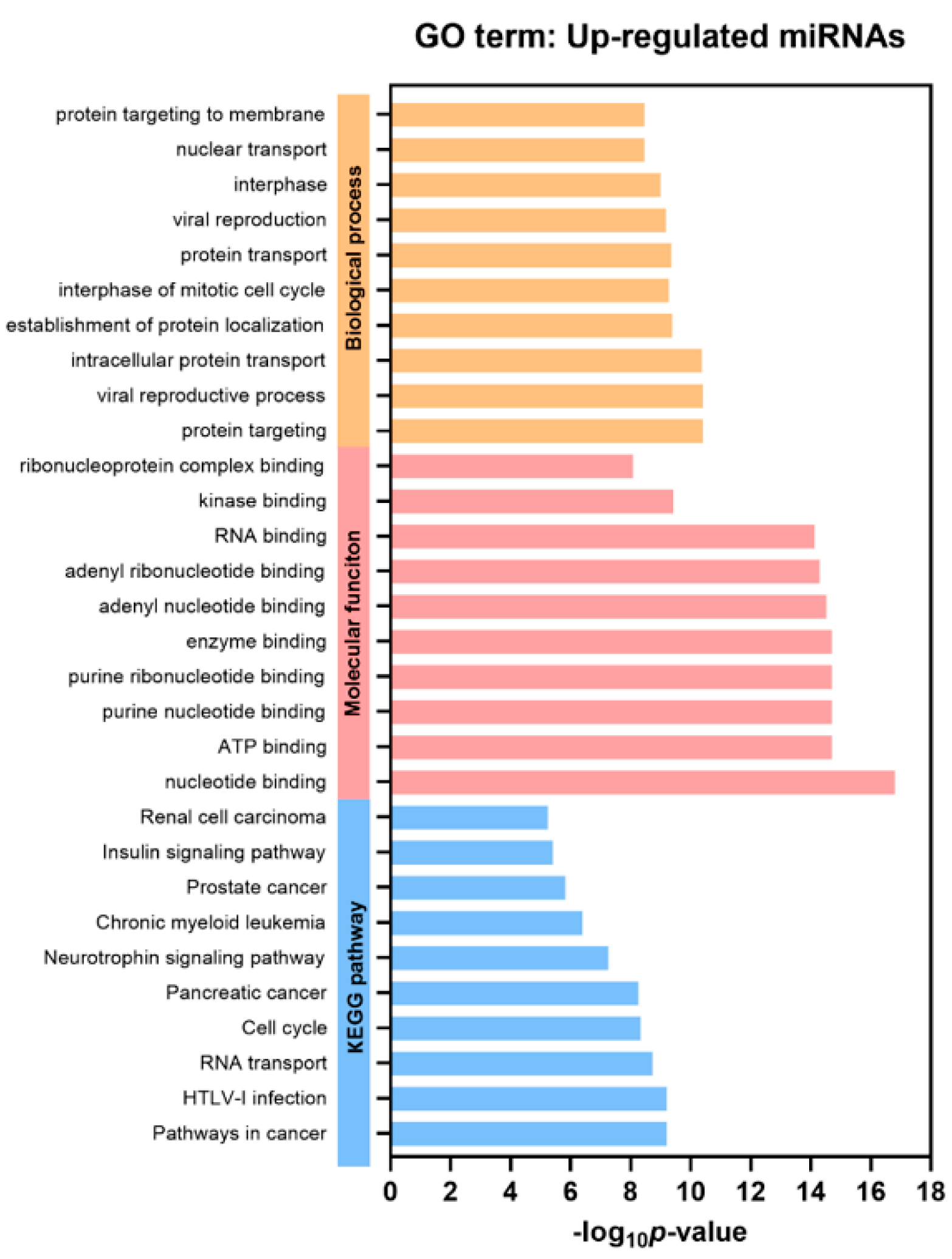
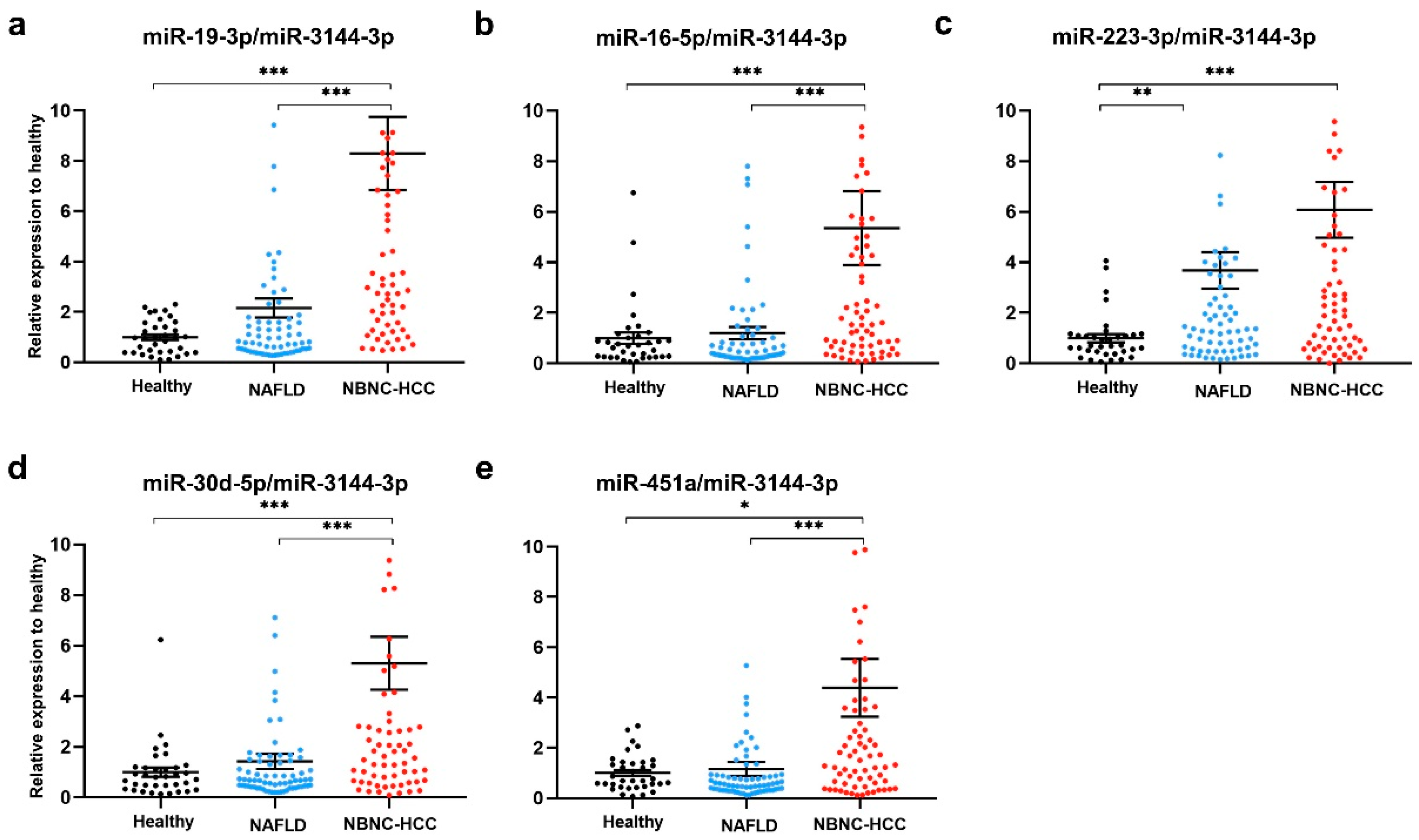
| Baseline Characteristics | Healthy Controls | Patients with NAFLD | Patients with NBNC-HCC | p |
|---|---|---|---|---|
| (n = 35) | (n = 70) | (n = 70) | ||
| Age (years) | 53.2 ± 5.3 | 50.7 ± 9.5 | 68.8 ± 11.4 | <0.001 |
| Gender (Male) | 4 (11.4) | 30 (42.9) | 54 (77.1) | <0.001 |
| Body mass index (kg/m2) | 22.8 ± 2.6 | 26.9 ± 4.1 | 24.3 ± 4.2 | <0.001 |
| Presence of metabolic syndrome | 33 (47.1) | 50 (71.4) | 0.006 | |
| Total bilirubin (mg/dL) | 0.7 ± 0.6 | 0.8 ± 0.5 | 0.845 | |
| Serum albumin (g/dL) | 4.0 ± 0.7 | 3.6 ± 0.5 | <0.001 * | |
| Aspartate aminotransferase (IU/L) | 24.5 ± 6.8 | 34.5 ± 17.9 | <0.001 * | |
| Alanine aminotransferase (IU/L) | 42.9 ± 42.8 | 60.8 ± 79.0 | 0.134 | |
| Alkaline phosphatase (IU/L) | 81.4 ± 45.2 | 146.7 ± 171.3 | <0.001 * | |
| Platelet count (109/L) | 225.4 ± 92.3 | 216.0 ± 104.7 | 0.157 | |
| Alpha fetoprotein (ng/mL) | 3.0 ± 2.9 | 3423.3 ± 12,469.8 | 0.025 * | |
| Controlled attenuation parameter (dB/m) | 196.5 ± 23.6 | 304.0 ± 42.9 | - | <0.001 * |
| Transient Elastography (kPa) | 3.8 ± 0.8 | 6.1 ± 1.7 | - | <0.001 * |
| BCLC stage (0-A/B/C) | - | 25(35.7)/27(38.6)/18(25.7) | - |
| Variables | Category | Overall Survival | |||
|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | ||||
| OR (95%CI) | p | OR (95%CI) | p | ||
| Age (years) | <60 vs. ≥60 | 0.62 (0.28–1.41) | 0.254 | ||
| Gender | Male vs. Female | 0.67 (0.32–1.42) | 0.295 | ||
| Total bilirubin (mg/dL) | <1.2 vs. ≥1.2 | 0.77 (0.49–1.27) | 0.303 | ||
| Serum albumin (g/dL) | <3.5 vs. ≥3.5 | 0.97 (0.46–2.04) | 0.931 | ||
| Aspartate aminotransferase (IU/L) | <55 vs. ≥55 | 1.64 (1.83–3.23) | 0.153 | ||
| Alanine aminotransferase (IU/L) | <50 vs. ≥50 | 1.00 (0.42–2.39) | 0.998 | ||
| Platelet count (109/L) | ≥100 vs. <100 | 1.05 (0.37–2.97) | 0.927 | ||
| Alpha fetoprotein (ng/mL) | <100 vs. ≥100 | 2.09 (1.08–4.08) | 0.029 * | 2.04 (1.01–4.13) | 0.048 * |
| Tumor size (cm.) | <5.0 vs. ≥5.0 | 1.63 (0.88–3.04) | 0.123 | ||
| BCLC stage | 0-A vs. B vs. C | 2.15 (1.38–3.36) | 0.001 * | 2.07 (1.29–3.32) | 0.002 * |
| EV-miR-19-3p | <3.5 vs. ≥3.5 | 2.39 (1.26–4.52) | 0.008 * | 2.71 (1.19–6.19) | 0.018 * |
| EV-miR-16-5p | <1.9 vs. ≥1.9 | 1.98 (1.07–3.69) | 0.030 * | 0.97 (0.44–2.13) | 0.944 |
| EV-miR-30d-5p | <2.0 vs. ≥2.0 | 1.56 (0.83–2.93) | 0.171 | ||
| EV-miR-451a | <1.7 vs. ≥1.7 | 1.74 (0.94–3.23) | 0.078 | ||
| EV-miR-223-3p | <2.5 vs. ≥2.5 | 1.71 (0.92–3.19) | 0.091 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonkaew, B.; Satthawiwat, N.; Pinjaroen, N.; Chuaypen, N.; Tangkijvanich, P. Circulating Extracellular Vesicle-Derived microRNAs as Novel Diagnostic and Prognostic Biomarkers for Non-Viral-Related Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 16043. https://doi.org/10.3390/ijms242216043
Boonkaew B, Satthawiwat N, Pinjaroen N, Chuaypen N, Tangkijvanich P. Circulating Extracellular Vesicle-Derived microRNAs as Novel Diagnostic and Prognostic Biomarkers for Non-Viral-Related Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2023; 24(22):16043. https://doi.org/10.3390/ijms242216043
Chicago/Turabian StyleBoonkaew, Bootsakorn, Nantawat Satthawiwat, Nutcha Pinjaroen, Natthaya Chuaypen, and Pisit Tangkijvanich. 2023. "Circulating Extracellular Vesicle-Derived microRNAs as Novel Diagnostic and Prognostic Biomarkers for Non-Viral-Related Hepatocellular Carcinoma" International Journal of Molecular Sciences 24, no. 22: 16043. https://doi.org/10.3390/ijms242216043
APA StyleBoonkaew, B., Satthawiwat, N., Pinjaroen, N., Chuaypen, N., & Tangkijvanich, P. (2023). Circulating Extracellular Vesicle-Derived microRNAs as Novel Diagnostic and Prognostic Biomarkers for Non-Viral-Related Hepatocellular Carcinoma. International Journal of Molecular Sciences, 24(22), 16043. https://doi.org/10.3390/ijms242216043







