Responsiveness of Candidate Genes on CoPv01CDRK/PhgPv01CDRK Loci in Common Bean Challenged by Anthracnose and Angular Leaf Spot Pathogens
Abstract
:1. Introduction
2. Results
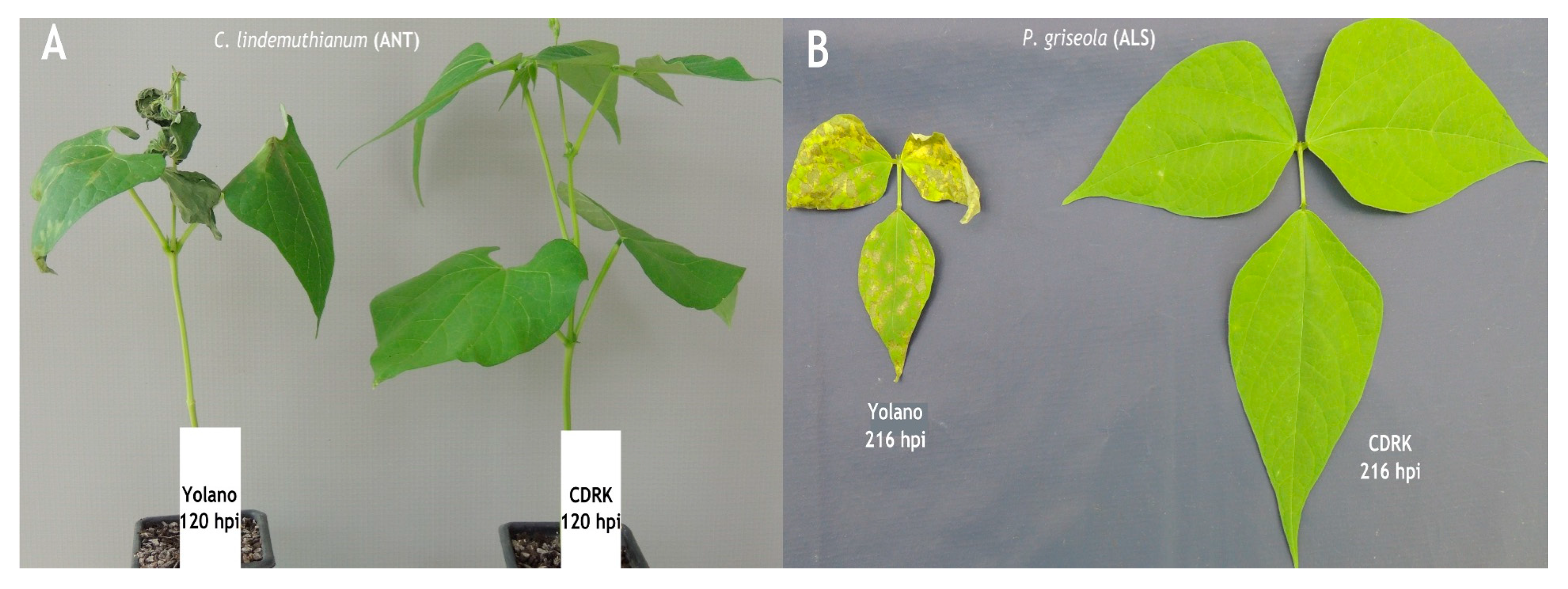
2.1. Phenotypic Evaluation of Cultivars
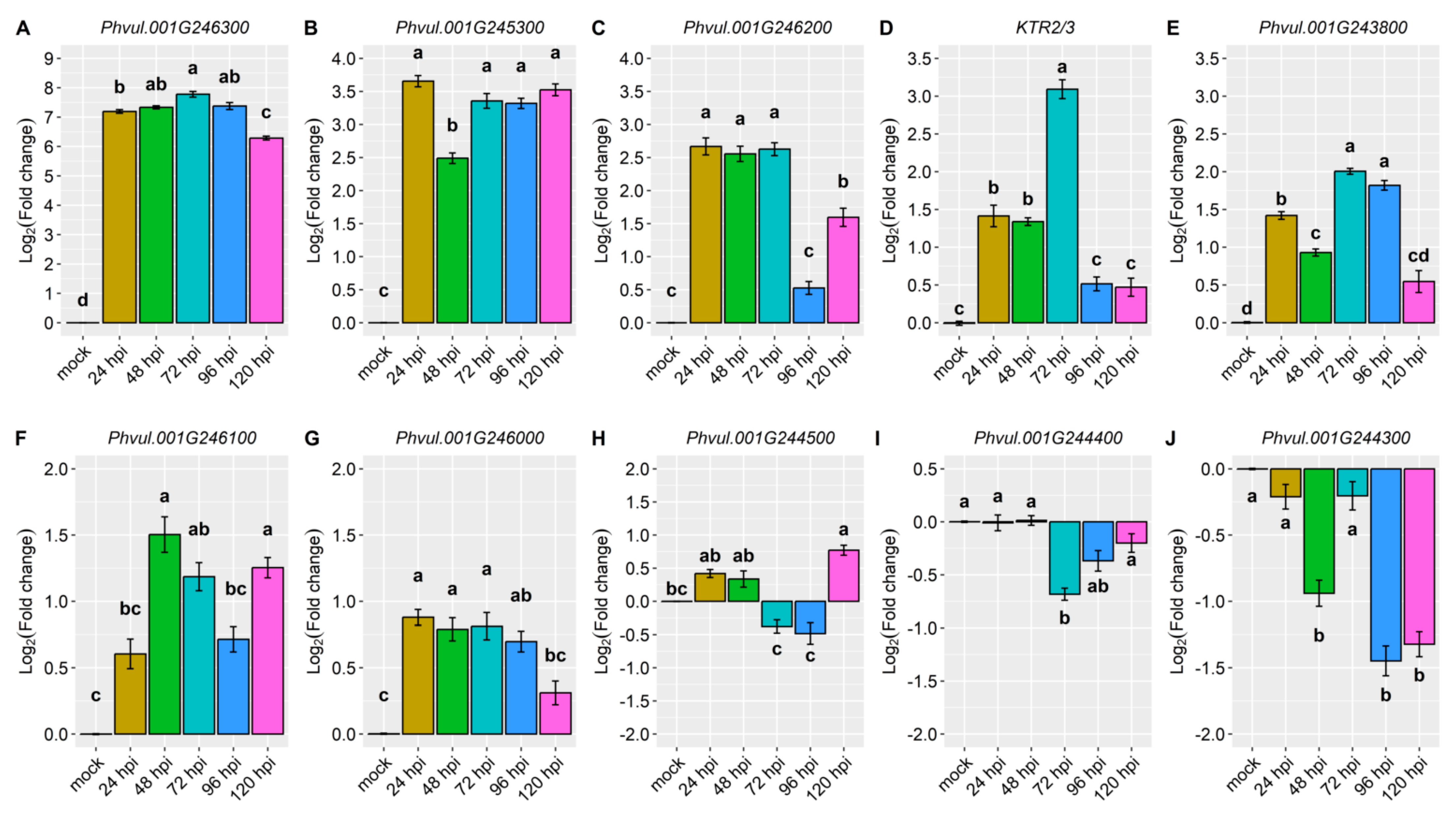
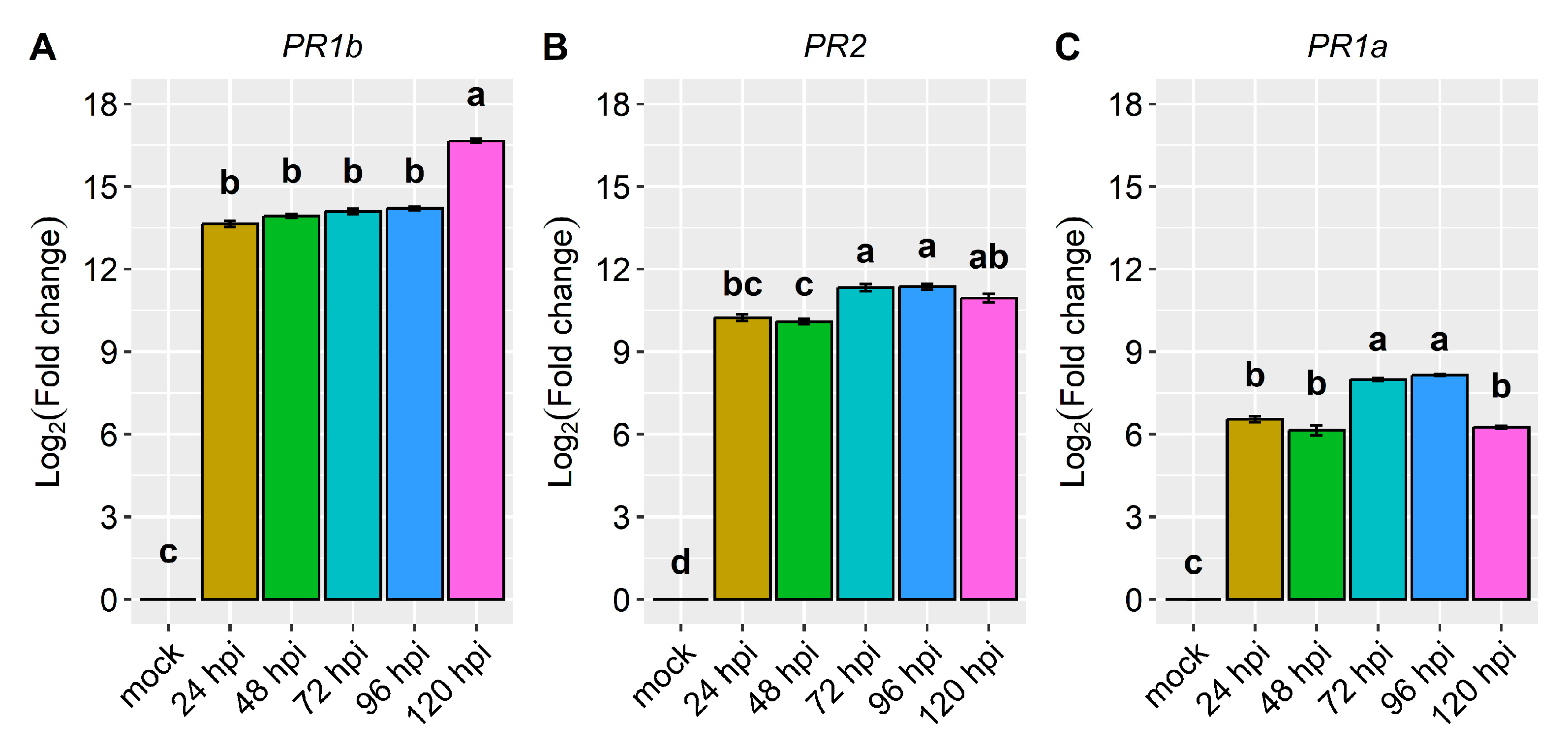
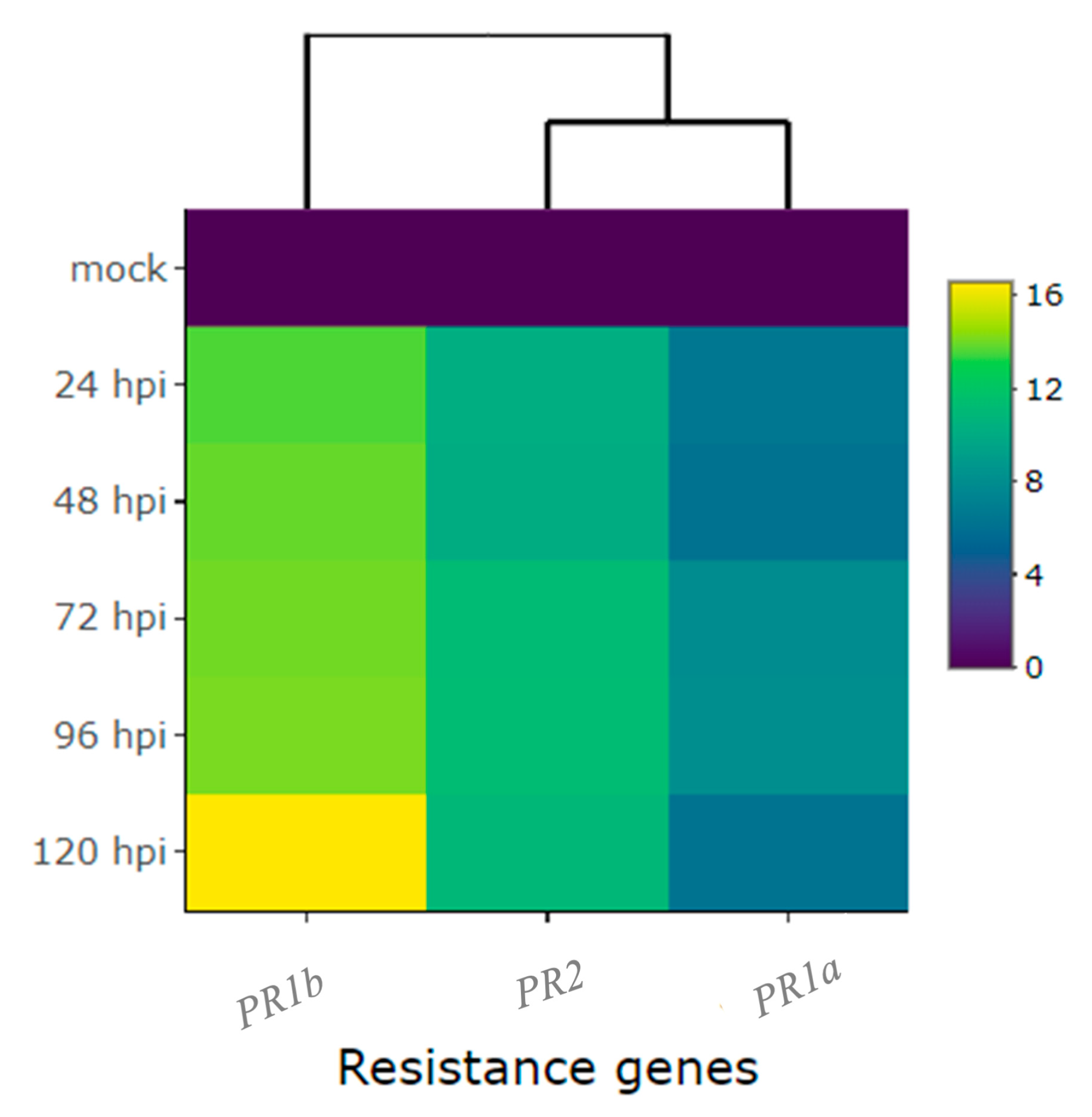
2.2. Differential Expression of Candidate and Defense Genes in the CDRK Cultivar Inoculated with Race 73 of C. lindemuthianum
2.3. Differential Expression of Candidate and Defense Genes in CDRK Cultivar Inoculated with P. griseola Race 63-39
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Experimental Design
4.2. Pathogenesis Assays
4.3. Total RNA Extraction and cDNA Synthesis
4.4. Target Genes and Primer Design
4.5. Quantitative PCR (qPCR) and Data Analysis
| Gene Model a | Reference Genes | Primers Forward (F) and Reverse (R) (5′-3′) | Amplicon (bp) | Tm (°C) | E b | R² c |
|---|---|---|---|---|---|---|
| Phvul.001G133200 | IDE | F: AAGCAGGTATCTTGGCCATCTC | 126 | 60.16 | 1.04 | 0.99 |
| R: AAAGCAAACTCCAAGCTCCAATC | 59.99 | |||||
| Phvul.008G011000 | ACT | F: GAAGTTCTCTTCCAACCATCC | 154 | 59.67 | 1 | 0.98 |
| R: TTTCCT TGCTCATTCTGTCCG | 58.38 | |||||
| Phvul.001G243800 | Co-1 | F: CCTCAAGGTGGGGCTTTTGAG | 118 | 61.16 | 1.04 | 0.99 |
| R: TCACCGAGAAACTCCCATTGC | 60.61 | |||||
| KTR2/3 | Co-x | F: ATGCACAGGGGAATGGGATG | 279 | 60.11 | 1.04 | 0.98 |
| R: GCCATAGCGAGTGAGAGTGCG | 63.42 | |||||
| Phvul.001G244300 | Co-AC | F: GAAACGTCTCCGCAGAATAGTG | 150 | 59.4 | 1.03 | 0.99 |
| R: GTCTTGTGTTGTTCCTTGGAGTTG | 60.44 | |||||
| Phvul.001G244400 | Co-AC | F: TACAGCAAGAGAGCGGTTAAAGG | 121 | 60.62 | 1.01 | 0.99 |
| R: CCCTTTGTCACTTTGTTTTGAAGC | 59.67 | |||||
| Phvul.001G244500 | Co-AC | F: CAATGCACAGCTCGCAACTC | 141 | 60.45 | 1.09 | 0.97 |
| R: GGAACTGTGAAAGCTCTGCTAAC | 59.81 | |||||
| Phvul.001G245300 | CoPv01CDRK/ PhgPv01CDRK | F: TCTGCTGGAAGGGTGGTAGTC | 93 | 61.17 | 1.07 | 0.98 |
| R: GGACGTTATGTGAACAAGGTTTGC | 61.08 | |||||
| Phvul.001G246000 | CoPv01CDRK/ PhgPv01CDRK | F: ATGAAGCGGATGGATGTCTTG | 132 | 58.43 | 1.01 | 0.97 |
| R: TCTACGAAGCTTAGGCAATTGAG | 58.57 | |||||
| Phvul.001G246100 | CoPv01CDRK/ PhgPv01CDRK | F: CACGGTATCCTCAGCGAAGAC | 119 | 60.53 | 1.05 | 0.99 |
| R: CAGCAGTCAGCACATACTGGAG | 60.99 | |||||
| Phvul.001G246200 | CoPv01CDRK/ PhgPv01CDRK | F: GAAGGAGGCTGTGACGTGTTC | 150 | 61.2 | 1.04 | 0.99 |
| R: CCATCGCCACCGTTGATACTC | 61.13 | |||||
| Phvul.001G246300 | CoPv01CDRK/ PhgPv01CDRK | F: CTTCTTCCCTTCACTTCGATACC | 87 | 58.57 | 1.09 | 0.99 |
| R: GTTGAGAGTGTTTGTGGCAGT | 58.98 | |||||
| Phvul.003G109100 | PR1a | F: GTCCTAACGGAGGATCACTCA | 148 | 58.62 | 1.06 | 0.99 |
| R: CAGGGATTGGCCAGAAGGTAT | 59.5 | |||||
| Phvul.006G196900 | PR1b | F: GGTTTGCCTATGATCCCAATGC | 115 | 59.96 | 1.03 | 0.99 |
| R: TGTTGTGAGCGTTGAGGAAGTC | 61.06 | |||||
| Phvul.009G256400 | PR2 | F: CAGAGGTTCTCATTTGCTGCTTTC | 98 | 60.62 | 1.07 | 0.99 |
| R: ATGCCATAACACACCCCGATTTG | 61.75 |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Padder, B.A.; Sharma, P.N.; Awale, H.E.; Kelly, J.D. Colletotrichum lindemuthianum, the causal agent of bean anthracnose. J. Plant Pathol. 2017, 99, 317–330. [Google Scholar] [CrossRef]
- Crous, P.W.; Liebenberg, M.M.; Braun, U.; Johannes, Z.; Groenewald, J.Z. Re-evaluating the taxonomic status of Phaeoisariopsis griseola, the causal agent of angular leaf spot of bean. Stud. Mycol. 2006, 55, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Miklas, P.N.; Kelly, J.D.; Beebe, S.E.; Blair, M.W. Common bean breeding for resistance against biotic and abiotic stresses: From classical to MAS breeding. Euphytica 2006, 147, 105–131. [Google Scholar] [CrossRef]
- Singh, S.P.; Schwartz, H.F. Breeding common bean for resistance to diseases: A review. Crop Sci. 2010, 50, 2199–2223. [Google Scholar] [CrossRef]
- Guzman, P. Characterization of variability in the fungus Phaeoisariopsis griseola suggests coevolution with the common bean (Phaseolus vulgaris). Phytopathology 1995, 85, 600. [Google Scholar] [CrossRef]
- Pastor-Corrales, M.A.; Jara, C.E. La evolución de Phaeoisariopsis griseola con el frijol común en América Latina. Fitop Colomb. 1995, 19, 15–24. [Google Scholar]
- Pastor-Corrales, M.A.; Jara, C.; Singh, S.P. Pathogenic variation in, sources of, and breeding for resistance to Phaeoisariopsis griseola causing angular leaf spot in common bean. Euphytica 1998, 103, 161–171. [Google Scholar] [CrossRef]
- Stenglein, S.A.; Balatti, P.A. Genetic diversity of Phaeoisariopsis griseola in Argentina as revealed by pathogenic and molecular markers. Physiol. Mol. Plant Pathol. 2006, 68, 158–167. [Google Scholar] [CrossRef]
- Gonçalves-Vidigal, M.C.; Cruz, A.S.; Garcia, A.; Kami, J.; Vidigal Filho, P.S.; Sousa, L.L.; McClean, P.; Gepts, P.; Pastor-Corrales, M.A. Linkage mapping of the Phg-1 and Co-14 genes for resistance to angular leaf spot and anthracnose in the common bean cultivar AND 277. Theor. Appl. Genet. 2011, 122, 893–903. [Google Scholar] [CrossRef]
- Zuiderveen, G.H.; Padder, B.A.; Kamfwa, K.; Song, Q.; Kelly, J.D. Genome-wide association study of anthracnose resistance in Andean beans (Phaseolus vulgaris). PLoS ONE 2016, 11, e0156391. [Google Scholar] [CrossRef]
- Chen, M.; Wu, J.; Wang, L.; Mantri, N.; Zhang, X.; Zhu, Z.; Wang, S. Mapping and genetic structure analysis of the anthracnose resistance locus Co-1HY in the common bean (Phaseolus vulgaris L.). PLoS ONE 2017, 12, e0169954. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.R.L.; Gonçalves-Vidigal, M.C.; Vaz Bisneta, M.; Valentini, G.; Vidigal Filho, P.S.; Martins, V.S.R.; Souza, T.L.P.O. Genetic fine-mapping of anthracnose disease-resistance allele Co-14 present in the Andean common bean cultivar AND 277. Crop Sci. 2023, 63, 750–763. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Pflieger, S.; Sevignac, M.; Thareau, V.; Blanchet, S.; Li, Y.; Jackson, S.A.; Jackson, S.A.; Geffroy, V. Fine mapping of Co-x, an anthracnose resistance gene to a highly virulent strain of Colletotrichum lindemuthianum in common bean. Theor. Appl. Genet. 2014, 127, 1653–1666. [Google Scholar] [CrossRef] [PubMed]
- Gilio, T.A.S.; Hurtado-Gonzales, O.P.; Gonçalves-Vidigal, M.C.; Valentini, G.; Elias, J.C.F.; Song, Q.; Pastor-Corrales, M.A. Fine mapping of an anthracnose-resistance locus in Andean common bean cultivar Amendoim Cavalo. PLoS ONE 2020, 15, e0239763. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Vidigal, M.C.; Gilio, T.A.S.; Valentini, G.; Vaz Bisneta, M.; Vidigal Filho, P.S.; Song, Q.; Oblessuc, P.R.; Melotto, M. New Andean source of resistance to anthracnose and angular leaf spot: Fine-mapping of disease-resistance genes in California Dark Red Kidney common bean cultivar. PLoS ONE 2020, 15, e0235215. [Google Scholar] [CrossRef] [PubMed]
- Oblessuc, P.R.; Baroni, R.M.; Garcia, A.A.F.; Chioratto, A.F.; Carbonell, S.A.M.; Camargo, L.E.A.; Benchimol, L.L. Mapping of angular leaf spot resistance QTL in common bean (Phaseolus vulgaris L.) under different environments. BMC Genet. 2012, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.; Manzanares, C.; Jara, C.; Lobaton, J.D.; Studer, B.; Raatz, B. Fine-mapping of a major QTL controlling angular leaf spot resistance in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2015, 128, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Sartorato, A.; Nietsche, S.; Barros, E.G.; Moreira, M.A. Inheritance of angular leaf spot resistance and RAPD markers linked to disease resistance gene in common beans. Ann. Rep. Bean Improv. Coop. 1999, 42, 21–22. [Google Scholar]
- Gonçalves-Vidigal, M.C.; Cruz, A.S.; Lacanallo, G.F.; Vidigal Filho, P.S.; Sousa, L.L.; Pacheco, C.M.N.A.; McClean, P.; Gepts, P.; Pastor-Corrales, M.A. Co-segregation analysis and mapping of the anthracnose Co-10 and angular leaf spot Phg-ON disease-resistance genes in the common bean cultivar Ouro Negro. Theor. Appl. Genet. 2013, 126, 2245–2255. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Perseguini, J.M.K.C.; Baroni, R.M.; Chiorato, A.F.; Carbonell, S.A.M.; Mondego, J.M.C.; Vidal, R.O.; Camargo, L.E.A.; Benchimol-Reis, L.L. Increasing the density of markers around a major QTL controlling resistance to angular leaf spot in common bean. Theor. Appl. Genet. 2013, 126, 2451–2465. [Google Scholar] [CrossRef]
- Nay, M.M.; Souza, T.L.P.O.; Raatz, B.; Mukankusi, C.M.; Gonçalves-Vidigal, M.C.; Abreu, A.F.B.; Melo, L.C.; Pastor-Corrales, M.A. A review of angular leaf spot resistance in common bean. Crop Sci. 2019, 59, 1376. [Google Scholar] [CrossRef]
- Mahiya-Farooq; Padder, B.A.; Bhat, N.N.; Shah, M.D.; Shikari, A.B.; Awale, H.E.; Kelly, J.D. Temporal expression of candidate genes at the Co-1 locus and their interaction with other defense related genes in common bean. Physiol. Mol. Plant Pathol. 2019, 108, 101424. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Gratias, A.; Diaz, J.C.A.; Thareau, V.; Pflieger, S.; Meziadi, C.; Blanchet, S.; Marande, W.; Bitocchi, E.; Papa, R.; et al. A common bean truncated CRINKLY4 kinase controls gene-for-gene resistance to the fungus Colletotrichum lindemuthianum. J. Exp. Bot. 2021, 72, 3569–3581. [Google Scholar] [CrossRef] [PubMed]
- McClean, P.E.; Myers, J.R.; Hammond, J.J. Coefficient of parentage and cluster analysis of North American dry bean cultivars. Crop Sci. 1993, 33, 190–197. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors, perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Vaz Bisneta, M.; Gonçalves-Vidigal, M.C. Integration of anthracnose resistance loci and RLK and NBS-LRR-encoding genes in the Phaseolus vulgaris L. genome. Crop Sci. 2020, 60, 2901–2918. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: A putative link of ABA and JA signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef]
- García-Andrade, J.; González, B.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Vera, P. The role of ABA in plant immunity is mediated through the PYR1 receptor. Int. J. Mol. Sci. 2020, 21, 5852. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.; Xavier, R.J. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kapos, P.; Zhang, Y. NLRs in plants. Curr. Opin. Immunol. 2015, 32, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Teng, Q.; Zhong, R.; Haghighat, M.; Richardson, E.A.; Ye, Z.-H. Mutations of Arabidopsis TBL32 and TBL33 affect Xylan acetylation and secondary wall deposition. PLoS ONE 2016, 11, e0146460. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Teng, Q.; Zhong, R.; Ye, Z.-H. The Arabidopsis DUF231 Domain-Containing Protein ESK1 Mediates 2-O- and 3-O-Acetylation of Xylosyl Residues in Xylan. Plant Cell Physiol. 2013, 54, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Padder, B.A.; Kamfwa, K.; Awale, H.E.; Kelly, J.D. Transcriptome profiling of the Phaseolus vulgaris—Colletotrichum lindemuthianum pathosystem. PLoS ONE 2016, 11, e0165823. [Google Scholar] [CrossRef] [PubMed]
- Shams, E.; Javan-Nikkhah, M.; Mirzadi Gohari, A. Dissecting molecular events and gene expression signatures involved in Colletotrichum lindemuthianum-Phaseolus vulgaris pathosystem in compatible and incompatible interactions. Eur. J. Plant Pathol. 2020, 156, 925–937. [Google Scholar] [CrossRef]
- Pastor-Corrales, M.A.; Otoya, M.M.; Molina, A.; Singh, S.P. Resistance to Colletotrichum lindemuthianum isolates from middle America and Andean South America in different common bean races. Plant Dis. 1995, 79, 63–67. [Google Scholar] [CrossRef]
- Mathur, R.S.; Barnett, H.l.; Lilly, V.G. Sporulation of Colletotrichum lindemuthianum in culture. Phytopathology 1950, 40, 104–114. [Google Scholar]
- Sanglard, D.A.; Balbi, B.P.; Barros, E.G.; Moreira, M.A. An efficient protocol for isolation, sporulation and maintenance of Pseudocercospora griseola. Ann. Rep. Bean Improv. Coop. 2009, 52, 62–63. [Google Scholar]
- Cárdenas, F.; Adams, M.W.; Andersen, A. The genetic system for reaction of field beans (Phaseolus vulgaris L.) to infection by three physiologic races of Colletotrichum lindemuthianum. Euphytica 1964, 13, 178–186. [Google Scholar] [CrossRef]
- de Almeida, C.P.; Arruda, N.; de Carvalho Paulino, J.F.; de Freitas, G.M.; Bonfante, G.F.J.; Bajay, M.M.; Deus, B.C.; Patrício, F.R.A.; Carbonell, S.A.M.; Chiorato, A.F.; et al. Genetic diversity of Pseudocercospora griseola resistance loci in common beans. Trop. Plant Pathol. 2021, 46, 129–138. [Google Scholar] [CrossRef]
- Castellanos, G.; Jara, C.; Mosquera, G. Bean Pathogens: Practical Guide for Lab and Greenhouse Work; Centro Internacional de Agricultura Tropical (CIAT): Cali, Colombia, 2015. [Google Scholar]
- Inglis, D.A.; Hagedorn, J.; Rand, R.E. Use of dry inoculum to evaluate beans for resistance to anthracnose and angular leaf spot. Plant Dis. 1988, 72, 771–774. [Google Scholar] [CrossRef]
- Borges, A.; Melotto, M.; Tsai, S.M.; Caldas, D.G.G. Changes in spatial and temporal gene expression during incompatible interaction between common bean and anthracnose pathogen. J. Plant Physiol. 2012, 169, 1216–1220. [Google Scholar] [CrossRef]
- Farrell, R.E. RNA Methodologies: Laboratory Guide for Isolation and Characterization, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1–855. [Google Scholar]
- Borges, A.; Tsai, S.M.; Caldas, D.G.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012, 31, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic. Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Bustin, S.; Huggett, J. qPCR primer design revisited. Biomol. Detect. Quantif. 2017, 14, 19–28. [Google Scholar] [CrossRef]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR; Meuer, S., Wittwer, C., Nakagawara, K.-I., Eds.; Springer: Heidelberg/Berlin, Germany, 2001; pp. 21–34. [Google Scholar] [CrossRef]
- Riedel, G.; Rüdrich, U.; Fekete-Drimusz, N.; Manns, M.P.; Vondran, F.W.R.; Bock, M. An extended ΔCT-method facilitating normalisation with multiple reference genes suited for quantitative RT-PCR analyses of human hepatocyte-like cells. PLoS ONE 2014, 9, e93031. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Dag, O.; Dolgun, A.; Konar, N.M. onewaytests: An R Package for One-Way Tests in Independent Groups Designs. R J. 2018, 10, 175–199. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistics Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef] [PubMed]

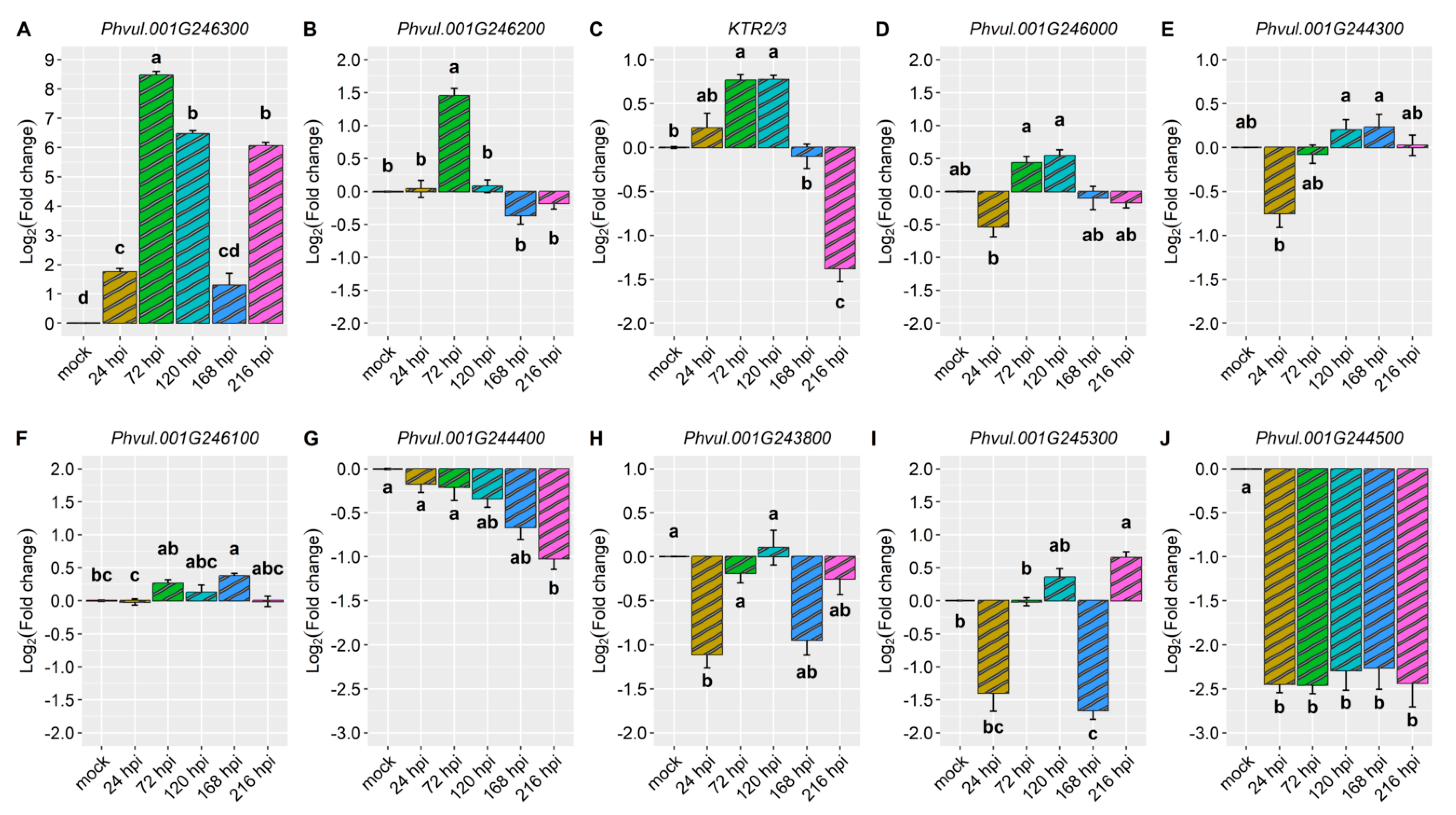
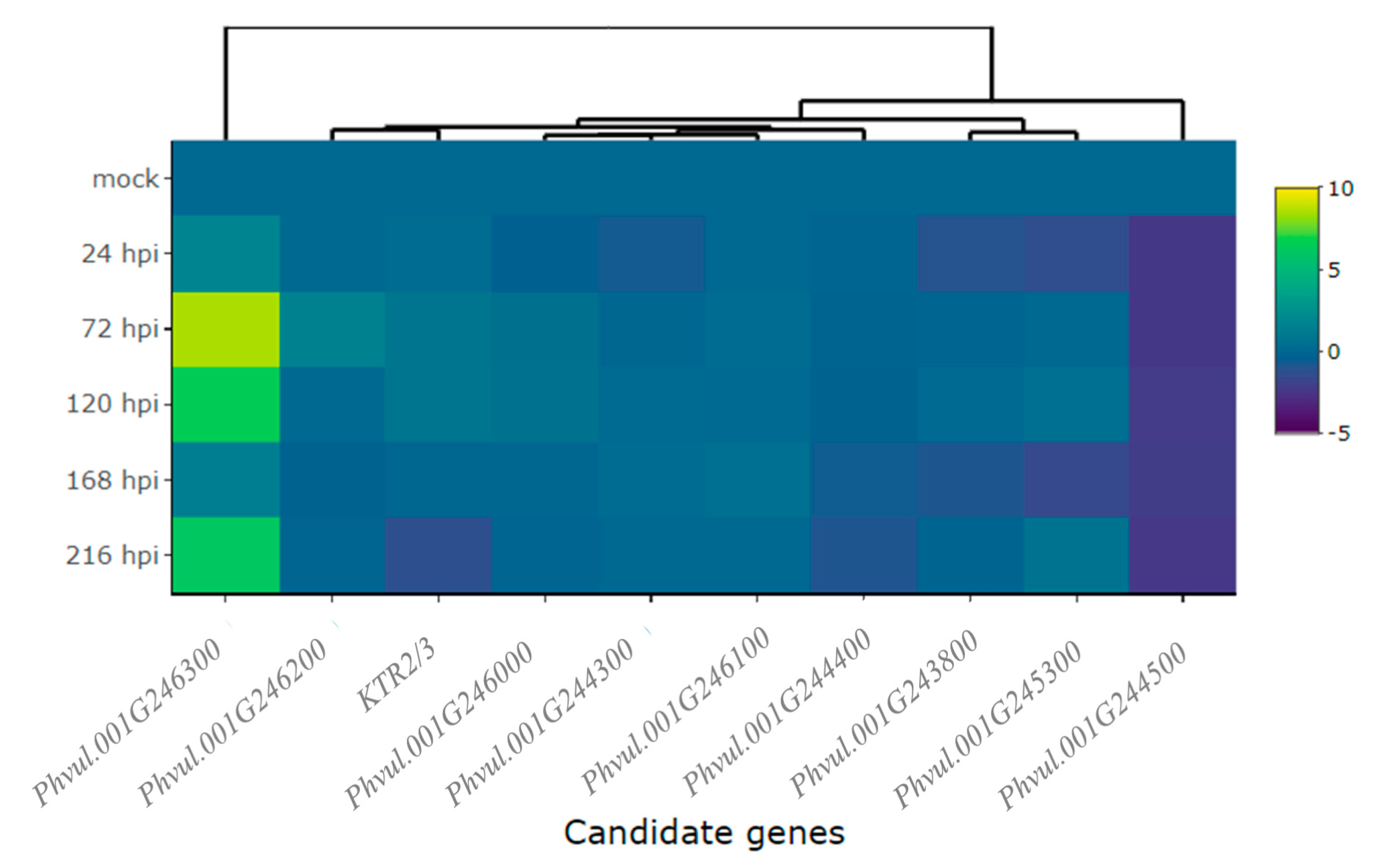
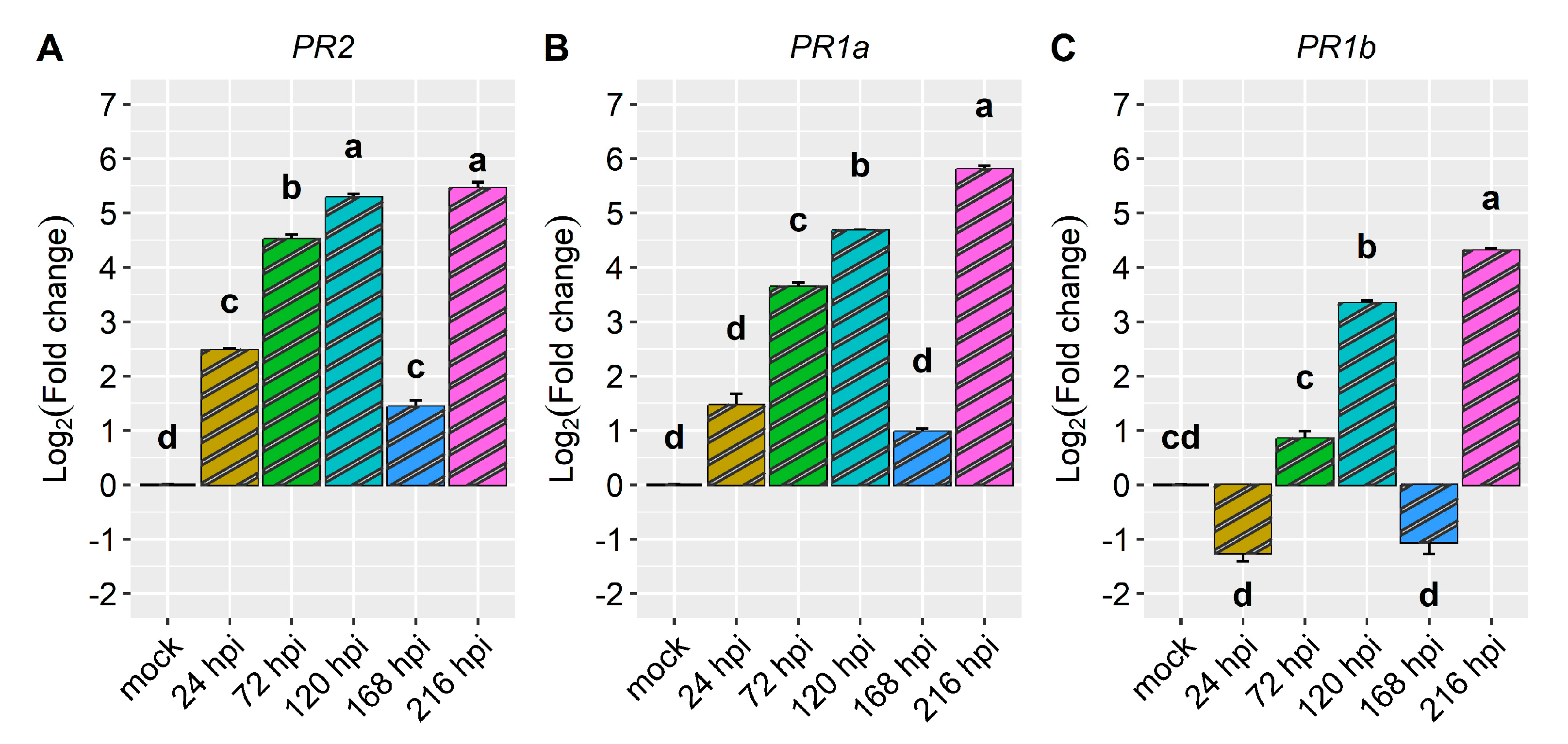

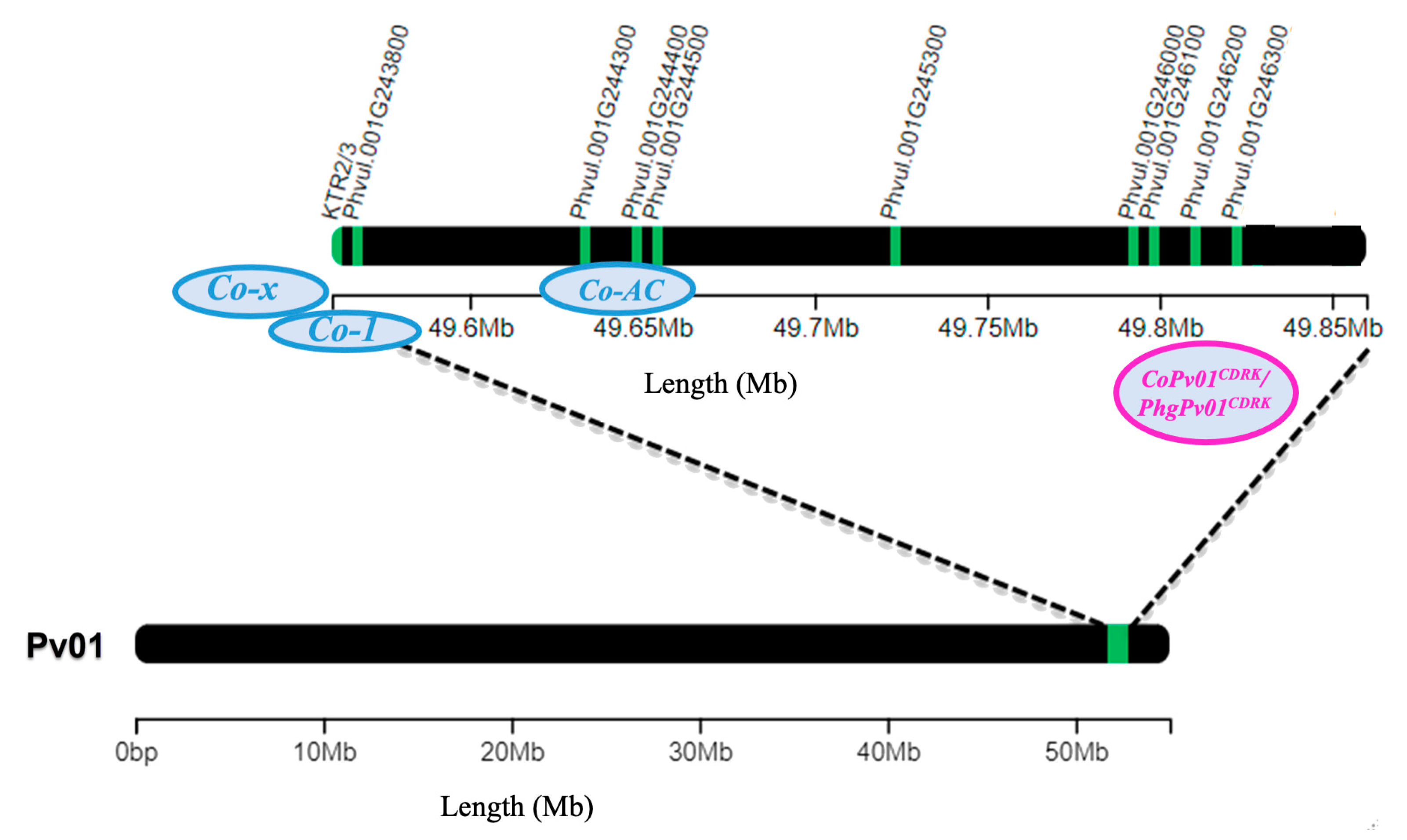
| Gene | Gene Model | C. lindemuthianum Race 73 (ANT) | P. griseola Race 63-39 (ALS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 hpi | 48 hpi | 72 hpi | 96 hpi | 120 hpi | 24 hpi | 72 hpi | 120 hpi | 168 hpi | 216 hpi | ||
| Co-x | KTR2/3 | 1.4 | 1.3 | 3.1 | 0.5 | 0.5 | 0.2 | 0.8 | 0.8 | −0.1 | −1.4 |
| Co-1 | Phvul.001G243800 | 1.4 | 0.9 | 2.0 | 1.8 | 0.6 | −1.1 | −0.2 | 0.1 | −0.9 | −0.3 |
| Co-AC | Phvul.001G244300 | −0.2 | −0.9 | −0.2 | −1.4 | −1.3 | −0.8 | −0.1 | 0.2 | 0.2 | 0.0 |
| Phvul.001G244400 | 0.0 | 0.0 | −0.7 | −0.4 | −0.2 | −0.2 | −0.2 | −0.3 | −0.7 | −1.0 | |
| Phvul.001G244500 | 0.4 | 0.3 | −0.4 | −0.5 | 0.8 | −2.4 | −2.5 | −2.3 | −2.3 | −2.4 | |
| CoPv01CDRK /PhgPv01CDRK | Phvul.001G245300 | 3.7 | 2.5 | 3.4 | 3.3 | 3.5 | −1.4 | 0.0 | 0.4 | −1.7 | 0.7 |
| Phvul.001G246000 | 0.9 | 0.8 | 0.8 | 0.7 | 0.3 | −0.5 | 0.4 | 0.5 | −0.1 | −0.2 | |
| Phvul.001G246100 | 0.6 | 1.5 | 1.2 | 0.7 | 1.3 | 0.0 | 0.3 | 0.1 | 0.4 | 0.0 | |
| Phvul.001G246200 | 2.7 | 2.6 | 2.6 | 0.5 | 1.6 | 0.0 | 1.5 | 0.1 | −0.4 | −0.2 | |
| Phvul.001G246300 | 7.2 | 7.3 | 7.8 | 7.4 | 6.3 | 1.7 | 8.5 | 6.5 | 1.3 | 6.1 | |
| Pathogenesis-related genes | PR1a | 6.6 | 6.2 | 8.0 | 8.1 | 6.3 | 1.5 | 3.6 | 4.7 | 1.0 | 5.8 |
| PR1b | 13.6 | 13.9 | 14.1 | 14.2 | 16.7 | −1.3 | 0.9 | 3.3 | −1.1 | 4.3 | |
| PR2 | 10.2 | 10.1 | 11.3 | 11.4 | 11.0 | 2.5 | 4.5 | 5.3 | 1.4 | 5.5 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lovatto, M.; Gonçalves-Vidigal, M.C.; Vaz Bisneta, M.; Calvi, A.C.; Mazucheli, J.; Vidigal Filho, P.S.; Miranda, E.G.R.; Melotto, M. Responsiveness of Candidate Genes on CoPv01CDRK/PhgPv01CDRK Loci in Common Bean Challenged by Anthracnose and Angular Leaf Spot Pathogens. Int. J. Mol. Sci. 2023, 24, 16023. https://doi.org/10.3390/ijms242216023
Lovatto M, Gonçalves-Vidigal MC, Vaz Bisneta M, Calvi AC, Mazucheli J, Vidigal Filho PS, Miranda EGR, Melotto M. Responsiveness of Candidate Genes on CoPv01CDRK/PhgPv01CDRK Loci in Common Bean Challenged by Anthracnose and Angular Leaf Spot Pathogens. International Journal of Molecular Sciences. 2023; 24(22):16023. https://doi.org/10.3390/ijms242216023
Chicago/Turabian StyleLovatto, Maike, Maria Celeste Gonçalves-Vidigal, Mariana Vaz Bisneta, Alexandre Catto Calvi, Josmar Mazucheli, Pedro Soares Vidigal Filho, Eduardo Gomes Rosa Miranda, and Maeli Melotto. 2023. "Responsiveness of Candidate Genes on CoPv01CDRK/PhgPv01CDRK Loci in Common Bean Challenged by Anthracnose and Angular Leaf Spot Pathogens" International Journal of Molecular Sciences 24, no. 22: 16023. https://doi.org/10.3390/ijms242216023
APA StyleLovatto, M., Gonçalves-Vidigal, M. C., Vaz Bisneta, M., Calvi, A. C., Mazucheli, J., Vidigal Filho, P. S., Miranda, E. G. R., & Melotto, M. (2023). Responsiveness of Candidate Genes on CoPv01CDRK/PhgPv01CDRK Loci in Common Bean Challenged by Anthracnose and Angular Leaf Spot Pathogens. International Journal of Molecular Sciences, 24(22), 16023. https://doi.org/10.3390/ijms242216023






