Microbiology and Biochemistry of Pesticides Biodegradation
Abstract
1. Introduction
2. Pesticides Used in Agriculture
2.1. Classification
2.2. Extremely Hazardous Pesticides
2.3. Highly Hazardous Pesticides
2.4. Moderately Hazardous Pesticides
2.5. Slightly Hazardous Pesticides
2.6. Unlikely to Present Acute Hazard Pesticides
3. Biodegradation of Pesticides
3.1. Microbial Diversity
3.1.1. Bacteria
3.1.2. Fungi
| WHO Pesticide Classification | Pesticide (% of Biodegradation Rate) | Fungi | Reference |
|---|---|---|---|
| Extremely Hazardous | Aldicarb (40–50%) Terbufos (50–100%) Methyl parathion (80–100%) | Ascochyta sp. CBS 237.37 Trametes versicolor Coriolus versicolor NBRC 9791 Bjerkandera adusta 8258 Pleurotus ostreatus 7989 Phanerochaete chrysosporium 3641 Fusarium sp. Yarrowia lipolytica Aspergillus niger AN400 Penicillium citrinum DL4M3 Penicillium citrinum DL9M3 Fusarium proliferatum DL11A Aspergillus sydowii Penicillium decaturense Aspergillus niger MRU01 Aspergillus niger NCIM 563 | [217,218,219,220,221,222,223,224,225,226] |
| Highly Hazardous | Cyfluthrin (10–70%) Carbofuran (96%) | Ascochyta sp. CBS 237.37 Trichoderma viride 2211 Aspergillus niger ZD11 Aspergillus nidulans var. dentatus Sepedonium maheswarium Trametes versicolor Mucor ramannianus Pichia anomala Trametes versicolor | [217,227,228,229,230,231,232,233] |
| Moderately Hazardous | DDT (50–100%) Lambda- Cyhalothrin (50–100%) Permethrin (90%) Chlorpyrifos (40–100%) Dimethoate (60–97%) 2,4-D (2–30%) | Ganoderma lingzhi Fomitopsis pinicola Gloeophyllum trabeum Cladosporium sp. Aspergillus sydowii Trichoderma sp. Cladosporium cladosporioides Hu-01 Rhodotorula glutinis Rhodotorula rubra Phanerochaete chrysosporium Trichoderma harzianum Trichoderma virens Byssochlamys spectabilis Aspergillus fumigates Aspergillus terreus TF1 Verticillium sp. Aspergillus sp. Trichoderma viride Trichoderma harzianum Aspergillus niger Aspergillus oryzae Penicillium citrinum Aspergillus fumigates Trametes versicolor Penicillium chrysogenum Aspergillus niger MRU01 Eurotim sp. F4 Emericella sp. F5 Trichosporon sp. Penicillium implicatum Aspergillus viridinutans | [96,120,137,216,225,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249] |
| Slightly Hazardous | Glyphosate (60–80%) Atrazine (70–100%) Metolachlor (30%) | Aspergillus flavus Aspergillus terricola Fusarium sp. Aspergillus niger Scopulariopsis sp. Trichoderma harzianum Fusarium oxysporum Penicillium notanum Aspergillus oryzae A-F02 Penicillium chrysogenum Fusarium dimerum Fusarium verticillioides Aspergillus fumigatus Penicillium citrinum Purpureocillium lilacinum Mucor spp. Sterilia spp. Trametes maxima Paecilomyces carneus Pleurotus ostreatus INCQS 40310 Trametes versicolor Bjerkandera adusta Pluteus cubensis SXS320 Gloelophyllum striatum MCA7 Agaricales MCA17 Polyporus sp. MCA128 Datronia stereoides MCA167 Datronia caperata MCA5 Metarhizium robertsii Trichoderma sp. Aspergillus section Flavi Pichia kudriavzevii Atz-EN-01 Penicillium sp. yz11-22N2 Saccharomyces cerevisiae Anthracophyllum discolor Glomus caledonium Aspergillus niger Candida xestobii Mortierella Kernia Chaetomium Trichosporon Candida tropicalis Penicillium oxalicum MET-F-1 | [165,167,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277] |
| Unlikely Hazardous | Trifuralin (80%) | Phanerochaete chrysosporium Trametes versicolor Penicillium simplicissimum Metacordyceps chlamydosporia Stachybotrys chartarum Alternia alternata | [211,278] |
3.1.3. Algae
| Algae | Degraded Pesticide | Reference |
|---|---|---|
| Chlorococcum humicola Gracilaria verrucosa | 2,4-D 1 | [283,284] |
| Chlorella vulgaris Scenedesmus bijugatus | Methyl Parathion 1 | [285] |
| Chlorococcum sp. | DDT 1 | [285] |
| Selenastrum capricornutum Synechococcus elongatus Chlorella vulgaris Chlorella sp. | Atrazine (60–80%) | [286,287,288] |
| Oscillatoria limnetica Skeletonema costatum Emiliania huxleyi Isochrysis galbana | Glyphosate 1 | [289,290] |
| Chlorella sp. Scenedesmus sp. | Chlorpyrifos 1 | [291] |
| Chlorella vulgaris | Carbofuran (100%) | [292] |
| Chlorella vulgaris | Dimethoate (100%) | [292] |
| Chlorella vulgaris | Metolachlor (100%) | [292] |
3.1.4. Actinomycetes
3.2. Metabolic Pathways
3.2.1. Extremely Hazardous Pesticides
Aldicarb
Terbufos
Methyl Parathion
3.2.2. Highly Hazardous Pesticides
Cyfluthrin
Tefluthrin
Carbofuran
3.2.3. Moderately Hazardous Pesticides
DDT
Lambda-Cyhalothrin
Permethrin
Chlorpyrifos
Dimethoate
2,4-Dichlorophenoxyacetic Acid
Dicamba
3.2.4. Slightly Hazardous Pesticides
Glyphosate
Atrazine
Metolachlor
3.3. Genetics of Pesticide Biodegradation
3.4. Application and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Uso de La Tierra En La Agricultura Según Las Cifras. Available online: https://www.fao.org/sustainability/news/detail/es/c/1279267/ (accessed on 27 June 2023).
- Clark, M.; Tilman, D. Comparative Analysis of Environmental Impacts of Agricultural Production Systems, Agricultural Input Efficiency, and Food Choice. Environ. Res. Lett. 2017, 12, 064016. [Google Scholar] [CrossRef]
- Zikankuba, V.L.; Mwanyika, G.; Ntwenya, J.E.; James, A. Pesticide Regulations and Their Malpractice Implications on Food and Environment Safety. Cogent Food Agric. 2019, 5, 1601544. [Google Scholar] [CrossRef]
- Clasificación Recomendada por la OMS de los Plaguicidas por el Peligro Que Presentan y Directrices para la Clasificación 2019. Available online: https://www.who.int/es/publications/i/item/9789240005662 (accessed on 27 June 2023).
- Carles, L.; Martin-Laurent, F.; Devers, M.; Spor, A.; Rouard, N.; Beguet, J.; Besse-Hoggan, P.; Batisson, I. Potential of Preventive Bioremediation to Reduce Environmental Contamination by Pesticides in an Agricultural Context: A Case Study with the Herbicide 2,4-D. J. Hazard. Mater. 2021, 416, 125740. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xiao, L.; Li, F.; Xiao, M.; Lin, D.; Long, X.; Wu, Z. Microbial Degradation of Pesticide Residues and an Emphasis on the Degradation of Cypermethrin and 3-Phenoxy Benzoic Acid: A Review. Molecules 2018, 23, 2313. [Google Scholar] [CrossRef] [PubMed]
- Briceño, G.; Fuentes, M.S.; Saez, J.M.; Diez, M.C.; Benimeli, C.S. Streptomyces Genus as Biotechnological Tool for Pesticide Degradation in Polluted Systems. Crit. Rev. Environ. Sci. Technol. 2018, 48, 773–805. [Google Scholar] [CrossRef]
- Inter-Organization Programme for the Sound Management of Chemicals; World Health Organization; Food and Agriculture Organization of the United Nations (Eds.) The International Code of Conduct on Pesticide Management; Inter-Organization Programme for the Sound Management of Chemicals: Paris, France; World Health Organization: Geneva, Switzerland; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014; ISBN 978-92-5-108548-6. [Google Scholar]
- Egbuna, C.; Sawicka, B. Natural Remedies for Pest, Disease and Weed Control; Academic Press: Amsterdam, The Netherlands, 2020; ISBN 978-0-12-819304-4. [Google Scholar]
- Raffa, C.M.; Chiampo, F. Bioremediation of Agricultural Soils Polluted with Pesticides: A Review. Bioengineering 2021, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization; Food and Agriculture Organization of the United Nations. Guidance on Pesticide Licensing Schemes: International Code of Conduct on Pesticide Management; Food and Agriculture Organization of the United Nations: Rome, Italy, 2021; ISBN 978-92-4-003042-8. [Google Scholar]
- Rice, N.C.; Rauscher, N.A.; Moffett, M.C.; Myers, T.M. Organoleptic Assessment and Median Lethal Dose Determination of Oral Aldicarb in Rats. Ann. N. Y. Acad. Sci. 2020, 1480, 136–145. [Google Scholar] [CrossRef]
- Zali Chedjeu, D.; Manfo Tsague, F.P.; Akono Nantia, E.; Zofou, D.; Nguedia Assob, J.C. Subchronic Toxicity of a Terbufos-Based Pesticide (Counter 15FC) in Adult Male Rats. J. Chem. Health Risks 2021, 11, 169–180. [Google Scholar] [CrossRef]
- Liu, P.; Song, X.; Yuan, W.; Wen, W.; Wu, X.; Li, J.; Chen, X. Effects of Cypermethrin and Methyl Parathion Mixtures on Hormone Levels and Immune Functions in Wistar Rats. Arch. Toxicol. 2006, 80, 449–457. [Google Scholar] [CrossRef]
- Mustapha, M.U.; Halimoon, N.; Johar, W.L.W.; Shukor, M.Y.A. An Overview on Biodegradation of Carbamate Pesticides by Soil Bacteria. Sci. Technol. 2019, 27, 547–563. [Google Scholar]
- Dixit, S.; Zia, M.K.; Siddiqui, T.; Ahsan, H.; Khan, F.H. Interaction of Human Alpha-2-Macroglobulin with Pesticide Aldicarb Using Spectroscopy and Molecular Docking. PPL 2021, 28, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hearon, S.E.; Johnson, N.M.; Phillips, T.D. Development of Broad-Acting Clays for the Tight Adsorption of Benzo[a]Pyrene and Aldicarb. Appl. Clay Sci. 2019, 168, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Tong, F.; Zhang, L.; Li, W.; Huang, W.; Zhou, Y. Fatal Poisoning by Terbufos Following Occupational Exposure. Clin. Toxicol. 2018, 56, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Pohanish, R.P.; Sittig, M. (Eds.) Sittig’s Handbook of Pesticides and Agricultural Chemicals, 2nd ed.; William Andrew Publishing: Norwich, NY, USA, 2015; ISBN 978-1-4557-3148-0. [Google Scholar]
- Tiwari, B.; Chakraborty, S.; Srivastava, A.K.; Mishra, A.K. Biodegradation and Rapid Removal of Methyl Parathion by the Paddy Field Cyanobacterium fischerella sp. Algal Res. 2017, 25, 285–296. [Google Scholar] [CrossRef]
- Mehta, M.; Mehta, B. Structural Correlation of Toxicological and Environmental Effects of Cypermethrin and Cyfluthrin-Type-II Pyrethroids. Int. J. Basic Appl. Sci. 2022, 11, 114–117. [Google Scholar] [CrossRef]
- Lewis, K.A.; Tzilivakis, J.; Warner, D.J.; Green, A. An International Database for Pesticide Risk Assessments and Management. Hum. Ecol. Risk Assess. Int. J. 2016, 22, 1050–1064. [Google Scholar] [CrossRef]
- Rodríguez, J.-L.; Ares, I.; Martínez, M.; Martínez-Larrañaga, M.-R.; Anadón, A.; Martínez, M.-A. Bioavailability and Nervous Tissue Distribution of Pyrethroid Insecticide Cyfluthrin in Rats. Food Chem. Toxicol. 2018, 118, 220–226. [Google Scholar] [CrossRef]
- Li, L.; Yang, D.; Song, Y.; Shi, Y.; Huang, B.; Bitsch, A.; Yan, J. The Potential Acute and Chronic Toxicity of Cyfluthrin on the Soil Model Organism, Eisenia fetida. Ecotoxicol. Environ. Saf. 2017, 144, 456–463. [Google Scholar] [CrossRef]
- Zhan, H.; Huang, Y.; Lin, Z.; Bhatt, P.; Chen, S. New Insights into the Microbial Degradation and Catalytic Mechanism of Synthetic Pyrethroids. Environ. Res. 2020, 182, 109138. [Google Scholar] [CrossRef]
- Wen, Y.; Wang, Z.; Gao, Y.; Zhao, X.; Gao, B.; Zhang, Z.; Li, L.; He, Z.; Wang, M. Novel Liquid Chromatography–Tandem Mass Spectrometry Method for Enantioseparation of Tefluthrin via a Box–Behnken Design and Its Stereoselective Degradation in Soil. J. Agric. Food Chem. 2019, 67, 11591–11597. [Google Scholar] [CrossRef]
- Yan, X.; Jin, W.; Wu, G.; Jiang, W.; Yang, Z.; Ji, J.; Qiu, J.; He, J.; Jiang, J.; Hong, Q. Hydrolase CehA and Monooxygenase CfdC Are Responsible for Carbofuran Degradation in Sphingomonas sp. Strain CDS-1. Appl. Environ. Microbiol. 2018, 84, e00805-18. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Gao, Q.; Zhang, L.; Wang, H.; Zhang, M.; Liu, X.; Zhou, Y.; Ke, Z.; Wu, C.; Qiu, J.; et al. Identification of the Key Amino Acid Sites of the Carbofuran Hydrolase CehA from a Newly Isolated Carbofuran-Degrading Strain Sphingobium sp. CFD-1. Ecotoxicol. Environ. Saf. 2020, 189, 109938. [Google Scholar] [CrossRef] [PubMed]
- Porto, M.F.; Milanez, B.; Soares, W.L.; Meyer, A. Double Standards and the International Trade of Pesticides: The Brazilian Case. Int. J. Occup. Environ. Health 2010, 16, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Çelik, A.; Mazmanci, B.; Çamlica, Y.; Aşkin, A.; Çömelekoǧlu, Ü. Cytogenetic Effects of Lambda-Cyhalothrin on Wistar Rat Bone Marrow. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2003, 539, 91–97. [Google Scholar] [CrossRef]
- Atashi, H.A.; Zaferani Arani, H.; Agatha, F.; Ghorani, S.M.; Teimouri Khorasani, M.S.; Moalem, M. Cardiac and Respiratory Arrest in a 12-year-old Girl with Acute Permethrin Oral Toxicity: A Case Report. Clin. Case Rep. 2022, 10, e05245. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Dani, V.; Dhawan, D.K. Protective Effects of Zinc on Lipid Peroxidation, Antioxidant Enzymes and Hepatic Histoarchitecture in Chlorpyrifos-Induced Toxicity. Chem.-Biol. Interact. 2005, 156, 131–140. [Google Scholar] [CrossRef]
- World Health Organization. Pesticide Residues in Food: 2021: Toxicological Evaluations: Joint Meeting of the FAO Panel of Experts on Pesticide Residues in Food and the Environment and the WHO Core Assessment Group on Pesticide Residues, Virtual Meeting, 6–17 September, 4 and 7 October 2021; World Health Organization: Geneva, Switzerland, 2022; ISBN 978-92-4-005462-2. [Google Scholar]
- Seiler, J.P. The Genetic Toxicology of Phenoxy Acids Other than 2,4,5-T. Mutat. Res. Rev. Genet. Toxicol. 1978, 55, 197–226. [Google Scholar] [CrossRef]
- Wolfgang Reuter Toxicology of Glyphosate, Isoxaflutole, Dicamba and Possible Combination Effects. Available online: https://www.testbiotech.org/content/toxicology-glyphosate-isoxaflutole-dicamba-and-possible-combination-effects (accessed on 27 June 2023).
- Walker, A.I.T.; Brown, V.K.H.; Kodama, J.K.; Thorpe, E.; Wilson, A.B. Toxicological Studies with the 1,3,5-Triazine Herbicide Cyanazine. Pestic. Sci. 1974, 5, 153–159. [Google Scholar] [CrossRef]
- Pan, X.; Lin, D.; Zheng, Y.; Zhang, Q.; Yin, Y.; Cai, L.; Fang, H.; Yu, Y. Biodegradation of DDT by Stenotrophomonas sp. DDT-1: Characterization and Genome Functional Analysis. Sci. Rep. 2016, 6, 21332. [Google Scholar] [CrossRef]
- Wong, M.H.; Leung, A.O.W.; Chan, J.K.Y.; Choi, M.P.K. A Review on the Usage of POP Pesticides in China, with Emphasis on DDT Loadings in Human Milk. Chemosphere 2005, 60, 740–752. [Google Scholar] [CrossRef]
- Matthews, G.A. A History of Pesticides; CABI: Wallingford, CT, USA; Boston, MA, USA, 2018; ISBN 978-1-78639-489-7. [Google Scholar]
- Schleier, J.J., III; Peterson, R.K.D. CHAPTER 3. Pyrethrins and Pyrethroid Insecticides. In Green Chemistry Series; Lopez, O., Fernandez-Bolanos, J., Eds.; Royal Society of Chemistry: Cambridge, UK, 2011; pp. 94–131. ISBN 978-1-84973-149-2. [Google Scholar]
- Shen, W.; Lou, B.; Xu, C.; Yang, G.; Yu, R.; Wang, X.; Li, X.; Wang, Q.; Wang, Y. Lethal Toxicity and Gene Expression Changes in Embryonic Zebrafish upon Exposure to Individual and Mixture of Malathion, Chlorpyrifos and Lambda-Cyhalothrin. Chemosphere 2020, 239, 124802. [Google Scholar] [CrossRef] [PubMed]
- Mundy, P.; Huff Hartz, K.; Fulton, C.; Lydy, M.; Brander, S.; Hung, T.; Fangue, N.; Connon, R. Exposure to Permethrin or Chlorpyrifos Causes Differential Dose- and Time-Dependent Behavioral Effects at Early Larval Stages of an Endangered Teleost Species. Endang. Species Res. 2021, 44, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.O.; El-Shamaa, I.S.; Abdel-Razik, N.I.; Elkomy, A.H.; Gewaily, M.S.; Abdo, S.E.; Soliman, A.A.; Paray, B.A.; Abdelkhalek, N. The Effect of Mannanoligosaccharide on the Growth Performance, Histopathology, and the Expression of Immune and Antioxidative Related Genes in Nile Tilapia Reared under Chlorpyrifos Ambient Toxicity. Fish Shellfish Immunol. 2020, 103, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei, S.; Hanachi, P.; Rafiee, G.; Seifori, P.; Walker, T.R. Toxicity of Polystyrene Microplastics on Juvenile Oncorhynchus mykiss (Rainbow Trout) after Individual and Combined Exposure with Chlorpyrifos. J. Hazard. Mater. 2021, 403, 123980. [Google Scholar] [CrossRef] [PubMed]
- Banazeer, A.; Afzal, M.B.S.; Shad, S.A. Characterization of Dimethoate Resistance in Oxycarenus hyalinipennis (Costa): Resistance Selection, Cross-Resistance to Three Insecticides and Mode of Inheritance. Phytoparasitica 2020, 48, 841–849. [Google Scholar] [CrossRef]
- Eken, A. Dimethoate Organophosphate Insecticide Toxicity and the Role of Oxidative Stress. In Toxicology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 59–68. ISBN 978-0-12-819092-0. [Google Scholar]
- Wu, G.; Ma, J.; Li, S.; Wang, S.; Jiang, B.; Luo, S.; Li, J.; Wang, X.; Guan, Y.; Chen, L. Cationic Metal-Organic Frameworks as an Efficient Adsorbent for the Removal of 2,4-Dichlorophenoxyacetic Acid from Aqueous Solutions. Environ. Res. 2020, 186, 109542. [Google Scholar] [CrossRef] [PubMed]
- Attademo, A.M.; Lajmanovich, R.C.; Peltzer, P.M.; Boccioni, A.P.C.; Martinuzzi, C.; Simonielo, F.; Repetti, M.R. Effects of the Emulsifiable Herbicide Dicamba on Amphibian Tadpoles: An Underestimated Toxicity Risk? Env. Sci. Pollut. Res. 2021, 28, 31962–31974. [Google Scholar] [CrossRef]
- Marouani, N.; Tebourbi, O.; Cherif, D.; Hallegue, D.; Yacoubi, M.T.; Sakly, M.; Benkhalifa, M.; Ben Rhouma, K. Effects of Oral Administration of 2,4-Dichlorophenoxyacetic Acid (2,4-D) on Reproductive Parameters in Male Wistar Rats. Environ. Sci. Pollut. Res. 2017, 24, 519–526. [Google Scholar] [CrossRef]
- Jin-Clark, Y.; Lydy, M.J.; Zhu, K.Y. Effects of Atrazine and Cyanazine on Chlorpyrifos Toxicity in Chironomus tentans (Diptera: Chironomidae). Environ. Toxicol. Chem. 2002, 21, 598–603. [Google Scholar] [CrossRef]
- Loux, M.M. Drinking Water Health Advisory: Pesticides: U.S. Environmental Protection Agency, Office of Drinking Water Health Advisories, Lewis Publishers, 121 South Main Street, P.O. Drawer 519, Chelsea, M. J. Environ. Qual. 1990, 19, 353. [Google Scholar] [CrossRef]
- Turkmen, R.; Dogan, I. Determination of Acute Oral Toxicity of Glyphosate Isopropylamine Salt in Rats. Env. Sci. Pollut. Res. 2020, 27, 19298–19303. [Google Scholar] [CrossRef]
- Suter Ii, G.W. Ecological Risk Assessment; CRC Press: Boca Raton, FL, USA, 2016; ISBN 978-0-429-19182-4. [Google Scholar]
- Nicol, É.; Genty, C.; Bouchonnet, S.; Bourcier, S. Structural Elucidation of Metolachlor Photoproducts by Liquid Chromatography/High-Resolution Tandem Mass Spectrometry: Elucidation of Metolachlor Photoproducts by LC/MS/MS. Rapid Commun. Mass Spectrom. 2015, 29, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, V.K.; Iyer, B.D. A New Spectrophotometric Method for the Determination of Glyphosate: Statistical Optimization and Application in Biodegradation Studies. Int. J. Environ. Sci. Technol. 2021, 18, 997–1008. [Google Scholar] [CrossRef]
- Richmond, M.E. Glyphosate: A Review of Its Global Use, Environmental Impact, and Potential Health Effects on Humans and Other Species. J. Environ. Stud. Sci. 2018, 8, 416–434. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Chauhan, A.; Datta, S.; Wani, A.B.; Singh, N.; Singh, J. Toxicity, Degradation and Analysis of the Herbicide Atrazine. Environ. Chem. Lett. 2018, 16, 211–237. [Google Scholar] [CrossRef]
- Liu, H.; Ye, W.; Zhan, X.; Liu, W. A Comparative Study of Rac- and S-Metolachlor Toxicity to Daphnia magna. Ecotoxicol. Environ. Saf. 2006, 63, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Perez, E.R.; Le Calvé, S.; Mirabel, P. Near-UV Molar Absorptivities of Alachlor, Mecroprop-p, Pendimethalin, Propanil and Trifluralin in Methanol. J. Photochem. Photobiol. A Chem. 2008, 193, 237–244. [Google Scholar] [CrossRef]
- Fernandes, T.C.C.; Mazzeo, D.E.C.; Marin-Morales, M.A. Origin of Nuclear and Chromosomal Alterations Derived from the Action of an Aneugenic Agent—Trifluralin Herbicide. Ecotoxicol. Environ. Saf. 2009, 72, 1680–1686. [Google Scholar] [CrossRef]
- Busi, R.; Goggin, D.E.; Onofri, A.; Boutsalis, P.; Preston, C.; Powles, S.B.; Beckie, H.J. Loss of Trifluralin Metabolic Resistance in Lolium rigidum. Plants Exposed to Prosulfocarb Recurrent Selection. Pest Manag. Sci. 2020, 76, 3926–3934. [Google Scholar] [CrossRef]
- Sviridov, A.V.; Shushkova, T.V.; Epiktetov, D.O.; Tarlachkov, S.V.; Ermakova, I.T.; Leontievsky, A.A. Biodegradation of Organophosphorus Pollutants by Soil Bacteria: Biochemical Aspects and Unsolved Problems. Appl. Biochem. Microbiol. 2021, 57, 836–844. [Google Scholar] [CrossRef]
- Matsumura, F. Degradation of Pesticides in the Environment by Microorganisms and Sunlight. In Biodegradation of Pesticides; Matsumura, F., Murti, C.R.K., Eds.; Springer: Boston, MA, USA, 1982; pp. 67–87. ISBN 978-1-4684-4090-4. [Google Scholar]
- Dar, M.A.; Baba, Z.A.; Kaushik, G. A Review on Phorate Persistence, Toxicity and Remediation by Bacterial Communities. Pedosphere 2022, 32, 171–183. [Google Scholar] [CrossRef]
- Maqbool, Z.; Hussain, S.; Imran, M.; Mahmood, F.; Shahzad, T.; Ahmed, Z.; Azeem, F.; Muzammil, S. Perspectives of Using Fungi as Bioresource for Bioremediation of Pesticides in the Environment: A Critical Review. Environ. Sci. Pollut. Res. 2016, 23, 16904–16925. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Ji, H.; Hou, X. Research Progress of Microbial Degradation of Organophosphorus Pesticides. Prog. Appl. Microbiol. 2017, 1, 29–35. [Google Scholar]
- Fareed, A.; Zaffar, H.; Rashid, A.; Maroof Shah, M.; Naqvi, T.A. Biodegradation of N-Methylated Carbamates by Free and Immobilized Cells of Newly Isolated Strain Enterobacter cloacae strain TA7. Bioremediat. J. 2017, 21, 119–127. [Google Scholar] [CrossRef]
- Naqvi, T.A.; Kanhar, N.A.; Shar, A.H.; Hussain, M.; Ahmed, S. Microcosm Studies for the Biodegradation of Carbaryl in Soil. Pak. J. Bot. 2011, 43, 1079–1084. [Google Scholar]
- Yang, C.; Xu, X.; Liu, Y.; Jiang, H.; Wu, Y.; Xu, P.; Liu, R. Simultaneous Hydrolysis of Carbaryl and Chlorpyrifos by Stenotrophomonas sp. Strain YC-1 with Surface-Displayed Carbaryl Hydrolase. Sci. Rep. 2017, 7, 13391. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Hernández, M.L.; Quintero-Ramírez, R.; Nava-Ocampo, A.A.; Bello-Ramírez, A.M. Study of the Mechanism of Flavobacterium sp. for Hydrolyzing Organophosphate Pesticides. Fundam. Clin. Pharmacol. 2003, 17, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hong, M.; Liu, D.; Li, Y.; Shou, P.; Yan, H.; Shi, G. Biodegradation of Methyl Parathion by Acinetobacter radioresistens USTB-04. J. Environ. Sci. 2007, 19, 1257–1260. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.-H.; Bai, W.-Q.; Zhong, Q.-Z.; Li, M.; He, F.-Q.; Li, B.-T. Isolation and Characterization of a Bacterial Strain of the Genus Ochrobactrum with Methyl Parathion Mineralizing Activity. J. Appl. Microbiol. 2006, 101, 986–994. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, C.; Yan, Y. Biodegradation of Methyl Parathion and P-Nitrophenol by a Newly Isolated Agrobacterium sp. Strain Yw12. Biodegradation 2012, 23, 107–116. [Google Scholar] [CrossRef]
- Pakala, S.B.; Gorla, P.; Pinjari, A.B.; Krovidi, R.K.; Baru, R.; Yanamandra, M.; Merrick, M.; Siddavattam, D. Biodegradation of Methyl Parathion and P-Nitrophenol: Evidence for the Presence of a p-Nitrophenol 2-Hydroxylase in a Gram-Negative Serratia sp. Strain DS001. Appl. Microbiol. Biotechnol. 2007, 73, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, N.; Birolli, W.G.; Meira, E.B.; Lucas, S.C.O.; De Matos, I.L.; Nitschke, M.; Romão, L.P.C.; Porto, A.L.M. Biotransformation and Biodegradation of Methyl Parathion by Brazilian Bacterial Strains Isolated from Mangrove Peat. Biocatal. Agric. Biotechnol. 2018, 13, 319–326. [Google Scholar] [CrossRef]
- Rong, X.; Zhao, G.; Fein, J.B.; Yu, Q.; Huang, Q. Role of Interfacial Reactions in Biodegradation: A Case Study in a Montmorillonite, Pseudomonas sp. Z1 and Methyl Parathion Ternary System. J. Hazard. Mater. 2019, 365, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Castrejón-Godínez, M.L.; Tovar-Sánchez, E.; Ortiz-Hernández, M.L.; Encarnación-Guevara, S.; Martínez-Batallar, Á.G.; Hernández-Ortiz, M.; Sánchez-Salinas, E.; Rodríguez, A.; Mussali-Galante, P. Proteomic Analysis of Burkholderia zhejiangensis CEIB S4–3 during the Methyl Parathion Degradation Process. Pestic. Biochem. Physiol. 2022, 187, 105197. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhan, H. Biodegradation of Synthetic Pyrethroid Insecticides. In Microbial Metabolism of Xenobiotic Compounds; Arora, P.K., Ed.; Microorganisms for Sustainability; Springer: Singapore, 2019; Volume 10, pp. 229–244. ISBN 9789811374616. [Google Scholar]
- Ortiz-Hernández, M.L.; Gama-Martínez, Y.; Fernández-López, M.; Castrejón-Godínez, M.L.; Encarnación, S.; Tovar-Sánchez, E.; Salazar, E.; Rodríguez, A.; Mussali-Galante, P. Transcriptomic Analysis of Burkholderia cenocepacia CEIB S5-2 during Methyl Parathion Degradation. Env. Sci. Pollut. Res. 2021, 28, 42414–42431. [Google Scholar] [CrossRef] [PubMed]
- Pino, N.; Peñuela, G. Simultaneous Degradation of the Pesticides Methyl Parathion and Chlorpyrifos by an Isolated Bacterial Consortium from a Contaminated Site. Int. Biodeterior. Biodegrad. 2011, 65, 827–831. [Google Scholar] [CrossRef]
- Fioravante, I.A.; Barbosa, F.A.R.; Augusti, R.; Magalhães, S.M.S. Removal of Methyl Parathion by Cyanobacteria Microcystis novacekii under Culture Conditions. J. Environ. Monit. 2010, 12, 1302. [Google Scholar] [CrossRef]
- Geed, S.R.; Samal, K.; Srivastava, H.; Kartheek, B. Study the Performance of Continuous Bioreactor for the Treatment of Wastewater Containing Methyl Parathion by Isolated Alcaligenes Species. J. Environ. Chem. Eng. 2019, 7, 103158. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Wan, J.; Sun, X.; Ma, W.; Ni, H. Molecular Cloning and Characterization of a Methyl Parathion Hydrolase from an Organophosphorus-Degrading Bacterium, Serratia marcescens MEW06. FEMS Microbiol. Lett. 2018, 365, fny279. [Google Scholar] [CrossRef]
- Jayasree, V.S.; Sobhana, K.S.; Poulose, P.; Babu, K.R.; Jasmine, S.; Ranjith, L.; Saravanan, R.; Jose Kingsly, H.; Sreenath, K.R.; Joshi, K.K.; et al. Biodegradation of the Pyrethroid Pesticide Cyfluthrin by the Halophilic Bacterium Photobacterium ganghwense Isolated from Coral Reef Ecosystem. Indian J. Fish. 2020, 67, 116–131. [Google Scholar] [CrossRef]
- Nguyen, T.P.O.; Helbling, D.E.; Bers, K.; Fida, T.T.; Wattiez, R.; Kohler, H.-P.E.; Springael, D.; De Mot, R. Genetic and Metabolic Analysis of the Carbofuran Catabolic Pathway in Novosphingobium sp. KN65.2. Appl. Microbiol. Biotechnol. 2014, 98, 8235–8252. [Google Scholar] [CrossRef] [PubMed]
- Saikia, N.; Das, S.K.; Patel, B.K.C.; Niwas, R.; Singh, A.; Gopal, M. Biodegradation of Beta-Cyfluthrin by Pseudomonas stutzeri Strain S1. Biodegradation 2005, 16, 581–589. [Google Scholar] [CrossRef]
- Hu, G.P.; Zhao, Y.; Song, F.Q.; Liu, B.; Vasseur, L.; Douglas, C.; You, M.S. Isolation, Identification and Cyfluthrin-Degrading Potential of a Novel Lysinibacillus sphaericus Strain, FLQ-11-1. Res. Microbiol. 2014, 165, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Dong, Y.H.; Chang, C.; Deng, Y.; Zhang, X.F.; Zhong, G.; Song, H.; Hu, M.; Zhang, L.-H. Characterization of a Novel Cyfluthrin-Degrading Bacterial Strain Brevibacterium aureum and Its Biochemical Degradation Pathway. Bioresour. Technol. 2013, 132, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.; Rathour, R.; Singh, R.; Thakur, I.S. Production and Characterization of Extracellular Polymeric Substances (EPS) Generated by a Carbofuran Degrading Strain Cupriavidus Sp. ISTL7. Bioresour. Technol. 2019, 282, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Duc, H.D. Enhancement of Carbofuran Degradation by Immobilized Bacillus sp. Strain DT1. Environ. Eng. Res. 2021, 27, 210158. [Google Scholar] [CrossRef]
- Park, H.; Seo, S.I.; Lim, J.-H.; Song, J.; Seo, J.-H.; Kim, P.I. Screening of Carbofuran-Degrading Bacteria Chryseobacterium sp. BSC2-3 and Unveiling the Change in Metabolome during Carbofuran Degradation. Metabolites 2022, 12, 219. [Google Scholar] [CrossRef] [PubMed]
- Laocharoen, S.; Plangklang, P.; Reungsang, A. Selection of Support Materials for Immobilization of Burkholderia cepacia PCL3 in Treatment of Carbofuran-Contaminated Water. Environ. Technol. 2013, 34, 2587–2597. [Google Scholar] [CrossRef]
- Yasmin, A.; Ambreen, S.; Shabir, S. Biotransformation of Dimethoate into Novel Metabolites by Bacterial Isolate Pseudomonas kilonensis MB490. J. Environ. Sci. Health Part B 2022, 57, 13–22. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, Y.; Zhang, B.; Yang, C.-H.; Zhang, X. Isolation and Characterization of a Chlorpyrifos and 3,5,6-Trichloro-2-Pyridinol Degrading Bacterium. FEMS Microbiol. Lett. 2005, 251, 67–73. [Google Scholar] [CrossRef]
- Aswathi, A.; Pandey, A.; Sukumaran, R.K. Rapid Degradation of the Organophosphate Pesticide—Chlorpyrifos by a Novel Strain of Pseudomonas nitroreducens AR-3. Bioresour. Technol. 2019, 292, 122025. [Google Scholar] [CrossRef] [PubMed]
- Purnomo, A.S.; Sariwati, A.; Kamei, I. Synergistic Interaction of a Consortium of the Brown-Rot Fungus Fomitopsis pinicola and the Bacterium Ralstonia pickettii for DDT Biodegradation. Heliyon 2020, 6, e04027. [Google Scholar] [CrossRef] [PubMed]
- Rizqi, H.D.; Purnomo, A.S.; Kamei, I. Interaction and Effects of Bacteria Addition on Dichlorodiphenyltrichloroethane Biodegradation by Daedalea dickinsii. Curr. Microbiol. 2021, 78, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Xu, T.; Xu, H.; Fang, H.; Yu, Y. Characterization and Genome Functional Analysis of the DDT-Degrading Bacterium Ochrobactrum sp. DDT-2. Sci. Total Environ. 2017, 592, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Zhu, L.; Xu, Q.; Wang, J.; Liu, W.; Jiang, J.; Meng, Y. Isolation and Degradation Ability of the DDT-Degrading Bacterial Strain KK. Env. Earth Sci. 2011, 62, 93–99. [Google Scholar] [CrossRef]
- Wang, X.; Oba, B.T.; Wang, H.; Luo, Q.; Liu, J.; Tang, L.; Yang, M.; Wu, H.; Sun, L. Degradation of DDT by a Novel Bacterium, Arthrobacter globiformis DC-1: Efficacy, Mechanism and Comparative Advantage. Water 2023, 15, 2723. [Google Scholar] [CrossRef]
- Grewal, J.; Bhattacharya, A.; Kumar, S.; Singh, D.K.; Khare, S.K. Biodegradation of 1,1,1-Trichloro-2,2- Bis (4-Chlorophenyl) Ethane (DDT) by Using Serratia marcescens NCIM 2919. J. Environ. Sci. Health Part B 2016, 51, 809–816. [Google Scholar] [CrossRef]
- Erdem, Z.; Cutright, T.J. Biotransformation of 1,1,1-Trichloro-2,2-Bis(p-Chlorophenyl) Ethane (4,4′-DDT) on a Sandy Loam Soil Using Aerobic Bacterium Corynebacterium sp. Env. Earth Sci. 2016, 75, 1267. [Google Scholar] [CrossRef]
- Suman, S. Tanuja Isolation and Characterization of a Bacterial Strain Enterobacter cloacae (Accession No. KX438060.1) Capable of Degrading DDTs Under Aerobic Conditions and Its Use in Bioremediation of Contaminated Soil. Microbiol. Insights 2021, 14, 117863612110242. [Google Scholar] [CrossRef]
- Chen, S.; Deng, Y.; Chang, C.; Lee, J.; Cheng, Y.; Cui, Z.; Zhou, J.; He, F.; Hu, M.; Zhang, L.-H. Pathway and Kinetics of Cyhalothrin Biodegradation by Bacillus thuringiensis Strain ZS-19. Sci. Rep. 2015, 5, 8784. [Google Scholar] [CrossRef]
- Ding, J.; Liu, Y.; Gao, Y.; Zhang, C.; Wang, Y.; Xu, B.; Yang, Y.; Wu, Q.; Huang, Z. Biodegradation of λ-Cyhalothrin through Cell Surface Display of Bacterial Carboxylesterase. Chemosphere 2022, 289, 133130. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, A.A.; Khalil, M.S.; Mohamed, M.S.M. Simultaneous Biodegradation of λ-Cyhalothrin Pesticide and Vicia faba Growth Promotion under Greenhouse Conditions. AMB Expr. 2022, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Long, X.; Zhang, S.; Qin, Q.; Gan, L.; Tian, Y. Screening Cyhalothrin Degradation Strains from Locust Epiphytic Bacteria and Studying Paracoccus acridae SCU-M53 Cyhalothrin Degradation Process. Env. Sci. Pollut. Res. 2018, 25, 11505–11515. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.; Liaqat, A.; Munir, A.; Ashraf, M.F.; Iqbal, S. Bioaugmentation of a Novel Bacterial Consortium in Cotton-Planted Soil for Degradation of Lambda-Cyhalothrin. Pedosphere 2023, in press. [Google Scholar] [CrossRef]
- Ramya, K.; Vasudevan, N. Biodegradation of Synthetic Pyrethroid Pesticides under Saline Conditions by a Novel Halotolerant Enterobacter ludwigii. DWT 2020, 173, 255–266. [Google Scholar] [CrossRef]
- Birolli, W.G.; Da Silva, B.F.; Rodrigues Filho, E. Biodegradation of the Pyrethroid Cypermethrin by Bacterial Consortia Collected from Orange Crops. Environ. Res. 2022, 215, 114388. [Google Scholar] [CrossRef] [PubMed]
- Birolli, W.G.; Dos Santos, A.; Pilau, E.; Rodrigues-Filho, E. New Role for a Commercially Available Bioinsecticide: Bacillus thuringiensis Berliner Biodegrades the Pyrethroid Cypermethrin. Environ. Sci. Technol. 2021, 55, 4792–4803. [Google Scholar] [CrossRef]
- Zhan, H.; Wang, H.; Liao, L.; Feng, Y.; Fan, X.; Zhang, L.; Chen, S. Kinetics and Novel Degradation Pathway of Permethrin in Acinetobacter baumannii ZH-14. Front. Microbiol. 2018, 9, 98. [Google Scholar] [CrossRef]
- Hadibarata, T.; Kristanti, R.A.; Bilal, M.; Yilmaz, M.; Sathishkumar, P. Biodegradation Mechanism of Chlorpyrifos by Halophilic Bacterium Hortaea sp. B15. Chemosphere 2023, 312, 137260. [Google Scholar] [CrossRef]
- Briceño, G.; Fuentes, M.S.; Palma, G.; Jorquera, M.A.; Amoroso, M.J.; Diez, M.C. Chlorpyrifos Biodegradation and 3,5,6-Trichloro-2-Pyridinol Production by Actinobacteria Isolated from Soil. Int. Biodeterior. Biodegrad. 2012, 73, 1–7. [Google Scholar] [CrossRef]
- Jha, S.K.; Chishti, Z.; Ahmad, Z.; Arshad, K.-R. Enterobacter sp. SWLC2 for Biodegradation of Chlorpyrifos in the Aqueous Medium: Modeling of the Process Using Artificial Neural Network Approaches. Comput. Electron. Agric. 2022, 193, 106680. [Google Scholar] [CrossRef]
- Islam, N.; Iyer, R. Functional Analysis of Chlorpyrifos Biodegradation in Agricultural Soils Augmented with a Three-Strain Bacterial Consortium. Water Air Soil Pollut. 2021, 232, 425. [Google Scholar] [CrossRef]
- Gaonkar, O.; Nambi, I.M.; Suresh Kumar, G. Biodegradation Kinetics of Dichlorvos and Chlorpyrifos by Enriched Bacterial Cultures from an Agricultural Soil. Bioremediat. J. 2019, 23, 259–276. [Google Scholar] [CrossRef]
- Farhan, M.; Ahmad, M.; Kanwal, A.; Butt, Z.A.; Khan, Q.F.; Raza, S.A.; Qayyum, H.; Wahid, A. Biodegradation of Chlorpyrifos Using Isolates from Contaminated Agricultural Soil, Its Kinetic Studies. Sci. Rep. 2021, 11, 10320. [Google Scholar] [CrossRef] [PubMed]
- Omeiri, M.; Khnayzer, R.; Yusef, H.; Tokajian, S.; Salloum, T. Biodegradation of Chlorpyrifos by Bacterial Strains Isolated from Lebanese Soil and Its Association with Plant Growth Improvement. Bioremediat. J. 2022, 1–20. [Google Scholar] [CrossRef]
- Kumar, G.; Lal, S.; Soni, S.K.; Maurya, S.K.; Shukla, P.K.; Chaudhary, P.; Bhattacherjee, A.K.; Garg, N. Mechanism and Kinetics of Chlorpyrifos Co-Metabolism by Using Environment Restoring Microbes Isolated from Rhizosphere of Horticultural Crops under Subtropics. Front. Microbiol. 2022, 13, 891870. [Google Scholar] [CrossRef] [PubMed]
- Tehri, N.; Khanna, S.; Vashishth, A. Biodegradation of Chlorpyrifos by Soil-Derived Aerobic Consortia and Bacterial Isolates. Appl. Biochem. Microbiol. 2023, 59, 138–144. [Google Scholar] [CrossRef]
- Shi, T.; Fang, L.; Qin, H.; Chen, Y.; Wu, X.; Hua, R. Rapid Biodegradation of the Organophosphorus Insecticide Chlorpyrifos by Cupriavidus nantongensis X1T. Int. J. Environ. Res. Public Health 2019, 16, 4593. [Google Scholar] [CrossRef]
- Lara-Moreno, A.; Morillo, E.; Merchán, F.; Madrid, F.; Villaverde, J. Chlorpyrifos Removal in an Artificially Contaminated Soil Using Novel Bacterial Strains and Cyclodextrin. Evaluation of Its Effectiveness by Ecotoxicity Studies. Agronomy 2022, 12, 1971. [Google Scholar] [CrossRef]
- Govarthanan, M.; Ameen, F.; Kamala-Kannan, S.; Selvankumar, T.; Almansob, A.; Alwakeel, S.S.; Kim, W. Rapid Biodegradation of Chlorpyrifos by Plant Growth-Promoting Psychrophilic Shewanella sp. BT05: An Eco-Friendly Approach to Clean up Pesticide-Contaminated Environment. Chemosphere 2020, 247, 125948. [Google Scholar] [CrossRef]
- Dubey, S.; Dhanya, M.S. Chlorpyrifos Degradation in Semi-Arid Soil by Pseudomonas fluorescens Strain CD5 Isolated from Manured Soil. Soil Sediment Contam. Int. J. 2023, 32, 460–477. [Google Scholar] [CrossRef]
- Levío-Raimán, M.; Bornhardt, C.; Diez, M.C. Biodegradation of Iprodione and Chlorpyrifos Using an Immobilized Bacterial Consortium in a Packed-Bed Bioreactor. Microorganisms 2023, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Conde-Avila, V.; Peña, C.; Pérez-Armendáriz, B.; Loera, O.; Martínez Valenzuela, C.; Leyva Morales, J.B.; Jesús Bastidas Bastidas, P.D.; Salgado-Lugo, H.; Ortega Martínez, L.D. Growth, Respiratory Activity and Chlorpyrifos Biodegradation in Cultures of Azotobacter vinelandii ATCC 12837. AMB Expr. 2021, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, N.P.; Ali, S.H.; Madathilkovilakathu, H.; Abdulhameed, S. Chlorpyrifos-Degrading Cyanobacterium—Coleofasciculus chthonoplastes Isolated from Paddy Field. Int. J. Environ. Stud. 2020, 77, 307–317. [Google Scholar] [CrossRef]
- Asamba, M.N.; Mugendi, E.N.; Oshule, P.S.; Essuman, S.; Chimbevo, L.M.; Atego, N.A. Molecular Characterization of Chlorpyrifos Degrading Bacteria Isolated from Contaminated Dairy Farm Soils in Nakuru County, Kenya. Heliyon 2022, 8, e09176. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Khan, M.A.; Sharma, R.; Malik, A.; Sharma, S. Potential of Formulated Dyadobacter jiangsuensis Strain 12851 for Enhanced Bioremediation of Chlorpyrifos Contaminated Soil. Ecotoxicol. Environ. Saf. 2021, 213, 112039. [Google Scholar] [CrossRef] [PubMed]
- Mali, H.; Shah, C.; Patel, D.H.; Trivedi, U.; Subramanian, R.B. Degradation Insight of Organophosphate Pesticide Chlorpyrifos through Novel Intermediate 2,6-Dihydroxypyridine by Arthrobacter sp. HM01. Bioresour. Bioprocess. 2022, 9, 31. [Google Scholar] [CrossRef]
- Khalid, S.; Hashmi, I. Biotreatment of Chlorpyrifos in a Bench Scale Bioreactor Using Psychrobacter alimentarius T14. Environ. Technol. 2016, 37, 316–325. [Google Scholar] [CrossRef]
- Santillan, J.Y.; Muzlera, A.; Molina, M.; Lewkowicz, E.S.; Iribarren, A.M. Microbial Degradation of Organophosphorus Pesticides Using Whole Cells and Enzyme Extracts. Biodegradation 2020, 31, 423–433. [Google Scholar] [CrossRef]
- Elzakey, E.M.; El-Sabbagh, S.M.; Eldeen, E.E.-S.N.; Adss, I.A.-A.; Nassar, A.M.K. Bioremediation of Chlorpyrifos Residues Using Some Indigenous Species of Bacteria and Fungi in Wastewater. Environ. Monit. Assess. 2023, 195, 779. [Google Scholar] [CrossRef]
- Li, X.; Yin, X.; Lian, B. The Degradation of Dimethoate and the Mineral Immobilizing Function for Cd2+ by Pseudomonas putida. Geomicrobiol. J. 2017, 34, 346–354. [Google Scholar] [CrossRef]
- Li, R.; Zheng, J.; Wang, R.; Song, Y.; Chen, Q.; Yang, X.; Li, S.; Jiang, J. Biochemical Degradation Pathway of Dimethoate by Paracoccus sp. Lgjj-3 Isolated from Treatment Wastewater. Int. Biodeterior. Biodegrad. 2010, 64, 51–57. [Google Scholar] [CrossRef]
- Derbalah, A.; Massoud, A.; El-Mehasseb, I.; Allah, M.S.; Ahmed, M.S.; Al-Brakati, A.; Elmahallawy, E.K. Microbial Detoxification of Dimethoate and Methomyl Residues in Aqueous Media. Water 2021, 13, 1117. [Google Scholar] [CrossRef]
- Xia, X.; Wu, W.; Chen, J.; Shan, H. The Gut Bacterium Serratia marcescens Mediates Detoxification of Organophosphate Pesticide in Riptortus pedestris by Microbial Degradation. J. Appl. Entomol. 2023, 147, 406–415. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, K.; Ni, H.; Zhuang, W.; Wang, H.; Zhu, J.; He, Q.; He, J. A Novel Amidohydrolase (DmhA) from Sphingomonas sp. That Can Hydrolyze the Organophosphorus Pesticide Dimethoate to Dimethoate Carboxylic Acid and Methylamine. Biotechnol. Lett. 2016, 38, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zeng, F.; Qiu, G.; Lu, X.; Liu, X.; Gao, H. Co-Metabolic Degradation of Dimethoate by Raoultella sp. X1. Biodegradation 2009, 20, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yang, F.; Yu, H.; Xie, Y.; Guo, Y.; Yao, W. Biodegradation of the Organophosphate Dimethoate by Lactobacillus plantarum during Milk Fermentation. Food Chem. 2021, 360, 130042. [Google Scholar] [CrossRef]
- Sandoval-Carrasco, C.A.; Ahuatzi-Chacón, D.; Galíndez-Mayer, J.; Ruiz-Ordaz, N.; Juárez-Ramírez, C.; Martínez-Jerónimo, F. Biodegradation of a Mixture of the Herbicides Ametryn, and 2,4-Dichlorophenoxyacetic Acid (2,4-D) in a Compartmentalized Biofilm Reactor. Bioresour. Technol. 2013, 145, 33–36. [Google Scholar] [CrossRef]
- Wu, X.; Wang, W.; Liu, J.; Pan, D.; Tu, X.; Lv, P.; Wang, Y.; Cao, H.; Wang, Y.; Hua, R. Rapid Biodegradation of the Herbicide 2,4-Dichlorophenoxyacetic Acid by Cupriavidus gilardii T-1. J. Agric. Food Chem. 2017, 65, 3711–3720. [Google Scholar] [CrossRef]
- Neetha, J.N.; Ujwal, P.; Kumar, K.G.; Chidananda, B.; Goveas, L.; Sandesh, K. Aerobic Biodegradation and Optimization of 2,4-Dichlorophenoxyacetic Acid by E. hormaechei Subsp. Xiangfangensis and Assessment of Biodegraded Metabolite Toxicity. Environ. Technol. Innov. 2021, 24, 102055. [Google Scholar] [CrossRef]
- Muhammad, J.B.; Shehu, D.; Usman, S.; Dankaka, S.M.; Gimba, M.Y.; Jagaba, A.H. Biodegradation Potential of 2,4 Dichlorophenoxyacetic Acid by Cupriavidus campinensis Isolated from Rice Farm Cultivated Soil. Case Stud. Chem. Environ. Eng. 2023, 8, 100434. [Google Scholar] [CrossRef]
- González, A.J.; Gallego, A.; Gemini, V.L.; Papalia, M.; Radice, M.; Gutkind, G.; Planes, E.; Korol, S.E. Degradation and Detoxification of the Herbicide 2,4-Dichlorophenoxyacetic Acid (2,4-D) by an Indigenous Delftia sp. Strain in Batch and Continuous Systems. Int. Biodeterior. Biodegrad. 2012, 66, 8–13. [Google Scholar] [CrossRef]
- Zabaloy, M.C.; Gómez, M.A. Isolation and Characterization of Indigenous 2,4-D Herbicide Degrading Bacteria from an Agricultural Soil in Proximity of Sauce Grande River, Argentina. Ann. Microbiol. 2014, 64, 969–974. [Google Scholar] [CrossRef]
- Vanitha, T.K.; Suresh, G.; Bhandi, M.M.; Mudiam, M.K.R.; Mohan, S.V. Microbial Degradation of Organochlorine Pesticide: 2,4-Dichlorophenoxyacetic Acid by Axenic and Mixed Consortium. Bioresour. Technol. 2023, 382, 129031. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhuang, L.; Zhou, S.; Yuan, Y.; Yuan, T.; Li, F. Humic Substance-mediated Reduction of Iron(III) Oxides and Degradation of 2,4- D by an Alkaliphilic Bacterium, Corynebacterium humireducens MFC-5. Microb. Biotechnol. 2013, 6, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Lin, R.; Shang, H.; Xu, Y.; Zhang, Z.; Wu, X.; Zong, F. Efficient Degradation of Phenoxyalkanoic Acid Herbicides by the Alkali-Tolerant Cupriavidus oxalaticus Strain X32. J. Agric. Food Chem. 2020, 68, 3786–3795. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.D. Anaerobic Degradation of 2,4-Dichlorophenoxyacetic Acid by Thauera sp. DKT. Biodegradation 2018, 29, 499–510. [Google Scholar] [CrossRef]
- Ghosh, S.; Purohit, H.J.; Qureshi, A. Managing Gene Expression in Pseudomonas simiae EGD-AQ6 for Chloroaromatic Compound Degradation. Arch. Microbiol. 2022, 204, 132. [Google Scholar] [CrossRef]
- Yao, L.; Jia, X.; Zhao, J.; Cao, Q.; Xie, X.; Yu, L.; He, J.; Tao, Q. Degradation of the Herbicide Dicamba by Two Sphingomonads via Different O-Demethylation Mechanisms. Int. Biodeterior. Biodegrad. 2015, 104, 324–332. [Google Scholar] [CrossRef]
- Chakraborty, S.; Behrens, M.; Herman, P.L.; Arendsen, A.F.; Hagen, W.R.; Carlson, D.L.; Wang, X.-Z.; Weeks, D.P. A Three-Component Dicamba O-Demethylase from Pseudomonas maltophilia, Strain DI-6: Purification and Characterization. Arch. Biochem. Biophys. 2005, 437, 20–28. [Google Scholar] [CrossRef]
- Yao, S.; Chen, L.; Yang, Z.; Yao, L.; Zhu, J.; Qiu, J.; Wang, G.; He, J. The Properties of 5-Methyltetrahydrofolate Dehydrogenase (MetF1) and Its Role in the Tetrahydrofolate-Dependent Dicamba Demethylation System in Rhizorhabdus dicambivorans Ndbn-20. J. Bacteriol. 2019, 201, 10–1128. [Google Scholar] [CrossRef]
- Ermakova, I.T.; Kiseleva, N.I.; Shushkova, T.; Zharikov, M.; Zharikov, G.A.; Leontievsky, A.A. Bioremediation of Glyphosate-Contaminated Soils. Appl. Microbiol. Biotechnol. 2010, 88, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Vo, V.T.; Nguyen, T.H.P.; Kiefer, R. Isolation and Optimization of a Glyphosate-Degrading Rhodococcus soli G41 for Bioremediation. Arch. Microbiol. 2022, 204, 252. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, I.T.; Shushkova, T.V.; Sviridov, A.V.; Zelenkova, N.F.; Vinokurova, N.G.; Baskunov, B.P.; Leontievsky, A.A. Organophosphonates Utilization by Soil Strains of Ochrobactrum anthropi and Achromobacter sp. Arch. Microbiol. 2017, 199, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Cortés, A.G.; Martinez-Ledezma, C.; López-Chuken, U.J.; Kaushik, G.; Nimesh, S.; Villarreal-Chiu, J.F. Polyphosphate Recovery by a Native Bacillus cereus Strain as a Direct Effect of Glyphosate Uptake. ISME J. 2019, 13, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Manogaran, M.; Shukor, M.Y.; Yasid, N.A.; Johari, W.L.W.; Ahmad, S.A. Isolation and Characterization of Glyphosate-Degrading Bacteria Isolated from Local Soils in Malaysia. Rend. Fis. Acc. Lincei 2017, 28, 471–479. [Google Scholar] [CrossRef]
- Kryuchkova, Y.V.; Burygin, G.L.; Gogoleva, N.E.; Gogolev, Y.V.; Chernyshova, M.P.; Makarov, O.E.; Fedorov, E.E.; Turkovskaya, O.V. Isolation and Characterization of a Glyphosate-Degrading Rhizosphere Strain, Enterobacter cloacae K7. Microbiol. Res. 2014, 169, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Hadi, F.; Mousavi, A.; Noghabi, K.A.; Tabar, H.G.; Salmanian, A.H. New Bacterial Strain of the Genus Ochrobactrum with Glyphosate-Degrading Activity. J. Environ. Sci. Health Part B 2013, 48, 208–213. [Google Scholar] [CrossRef]
- Zhao, H.; Tao, K.; Zhu, J.; Liu, S.; Gao, H.; Zhou, X. Bioremediation Potential of Glyphosate-Degrading Pseudomonas spp. Strains Isolated from Contaminated Soil. J. Gen. Appl. Microbiol. 2015, 61, 165–170. [Google Scholar] [CrossRef][Green Version]
- Rossi, F.; Carles, L.; Donnadieu, F.; Batisson, I.; Artigas, J. Glyphosate-Degrading Behavior of Five Bacterial Strains Isolated from Stream Biofilms. J. Hazard. Mater. 2021, 420, 126651. [Google Scholar] [CrossRef]
- Masotti, F.; Garavaglia, B.S.; Piazza, A.; Burdisso, P.; Altabe, S.; Gottig, N.; Ottado, J. Bacterial Isolates from Argentine Pampas and Their Ability to Degrade Glyphosate. Sci. Total Environ. 2021, 774, 145761. [Google Scholar] [CrossRef]
- Zhang, W.; Li, J.; Zhang, Y.; Wu, X.; Zhou, Z.; Huang, Y.; Zhao, Y.; Mishra, S.; Bhatt, P.; Chen, S. Characterization of a Novel Glyphosate-Degrading Bacterial Species, Chryseobacterium sp. Y16C, and Evaluation of Its Effects on Microbial Communities in Glyphosate-Contaminated Soil. J. Hazard. Mater. 2022, 432, 128689. [Google Scholar] [CrossRef] [PubMed]
- Lipok, J.; Wieczorek, D.; Jewgiński, M.; Kafarski, P. Prospects of in Vivo 31P NMR Method in Glyphosate Degradation Studies in Whole Cell System. Enzym. Microb. Technol. 2009, 44, 11–16. [Google Scholar] [CrossRef]
- Pérez Rodríguez, M.; Melo, C.; Jiménez, E.; Dussán, J. Glyphosate Bioremediation through the Sarcosine Oxidase Pathway Mediated by Lysinibacillus sphaericus in Soils Cultivated with Potatoes. Agriculture 2019, 9, 217. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.-J.; Zhang, W.; Zhang, Y.; Lei, Q.; Wu, S.; Huang, Y.; Mishra, S.; Bhatt, P.; Chen, S. Effects of Free or Immobilized Bacterium Stenotrophomonas acidaminiphila Y4B on Glyphosate Degradation Performance and Indigenous Microbial Community Structure. J. Agric. Food Chem. 2022, 70, 13945–13958. [Google Scholar] [CrossRef] [PubMed]
- Hernández Guijarro, K.; De Gerónimo, E.; Erijman, L. Glyphosate Biodegradation Potential in Soil Based on Glycine Oxidase Gene (thiO) from Bradyrhizobium. Curr. Microbiol. 2021, 78, 1991–2000. [Google Scholar] [CrossRef] [PubMed]
- Firdous, S.; Iqbal, S.; Anwar, S. Optimization and Modeling of Glyphosate Biodegradation by a Novel Comamonas odontotermitis P2 through Response Surface Methodology. Pedosphere 2020, 30, 618–627. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Singh, J. Kinetic Study of the Biodegradation of Glyphosate by Indigenous Soil Bacterial Isolates in Presence of Humic Acid, Fe(III) and Cu(II) Ions. J. Environ. Chem. Eng. 2019, 7, 103098. [Google Scholar] [CrossRef]
- Grube, M.; Kalnenieks, U.; Muter, O. Metabolic Response of Bacteria to Elevated Concentrations of Glyphosate-Based Herbicide. Ecotoxicol. Environ. Saf. 2019, 173, 373–380. [Google Scholar] [CrossRef]
- Elarabi, N.I.; Abdelhadi, A.A.; Ahmed, R.H.; Saleh, I.; Arif, I.A.; Osman, G.; Ahmed, D.S. Bacillus aryabhattai FACU: A Promising Bacterial Strain Capable of Manipulate the Glyphosate Herbicide Residues. Saudi J. Biol. Sci. 2020, 27, 2207–2214. [Google Scholar] [CrossRef]
- Kaczynski, P.; Lozowicka, B.; Wolejko, E.; Iwaniuk, P.; Konecki, R.; Dragowski, W.; Lozowicki, J.; Amanbek, N.; Rusilowska, J.; Pietraszko, A. Complex Study of Glyphosate and Metabolites Influence on Enzymatic Activity and Microorganisms Association in Soil Enriched with Pseudomonas fluorescens and Sewage Sludge. J. Hazard. Mater. 2020, 393, 122443. [Google Scholar] [CrossRef]
- Xu, B.; Sun, Q.-J.; Lan, J.C.-W.; Chen, W.-M.; Hsueh, C.-C.; Chen, B.-Y. Exploring the Glyphosate-Degrading Characteristics of a Newly Isolated, Highly Adapted Indigenous Bacterial Strain, Providencia rettgeri GDB 1. J. Biosci. Bioeng. 2019, 128, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Abo Serih, N.; Salim, R.; Fikry, A.; Hammad, M.; El-Sayed, G. In-Vitro Biodegradation of Glyphosate Using Genetically Improved Bacterial Isolates from highly Polluted Wastewater. Egypt. J. Chem. 2022, 65, 669–681. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Patil, A.; Ahmad, S.; Kothari, R.; Yadav, S. Optimization and Elucidation of Organophosphorus and Pyrethroid Degradation Pathways by a Novel Bacterial Consortium C3 Using RSM and GC-MS-Based Metabolomics. J. Taiwan Inst. Chem. Eng. 2023, 144, 104744. [Google Scholar] [CrossRef]
- Góngora-Echeverría, V.R.; García-Escalante, R.; Rojas-Herrera, R.; Giácoman-Vallejos, G.; Ponce-Caballero, C. Pesticide Bioremediation in Liquid Media Using a Microbial Consortium and Bacteria-Pure Strains Isolated from a Biomixture Used in Agricultural Areas. Ecotoxicol. Environ. Saf. 2020, 200, 110734. [Google Scholar] [CrossRef] [PubMed]
- Stosiek, N.; Terebieniec, A.; Ząbek, A.; Młynarz, P.; Cieśliński, H.; Klimek-Ochab, M. N-Phosphonomethylglycine Utilization by the Psychrotolerant Yeast Solicoccozyma terricola M 3.1.4. Bioorganic Chem. 2019, 93, 102866. [Google Scholar] [CrossRef] [PubMed]
- Esikova, T.Z.; Anokhina, T.O.; Suzina, N.E.; Shushkova, T.V.; Wu, Y.; Solyanikova, I.P. Characterization of a New Pseudomonas putida Strain Ch2, a Degrader of Toxic Anthropogenic Compounds Epsilon-Caprolactam and Glyphosate. Microorganisms 2023, 11, 650. [Google Scholar] [CrossRef]
- Firdous, S.; Iqbal, S.; Anwar, S.; Jabeen, H. Identification and Analysis of 5-Enolpyruvylshikimate-3-Phosphate Synthase (EPSPS) Gene from Glyphosate-Resistant Ochrobactrum intermedium Sq20: Identification of EPSPS from Ochrobactrum Intermedium Sq20. Pest. Manag. Sci 2018, 74, 1184–1196. [Google Scholar] [CrossRef]
- Shahid, M.; Khan, M.S. Glyphosate Induced Toxicity to Chickpea Plants and Stress Alleviation by Herbicide Tolerant Phosphate Solubilizing Burkholderia cepacia PSBB1 Carrying Multifarious Plant Growth Promoting Activities. 3 Biotech 2018, 8, 131. [Google Scholar] [CrossRef]
- Háhn, J.; Kriszt, B.; Tóth, G.; Jiang, D.; Fekete, M.; Szabó, I.; Göbölös, B.; Urbányi, B.; Szoboszlay, S.; Kaszab, E. Glyphosate and Glyphosate-Based Herbicides (GBHs) Induce Phenotypic Imipenem Resistance in Pseudomonas aeruginosa. Sci. Rep. 2022, 12, 18258. [Google Scholar] [CrossRef]
- Zhumakayev, A.R.; Vörös, M.; Szekeres, A.; Rakk, D.; Vágvölgyi, C.; Szűcs, A.; Kredics, L.; Škrbić, B.D.; Hatvani, L. Comprehensive Characterization of Stress Tolerant Bacteria with Plant Growth-Promoting Potential Isolated from Glyphosate-Treated Environment. World J. Microbiol. Biotechnol. 2021, 37, 94. [Google Scholar] [CrossRef] [PubMed]
- Hertel, R.; Schöne, K.; Mittelstädt, C.; Meißner, J.; Zschoche, N.; Collignon, M.; Kohler, C.; Friedrich, I.; Schneider, D.; Hoppert, M.; et al. Characterization of Glyphosate-resistant Burkholderia anthina and Burkholderia cenocepacia Isolates from a Commercial Roundup® Solution. Env. Microbiol. Rep. 2022, 14, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Obojska, A.; Ternan, N.G.; Lejczak, B.; Kafarski, P.; McMullan, G. Organophosphonate Utilization by the Thermophile Geobacillus caldoxylosilyticus T20. Appl. Env. Microbiol. 2002, 68, 2081–2084. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, L.; Jin, C.; Meng, Z.; Yang, N. Study on the Isolation of Two Atrazine-Degrading Bacteria and the Development of a Microbial Agent. Microorganisms 2019, 7, 80. [Google Scholar] [CrossRef] [PubMed]
- Topp, E. A Comparison of Three Atrazine-Degrading Bacteria for Soil Bioremediation. Biol. Fertil Soils 2001, 33, 529–534. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.; Zhang, K.; Wang, X.; Ma, C.; Tang, H.; Xu, P. Atrazine Degradation by a Simple Consortium of Klebsiella sp. A1 and Comamonas sp. A2 in Nitrogen Enriched Medium. Biodegradation 2010, 21, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Wu, Y.; Zhai, Q.; Tang, Y.; Liu, X.; Xu, X.; Liang, S.; Zhang, H. Immobilization of Bacterial Mixture of Klebsiella variicola FH-1 and Arthrobacter sp. NJ-1 Enhances the Bioremediation of Atrazine-Polluted Soil Environments. Front. Microbiol. 2023, 14, 1056264. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Hu, S.; Han, S.; Shi, H.; Yang, Y.; Li, H.; Jiao, Y.; Zhang, Q.; Akindolie, M.S.; Ji, M.; et al. Efficient Removal of Atrazine by Iron-Modified Biochar Loaded Acinetobacter lwoffii DNS32. Sci. Total Environ. 2019, 682, 59–69. [Google Scholar] [CrossRef]
- Qu, M.; Li, N.; Li, H.; Yang, T.; Liu, W.; Yan, Y.; Feng, X.; Zhu, D. Phytoextraction and Biodegradation of Atrazine by Myriophyllum spicatum and Evaluation of Bacterial Communities Involved in Atrazine Degradation in Lake Sediment. Chemosphere 2018, 209, 439–448. [Google Scholar] [CrossRef]
- Ma, L.; Chen, S.; Yuan, J.; Yang, P.; Liu, Y.; Stewart, K. Rapid Biodegradation of Atrazine by Ensifer sp. Strain and Its Degradation Genes. Int. Biodeterior. Biodegrad. 2017, 116, 133–140. [Google Scholar] [CrossRef]
- Gali, S.B.; Sufyan, A.J.; Babandi, A.; Ibrahim, S.; Shehu, D.; Ya’u, M.; Mashi, J.A.; Babagana, K.; Abdullahi, N.; Ibrahim, A.; et al. Characterizing Atrazine Degradation by Molybdenum-Reducing Pseudomonas sp. J. Biochem. Microbiol. Biotechnol. 2023, 11, 11–16. [Google Scholar] [CrossRef]
- Liang, Y.; Ding, L.; Song, Q.; Zhao, B.; Wang, S.; Liu, S. Biodegradation of Atrazine by Three Strains: Identification, Enzymes Activities, and Biodegradation Mechanism. Environ. Pollut. Bioavailab. 2022, 34, 549–563. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Wong, M.; Geng, B. Atrazine Biodegradation in Water by Co-Immobilized Citricoccus sp. Strain TT3 with Chlorella vulgaris under a Harsh Environment. Algal Res. 2023, 70, 102994. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, X.; Wang, Z.; Cao, B.; Deng, S.; Bi, M.; Zhang, Y. Enhanced Biodegradation of Atrazine by Arthrobacter sp. DNS10 during Co-Culture with a Phosphorus Solubilizing Bacteria: Enterobacter sp. P1. Ecotoxicol. Environ. Saf. 2019, 172, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Sun, S.; Rong, Y.; Mao, L.; Yang, S.; Qian, L.; Li, R.; Zheng, Y. Enhanced Phytoremediation of Atrazine-Contaminated Soil by Vetiver (Chrysopogon zizanioides L.) and Associated Bacteria. Environ. Sci. Pollut. Res. 2023, 30, 44415–44429. [Google Scholar] [CrossRef] [PubMed]
- Madariaga-Navarrete, A.; Rodríguez-Pastrana, B.R.; Villagómez-Ibarra, J.R.; Acevedo-Sandoval, O.A.; Perry, G.; Islas-Pelcastre, M. Bioremediation Model for Atrazine Contaminated Agricultural Soils Using Phytoremediation (Using Phaseolus vulgaris L.) and a Locally Adapted Microbial Consortium. J. Environ. Sci. Health Part B 2017, 52, 367–375. [Google Scholar] [CrossRef] [PubMed]
- El-Bestawy, E.; Sabir, J.; Mansy, A.H.; Zabermawi, N. Isolation, Identification and Acclimatization of Atrazine-Resistant Soil Bacteria. Ann. Agric. Sci. 2013, 58, 119–130. [Google Scholar] [CrossRef]
- James, A.; Singh, D.K. Atrazine Detoxification by Intracellular Crude Enzyme Extracts Derived from Epiphytic Root Bacteria Associated with Emergent Hydrophytes. J. Environ. Sci. Health Part B 2021, 56, 577–586. [Google Scholar] [CrossRef]
- Fang, H.; Lian, J.; Wang, H.; Cai, L.; Yu, Y. Exploring Bacterial Community Structure and Function Associated with Atrazine Biodegradation in Repeatedly Treated Soils. J. Hazard. Mater. 2015, 286, 457–465. [Google Scholar] [CrossRef]
- Zhu, J.; Fu, L.; Meng, Z.; Jin, C. Characteristics of an Atrazine Degrading Bacterium and the Construction of a Microbial Agent for Effective Atrazine Degradation. Water Environ. J. 2021, 35, 7–17. [Google Scholar] [CrossRef]
- Billet, L.; Devers-Lamrani, M.; Serre, R.-F.; Julia, E.; Vandecasteele, C.; Rouard, N.; Martin-Laurent, F.; Spor, A. Complete Genome Sequences of Four Atrazine-Degrading Bacterial Strains, Pseudomonas sp. Strain ADPe, Arthrobacter sp. Strain TES, Variovorax sp. Strain 38R, and Chelatobacter sp. Strain SR38. Microbiol. Resour. Announc. 2021, 10, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Carles, L.; Joly, M.; Bonnemoy, F.; Leremboure, M.; Donnadieu, F.; Batisson, I.; Besse-Hoggan, P. Biodegradation and Toxicity of a Maize Herbicide Mixture: Mesotrione, Nicosulfuron and S-Metolachlor. J. Hazard. Mater. 2018, 354, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Zhang, X.; Zhao, X.; Chen, X.; Li, Y. The Metolachlor Degradation Kinetics and Bacterial Community Evolution in the Soil Bioelectrochemical Remediation. Chemosphere 2020, 248, 125915. [Google Scholar] [CrossRef] [PubMed]
- Kanissery, R.G.; Welsh, A.; Gomez, A.; Connor, L.; Sims, G.K. Identification of Metolachlor Mineralizing Bacteria in Aerobic and Anaerobic Soils Using DNA-Stable Isotope Probing. Biodegradation 2018, 29, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Bellinaso, M.D.L.; Greer, C.W.; Peralba, M.D.C.; Henriques, J.A.P.; Gaylarde, C.C. Biodegradation of the Herbicide Trifluralin by Bacteria Isolated from Soil. FEMS Microbiol. Ecol. 2003, 43, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Zablotowicz, R.M.; Locke, M.A.; Hoagland, R.E.; Knight, S.S.; Cash, B. Fluorescent Pseudomonas Isolates from Mississippi Delta Oxbow Lakes: In Vitro Herbicide Biotransformations. Environ. Toxicol. 2001, 16, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Erguven, G.O.; Bayhan, H.; Ikizoglu, B.; Kanat, G.; Nuhoglu, Y. The Capacity of Some Newly Bacteria and Fungi for Biodegradation of Herbicide Trifluralin under Agitated Culture Media. Cell. Mol. Biol. 2016, 62, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Bellinaso, M.D.L.; Henriques, J.A.; Gaylarde, C.C.; Greer, C.W. Genes Similar to Naphthalene Dioxygenase Genes in Trifluralin-Degrading Bacteria. Pest. Manag. Sci. 2004, 60, 474–478. [Google Scholar] [CrossRef]
- Mata-Sandoval, J.C.; Karns, J.; Torrents, A. Influence of Rhamnolipids and Triton X-100 on the Biodegradation of Three Pesticides in Aqueous Phase and Soil Slurries. J. Agric. Food Chem. 2001, 49, 3296–3303. [Google Scholar] [CrossRef]
- Faramarzi, M.; Avarseji, Z.; Gholamalipuor Alamdari, E.; Taliei, F. Biodegradation of the Trifluralin Herbicide by Pseudomonas fluorescens. Int. J. Environ. Sci. Technol. 2023, 20, 3591–3598. [Google Scholar] [CrossRef]
- Gouma, S.; Fragoeiro, S.; Bastos, A.C.; Magan, N. Bacterial and Fungal Bioremediation Strategies. In Microbial Biodegradation and Bioremediation; Elsevier: Amsterdam, The Netherlands, 2014; pp. 301–323. ISBN 978-0-12-800021-2. [Google Scholar]
- Ellouze, M.; Sayadi, S. White-Rot Fungi and Their Enzymes as a Biotechnological Tool for Xenobiotic Bioremediation. In Management of Hazardous Wastes; Saleh, H.E.-D.M., Abdel Rahman, R.O., Eds.; InTech: London, UK, 2016; ISBN 978-953-51-2616-4. [Google Scholar]
- Kaur, P.; Balomajumder, C. Simultaneous Biodegradation of Mixture of Carbamates by Newly Isolated Ascochyta sp. CBS 237.37. Ecotoxicol. Environ. Saf. 2019, 169, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rodríguez, C.E.; Madrigal-León, K.; Masís-Mora, M.; Pérez-Villanueva, M.; Chin-Pampillo, J.S. Removal of Carbamates and Detoxification Potential in a Biomixture: Fungal Bioaugmentation versus Traditional Use. Ecotoxicol. Environ. Saf. 2017, 135, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Jauregui, J.; Valderrama, B.; Albores, A.; Vazquez-Duhalt, R. Microsomal Transformation of Organophosphorus Pesticides by White Rot Fungi. Biodegradation 2003, 14, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Usharani, K.; Muthukumar, M. Optimization of Aqueous Methylparathion Biodegradation by Fusarium sp in Batch Scale Process Using Response Surface Methodology. Int. J. Environ. Sci. Technol. 2013, 10, 591–606. [Google Scholar] [CrossRef][Green Version]
- Wang, X.-X.; Chi, Z.; Ru, S.-G.; Chi, Z.-M. Genetic Surface-Display of Methyl Parathion Hydrolase on Yarrowia lipolytica for Removal of Methyl Parathion in Water. Biodegradation 2012, 23, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Marinho, G.; Rodrigues, K.; Araujo, R.; Pinheiro, Z.B.; Silva, G.M.M. Glucose Effect on Degradation Kinetics of Methyl Parathion by Filamentous Fungi Species Aspergillus niger AN400. Eng. Sanit. Ambient. 2011, 16, 225–230. [Google Scholar] [CrossRef]
- Rodrigues, G.N.; Alvarenga, N.; Vacondio, B.; de Vasconcellos, S.P.; Passarini, M.R.Z.; Seleghim, M.H.R.; Porto, A.L.M. Biotransformation of Methyl Parathion by Marine-Derived Fungi Isolated from Ascidian Didemnum ligulum. Biocatal. Agric. Biotechnol. 2016, 7, 24–30. [Google Scholar] [CrossRef]
- Alvarenga, N.; Birolli, W.G.; Seleghim, M.H.R.; Porto, A.L.M. Biodegradation of Methyl Parathion by Whole Cells of Marine-Derived Fungi Aspergillus sydowii and Penicillium decaturense. Chemosphere 2014, 117, 47–52. [Google Scholar] [CrossRef]
- Mohapatra, D.; Rath, S.K.; Mohapatra, P.K. Accelerated Degradation of Four Organophosphorus Insecticides by Malathion Tolerant Aspergillus niger MRU01 a Soil Fungus. Geomicrobiol. J. 2023, 1–10. [Google Scholar] [CrossRef]
- Shah, P.C.; Kumar, V.R.; Dastager, S.G.; Khire, J.M. Phytase Production by Aspergillus niger NCIM 563 for a Novel Application to Degrade Organophosphorus Pesticides. AMB Express 2017, 7, 66. [Google Scholar] [CrossRef]
- Saikia, N.; Gopal, M. Biodegradation of β-Cyfluthrin by Fungi. J. Agric. Food Chem. 2004, 52, 1220–1223. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Q.; Wang, Z.Y.; Li, H.; Wu, P.C.; Hu, J.M.; Luo, N.; Cao, L.X.; Liu, Y.H. Purification and Characterization of a Novel Pyrethroid Hydrolase from Aspergillus niger ZD11. J. Agric. Food Chem. 2005, 53, 7415–7420. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, I.; Mittal, A. Dissipation of β-Cyfluthrin by Two Fungi Aspergillus nidulans Var. dentatus and Sepedonium maheswarium. Toxicol. Environ. Chem. 2007, 89, 319–326. [Google Scholar] [CrossRef]
- Mir-Tutusaus, J.A.; Masís-Mora, M.; Corcellas, C.; Eljarrat, E.; Barceló, D.; Sarrà, M.; Caminal, G.; Vicent, T.; Rodríguez-Rodríguez, C.E. Degradation of Selected Agrochemicals by the White Rot Fungus Trametes versicolor. Sci. Total Environ. 2014, 500–501, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Jeon, J.; Kim, S.-D.; Kang, S.; Han, J.; Hur, H.-G. Fungal Biodegradation of Carbofuran and Carbofuran Phenol by the Fungus Mucor ramannianus: Identification of Metabolites. Water Sci. Technol. 2007, 55, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, S.; Hu, M.; Hao, W.; Geng, P.; Zhang, Y. Biodegradation of Carbofuran by Pichia anomala Strain HQ-C-01 and Its Application for Bioremediation of Contaminated Soils. Biol. Fertil. Soils 2011, 47, 917–923. [Google Scholar] [CrossRef]
- Madrigal-Zúñiga, K.; Ruiz-Hidalgo, K.; Chin-Pampillo, J.S.; Masís-Mora, M.; Castro-Gutiérrez, V.; Rodríguez-Rodríguez, C.E. Fungal Bioaugmentation of Two Rice Husk-Based Biomixtures for the Removal of Carbofuran in on-Farm Biopurification Systems. Biol. Fertil. Soils 2016, 52, 243–250. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, A.; Chaudhary, P.; Gangola, S. Chlorpyrifos Degradation Using Binary Fungal Strains Isolated from Industrial Waste Soil. Biologia 2021, 76, 3071–3080. [Google Scholar] [CrossRef]
- Fang, H.; Xiang, Y.Q.; Hao, Y.J.; Chu, X.Q.; Pan, X.D.; Yu, J.Q.; Yu, Y.L. Fungal Degradation of Chlorpyrifos by Verticillium sp. DSP in Pure Cultures and Its Use in Bioremediation of Contaminated Soil and Pakchoi. Int. Biodeterior. Biodegrad. 2008, 61, 294–303. [Google Scholar] [CrossRef]
- Yadav, M.; Srivastva, N.; Shukla, A.K.; Singh, R.S.; Upadhyay, S.N.; Dubey, S.K. Efficacy of Aspergillus sp. for Degradation of Chlorpyrifos in Batch and Continuous Aerated Packed Bed Bioreactors. Appl. Biochem. Biotechnol. 2015, 175, 16–24. [Google Scholar] [CrossRef]
- Abd-Alrahman, H.; Mostafa, A.A. Mycoremediation of Organophosphorous Insecticide Chlorpyrifos by Fungal Soil Isolates. J. Pure Appl. Microbiol. 2014, 8, 2945–2951. [Google Scholar]
- Gao, Y.; Chen, S.; Hu, M.; Hu, Q.; Luo, J.; Li, Y. Purification and Characterization of a Novel Chlorpyrifos Hydrolase from Cladosporium cladosporioides Hu-01. PLoS ONE 2012, 7, e38137. [Google Scholar] [CrossRef] [PubMed]
- Bempelou, E.D.; Vontas, J.G.; Liapis, K.S.; Ziogas, V.N. Biodegradation of Chlorpyrifos and 3,5,6-Trichloro-2-Pyridinol by the Epiphytic Yeasts Rhodotorula glutinis and Rhodotorula rubra. Ecotoxicology 2018, 27, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Castellana, G.; Loffredo, E. Simultaneous Removal of Endocrine Disruptors from a Wastewater Using White Rot Fungi and Various Adsorbents. Water Air Soil. Pollut. 2014, 225, 1872. [Google Scholar] [CrossRef]
- Xingjia, Y.; Bin, L. Dimethoate Degradation and Calcium Phosphate Formation Induced by Aspergillus niger. Afr. J. Microbiol. Res. 2012, 6, 7603–7609. [Google Scholar] [CrossRef]
- Ferreira-Guedes, S.; Mendes, B.; Leitão, A.L. Degradation of 2,4-Dichlorophenoxyacetic Acid by a Halotolerant Strain of Penicillium chrysogenum: Antibiotic Production. Environ. Technol. 2012, 33, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Boelan, E.G.; Purnomo, A.S. Biodegradation of 1,1,1-Trichloro-2,2-Bis (4-Chlorophenyl) Ethane (DDT) by Mixed Cultures of White-Rot Fungus Ganoderma lingzhi and Bacterium Pseudomonas aeruginosa. HAYATI J. Biosci. 2019, 26, 90. [Google Scholar] [CrossRef]
- Rizqi, H.D.; Purnomo, A.S.; Ulfi, A. The Effect of Bacteria Addition on DDT Biodegradation by Brown Rot Fungus Gloeophyllum trabeum. Heliyon 2023, 9, e18216. [Google Scholar] [CrossRef]
- Chen, S.; Hu, Q.; Hu, M.; Luo, J.; Weng, Q.; Lai, K. Isolation and Characterization of a Fungus Able to Degrade Pyrethroids and 3-Phenoxybenzaldehyde. Bioresour. Technol. 2011, 102, 8110–8116. [Google Scholar] [CrossRef]
- Willian, G.A. Biodegradation of Chlorpyrifos by Whole Cells of Marine-Derived Fungi Aspergillus sydowii and Trichoderma sp. J. Microb. Biochem. Technol. 2015, 7, 133–139. [Google Scholar] [CrossRef]
- Maya, K.; Upadhyay, S.N.; Singh, R.S.; Dubey, S.K. Degradation Kinetics of Chlorpyrifos and 3,5,6-Trichloro-2-Pyridinol (TCP) by Fungal Communities. Bioresour. Technol. 2012, 126, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Li, Y.; Zheng, W.; Peng, X.; Li, W.; Yan, Y. Mineralization of Chlorpyrifos by Co-Culture of Serratia and Trichosporon spp. Biotechnol. Lett. 2007, 29, 1469–1473. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wareth, M.T.A.; Abd El-Hamid, R.M. Mycoremediation of Chlorpyrifos and Lambda-Cyhalothrin by Two Species of Filamentous Fungi. Int. J. Environ. Stud. 2016, 73, 974–987. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Zhao, X.; Zhang, X.; Zhao, Q.; Wang, X.; Li, Y. Restructured Fungal Community Diversity and Biological Interactions Promote Metolachlor Biodegradation in Soil Microbial Fuel Cells. Chemosphere 2019, 221, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Esparza-Naranjo, S.B.; Da Silva, G.F.; Duque-Castaño, D.C.; Araújo, W.L.; Peres, C.K.; Boroski, M.; Bonugli-Santos, R.C. Potential for the Biodegradation of Atrazine Using Leaf Litter Fungi from a Subtropical Protection Area. Curr. Microbiol. 2021, 78, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Marinho, G.; Barbosa, B.C.A.; Rodrigues, K.; Aquino, M.; Pereira, L. Potential of the Filamentous Fungus Aspergillus niger AN 400 to Degrade Atrazine in Wastewaters. Biocatal. Agric. Biotechnol. 2017, 9, 162–167. [Google Scholar] [CrossRef]
- Castro, J.V.; Peralba, M.C.R.; Ayub, M.A.Z. Biodegradation of the Herbicide Glyphosate by Filamentous Fungi in Platform Shaker and Batch Bioreactor. J. Environ. Sci. Health Part B 2007, 42, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, B.; Wieczorek, P.; Krzysko-Lupicka, T.; Golab, Z.; Lejczak, B.; Kavfarski, P. Organophosphonate Utilization by the Wild-Type Strain of Penicillium notatum. Appl. Env. Microbiol. 1995, 61, 2905–2910. [Google Scholar] [CrossRef]
- Fu, G.; Chen, Y.; Li, R.; Yuan, X.; Liu, C.; Li, B.; Wan, Y. Pathway and Rate-Limiting Step of Glyphosate Degradation by Aspergillus oryzae A-F02. Prep. Biochem. Biotechnol. 2017, 47, 782–788. [Google Scholar] [CrossRef]
- Klimek, M.; Lejczak, B.; Kafarski, P.; Forlani, G. Metabolism of the Phosphonate Herbicide Glyphosate by a Non-Nitrate-Utilizing Strain of Penicillium chrysogenum. Pest. Manag. Sci. 2001, 57, 815–821. [Google Scholar] [CrossRef]
- Guo, J.; Song, X.; Li, R.; Zhang, Q.; Zheng, S.; Li, Q.; Tao, B. Isolation of a Degrading Strain of Fusarium verticillioides and Bioremediation of Glyphosate Residue. Pestic. Biochem. Physiol. 2022, 182, 105031. [Google Scholar] [CrossRef]
- Bravim, N.P.B.; Alves, A.F.; Orlanda, J.F.F. Biodegradation of Atrazine, Glyphosate and Pendimetaline Employing Fungal Consortia. RSD 2020, 9, e1549119679. [Google Scholar] [CrossRef]
- Spinelli, V.; Ceci, A.; Dal Bosco, C.; Gentili, A.; Persiani, A.M. Glyphosate-Eating Fungi: Study on Fungal Saprotrophic Strains’ Ability to Tolerate and Utilise Glyphosate as a Nutritional Source and on the Ability of Purpureocillium lilacinum to Degrade It. Microorganisms 2021, 9, 2179. [Google Scholar] [CrossRef] [PubMed]
- Aluffi, M.E.; Carranza, C.S.; Benito, N.; Magnoli, K.; Magnoli, C.E.; Barberis, C.L. Isolation of Culturable Mycota from Argentinean Soils Exposed or Not-Exposed to Pesticides and Determination of Glyphosate Tolerance of Fungal Species in Media Supplied with the Herbicide. Rev. Argent. Microbiol. 2020, 52, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Chan-Cupul, W.; Heredia-Abarca, G.; Rodríguez-Vázquez, R. Atrazine Degradation by Fungal Co-Culture Enzyme Extracts under Different Soil Conditions. J. Environ. Sci. Health Part B 2016, 51, 298–308. [Google Scholar] [CrossRef] [PubMed]
- de Lopes, R.O.; Pereira, P.M.; Pereira, A.R.B.; Fernandes, K.V.; Carvalho, J.F.; da França, A.S.D.; Valente, R.H.; da Silva, M.; Ferreira-Leitão, V.S. Atrazine, Desethylatrazine (DEA) and Desisopropylatrazine (DIA) Degradation by Pleurotus ostreatus INCQS 40310. Biocatal. Biotransform. 2020, 38, 415–430. [Google Scholar] [CrossRef]
- Bastos, A.C.; Magan, N. Trametes versicolor: Potential for Atrazine Bioremediation in Calcareous Clay Soil, under Low Water Availability Conditions. Int. Biodeterior. Biodegrad. 2009, 63, 389–394. [Google Scholar] [CrossRef]
- Dhiman, N.; Jasrotia, T.; Sharma, P.; Negi, S.; Chaudhary, S.; Kumar, R.; Mahnashi, M.H.; Umar, A.; Kumar, R. Immobilization Interaction between Xenobiotic and Bjerkandera adusta for the Biodegradation of Atrazine. Chemosphere 2020, 257, 127060. [Google Scholar] [CrossRef]
- Henn, C.; Monteiro, D.A.; Boscolo, M.; da Silva, R.; Gomes, E. Biodegradation of Atrazine and Ligninolytic Enzyme Production by Basidiomycete Strains. BMC Microbiol. 2020, 20, 266. [Google Scholar] [CrossRef]
- Szewczyk, R.; Różalska, S.; Mironenka, J.; Bernat, P. Atrazine Biodegradation by Mycoinsecticide Metarhizium robertsii: Insights into Its Amino Acids and Lipids Profile. J. Environ. Manag. 2020, 262, 110304. [Google Scholar] [CrossRef]
- Pelcastre, M.I.; Ibarra, J.R.V.; Navarrete, A.M.; Rosas, J.C.; Ramirez, C.A.G.; Sandoval, O.A.A. Bioremediation Perspectives Using Autochthonous Species of Trichoderma sp. for Degradation of Atrazine in Agricultural Soil from the Tulancingo Valley, Hidalgo, Mexico. Trop. Subtrop. Agroecosystems 2013, 16, 265–276. [Google Scholar]
- Yu, J.; He, H.; Yang, W.L.; Yang, C.; Zeng, G.; Wu, X. Magnetic Bionanoparticles of Penicillium sp. Yz11-22N2 Doped with Fe3O4 and Encapsulated within PVA-SA Gel Beads for Atrazine Removal. Bioresour. Technol. 2018, 260, 196–203. [Google Scholar] [CrossRef]
- Wu, X.; He, H.; Yang, W.L.; Yu, J.; Yang, C. Efficient Removal of Atrazine from Aqueous Solutions Using Magnetic Saccharomyces cerevisiae Bionanomaterial. Appl. Microbiol. Biotechnol. 2018, 102, 7597–7610. [Google Scholar] [CrossRef] [PubMed]
- Elgueta, S.; Santos, C.; Lima, N.; Diez, M.C. Immobilization of the White-Rot Fungus Anthracophyllum discolor to Degrade the Herbicide Atrazine. AMB Express 2016, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, S.; Shan, X.; Chen, B.-D.; Zhu, Y.-G.; Bell, J.N.B. Effect of Arbuscular Mycorrhizal Fungus (Glomus caledonium) on the Accumulation and Metabolism of Atrazine in Maize (Zea mays L.) and Atrazine Dissipation in Soil. Environ. Pollut. 2007, 146, 452–457. [Google Scholar] [CrossRef]
- Herrera-Gallardo, B.E.; Guzmán-Gil, R.; Colín-Luna, J.A.; García-Martínez, J.C.; León-Santiesteban, H.H.; González-Brambila, O.M.; González-Brambila, M.M. Atrazine Biodegradation in Soil by Aspergillus niger. Can. J. Chem. Eng. 2021, 99, 932–946. [Google Scholar] [CrossRef]
- Munoz, A.; Koskinen, W.C.; Cox, L.; Sadowsky, M.J. Biodegradation and Mineralization of Metolachlor and Alachlor by Candida xestobii. J. Agric. Food Chem. 2011, 59, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Orozco, A.; Galíndez-Nájera, S.P.; Ruiz-Ordaz, N.; Galíndez-Mayer, J.; Martínez-Jerónimo, F.F. Biodegradation of a Commercial Mixture of the Herbicides Atrazine and S-Metolachlor in a Multi-Channel Packed Biofilm Reactor. Environ. Sci. Pollut. Res. 2017, 24, 25656–25665. [Google Scholar] [CrossRef]
- Chang, X.; Liang, J.; Sun, Y.; Zhao, L.; Zhou, B.; Li, X.; Li, Y. Isolation, Degradation Performance and Field Application of the Metolachlor-Degrading Fungus Penicillium oxalicum MET-F-1. Appl. Sci. 2020, 10, 8556. [Google Scholar] [CrossRef]
- Sanyal, D.; Kulshrestha, G. Metabolism of Metolachlor by Fungal Cultures. J. Agric. Food Chem. 2002, 50, 499–505. [Google Scholar] [CrossRef]
- Nykiel-Szymańska, J.; Bernat, P.; Słaba, M. Biotransformation and Detoxification of Chloroacetanilide Herbicides by Trichoderma spp. with Plant Growth-Promoting Activities. Pestic. Biochem. Physiol. 2020, 163, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Fragoeiro, S.; Magan, N. Enzymatic Activity, Osmotic Stress and Degradation of Pesticide Mixtures in Soil Extract Liquid Broth Inoculated with Phanerochaete chrysosporium and Trametes versicolor. Environ. Microbiol. 2005, 7, 348–355. [Google Scholar] [CrossRef] [PubMed]
- García-Galán, M.J.; Monllor-Alcaraz, L.S.; Postigo, C.; Uggetti, E.; López De Alda, M.; Díez-Montero, R.; García, J. Microalgae-Based Bioremediation of Water Contaminated by Pesticides in Peri-Urban Agricultural Areas. Environ. Pollut. 2020, 265, 114579. [Google Scholar] [CrossRef]
- Lutzu, G.A.; Ciurli, A.; Chiellini, C.; Di Caprio, F.; Concas, A.; Dunford, N.T. Latest Developments in Wastewater Treatment and Biopolymer Production by Microalgae. J. Environ. Chem. Eng. 2021, 9, 104926. [Google Scholar] [CrossRef]
- Singhal, M.; Swapnali, J.; Swaroop, S.S.; Mahipal Singh, S.; Rajeev, K. Microalgae Based Sustainable Bioremediation of Water Contaminated by Pesticides. Biointerface Res. Appl. Chem. 2021, 12, 149–169. [Google Scholar] [CrossRef]
- Muñoz, R.; Guieysse, B. Algal–Bacterial Processes for the Treatment of Hazardous Contaminants: A Review. Water Res. 2006, 40, 2799–2815. [Google Scholar] [CrossRef] [PubMed]
- Narchonai, G.; Arutselvan, C.; LewisOscar, F.; Thajuddin, N. Enhancing Starch Accumulation/Production in Chlorococcum humicola through Sulphur Limitation and 2,4-D Treatment for Butanol Production. Biotechnol. Rep. 2020, 28, e00528. [Google Scholar] [CrossRef]
- Nie, J.; Sun, Y.; Zhou, Y.; Kumar, M.; Usman, M.; Li, J.; Shao, J.; Wang, L.; Tsang, D.C.W. Bioremediation of Water Containing Pesticides by Microalgae: Mechanisms, Methods, and Prospects for Future Research. Sci. Total Environ. 2020, 707, 136080. [Google Scholar] [CrossRef]
- Fu, P.; Secundo, F. Algae and Their Bacterial Consortia for Soil Bioremediation. Chem. Eng. Trans. 2016, 49, 427–432. [Google Scholar] [CrossRef]
- Friesen-Pankratz, B.B.; Doebel, C.C.; Farenhorst, A.A.; Gordon Goldsborough, L. Interactions Between Algae (Selenastrum capricornutum) and Pesticides: Implications for Managing Constructed Wetlands for Pesticide Removal. J. Environ. Sci. Health Part B 2003, 38, 147–155. [Google Scholar] [CrossRef]
- Hu, N.; Xu, Y.; Sun, C.; Zhu, L.; Sun, S.; Zhao, Y.; Hu, C. Removal of Atrazine in Catalytic Degradation Solutions by Microalgae Chlorella sp. and Evaluation of Toxicity of Degradation Products via Algal Growth and Photosynthetic Activity. Ecotoxicol. Environ. Saf. 2021, 207, 111546. [Google Scholar] [CrossRef] [PubMed]
- González-Barreiro, O.; Rioboo, C.; Herrero, C.; Cid, A. Removal of Triazine Herbicides from Freshwater Systems Using Photosynthetic Microorganisms. Environ. Pollut. 2006, 144, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Salman, J.M.; Abdul-Adel, E. Potential Use of Cyanophyta Species Oscillatoria limnetica in Bioremediation of Organophosphorus Herbicide Glyphosate. Mesop. Environ. J. 2015, 1, 15–26. [Google Scholar]
- Wang, C.; Lin, X.; Li, L.; Lin, S. Differential Growth Responses of Marine Phytoplankton to Herbicide Glyphosate. PLoS ONE 2016, 11, e0151633. [Google Scholar] [CrossRef] [PubMed]
- Avila, R.; Peris, A.; Eljarrat, E.; Vicent, T.; Blánquez, P. Biodegradation of Hydrophobic Pesticides by Microalgae: Transformation Products and Impact on Algae Biochemical Methane Potential. Sci. Total Environ. 2021, 754, 142114. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.H.; Abdullah, A.M.; Badr El Din, N.I.; Mishaqa, E.S.I. Biosorption Potential of the Microchlorophyte Chlorella vulgaris for Some Pesticides. J. Fertil. Pestic. 2017, 8, 1000177. [Google Scholar] [CrossRef]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine Actinomycetes, New Sources of Biotechnological Products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Singh, D.; Kumari, A.; Sharma, G.; Rajput, S.; Arora, S.; Kaur, R. Pesticide Residues Degradation Strategies in Soil and Water: A Review. Int. J. Environ. Sci. Technol. 2023, 20, 3537–3560. [Google Scholar] [CrossRef]
- Alvarez, A.; Saez, J.M.; Davila Costa, J.S.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current Research and Perspectives for Bioremediation of Pesticides and Heavy Metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef]
- Ariffin, F.; Rahman, S.A. Biodegradation of Carbofuran; A Review. J. Environ. Microbiol. Toxicol. 2020, 8, 50–57. [Google Scholar] [CrossRef]
- Ichinose, H. Molecular and Functional Diversity of Fungal Cytochrome P450s. Biol. Pharm. Bull. 2012, 35, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Kim, J.-E.; Lee, Y.-W.; Son, H. Fungal Cytochrome P450s and the P450 Complement (CYPome) of Fusarium graminearum. Toxins 2018, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Pietikäinen, J.; Pettersson, M.; Bååth, E. Comparison of Temperature Effects on Soil Respiration and Bacterial and Fungal Growth Rates. FEMS Microbiol. Ecol. 2005, 52, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Klimek, B. Scots pine roots modify the short-term effects of temperature and moisture on soil bacteria and fungi. Appl. Ecol. Env. Res. 2013, 11, 173–188. [Google Scholar] [CrossRef]
- Fabian, J.; Zlatanovic, S.; Mutz, M.; Premke, K. Fungal–Bacterial Dynamics and Their Contribution to Terrigenous Carbon Turnover in Relation to Organic Matter Quality. ISME J. 2017, 11, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Krauss, G.-J.; Solé, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Bärlocher, F. Fungi in Freshwaters: Ecology, Physiology and Biochemical Potential. FEMS Microbiol. Rev. 2011, 35, 620–651. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, H.; Kaur, S.; Phale, P.S. Conserved Metabolic and Evolutionary Themes in Microbial Degradation of Carbamate Pesticides. Front. Microbiol. 2021, 12, 648868. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Pang, S.; Zhang, W.; Lin, Z.; Bhatt, P.; Chen, S. Insights into the Microbial Degradation and Biochemical Mechanisms of Carbamates. Chemosphere 2021, 279, 130500. [Google Scholar] [CrossRef]
- Kattiparambil Manoharan, R.; Sankaran, S. Photocatalytic Degradation of Organic Pollutant Aldicarb by Non-Metal-Doped Nanotitania: Synthesis and Characterization. Env. Sci. Pollut. Res. 2018, 25, 20510–20517. [Google Scholar] [CrossRef]
- Kumar, S.S.; Ghosh, P.; Malyan, S.K.; Sharma, J.; Kumar, V. A Comprehensive Review on Enzymatic Degradation of the Organophosphate Pesticide Malathion in the Environment. J. Environ. Sci. Health Part C 2019, 37, 288–329. [Google Scholar] [CrossRef]
- Córdoba Gamboa, L.; Solano Diaz, K.; Ruepert, C.; Van Wendel De Joode, B. Passive Monitoring Techniques to Evaluate Environmental Pesticide Exposure: Results from the Infant’s Environmental Health Study (ISA). Environ. Res. 2020, 184, 109243. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, G.K.; Singh, S.; Kumar, V.; Dhanjal, D.S.; Datta, S.; Singh, J. Toxicity, Monitoring and Biodegradation of Organophosphate Pesticides: A Review. Crit. Rev. Environ. Sci. Technol. 2019, 49, 1135–1187. [Google Scholar] [CrossRef]
- Tsipi, D.; Botitsi, H.; Economou, A. (Eds.) Mass Spectrometry for the Analysis of Pesticide Residues and Their Metabolites; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2015; ISBN 978-1-119-06998-0. [Google Scholar]
- Wu, R.-J.; Chen, C.-C.; Chen, M.-H.; Lu, C.-S. Titanium Dioxide-Mediated Heterogeneous Photocatalytic Degradation of Terbufos: Parameter Study and Reaction Pathways. J. Hazard. Mater. 2009, 162, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Mishra, A.; Melo, J.S. Biodegradation of Methyl Parathion and Its Application in Biosensors. Austin J. Environ. Toxicol. 2018, 4, 1024. [Google Scholar]
- Zhao, F.; He, J.; Li, X.; Bai, Y.; Ying, Y.; Ping, J. Smart Plant-Wearable Biosensor for in-Situ Pesticide Analysis. Biosens. Bioelectron. 2020, 170, 112636. [Google Scholar] [CrossRef] [PubMed]
- Bhende, R.S.; Jhariya, U.; Srivastava, S.; Bombaywala, S.; Das, S.; Dafale, N.A. Environmental Distribution, Metabolic Fate, and Degradation Mechanism of Chlorpyrifos: Recent and Future Perspectives. Appl. Biochem. Biotechnol. 2022, 194, 2301–2335. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, B.; Wang, M.-Q.; Gao, J.-J.; Li, Z.-J.; Tian, Y.-S.; Peng, R.-H.; Yao, Q.-H. Metabolic Engineering of Escherichia coli for Methyl Parathion Degradation. Front. Microbiol. 2022, 13, 679126. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, Y.; Duan, Y.; Li, Z.; Cui, Z.; Liu, W. Crystal Structure of P-Nitrophenol 4-Monooxygenase PnpA from Pseudomonas putida DLL-E4: The Key Enzyme Involved in p-Nitrophenol Degradation. Biochem. Biophys. Res. Commun. 2018, 504, 715–720. [Google Scholar] [CrossRef]
- Li, H.; Ma, Y.; Yao, T.; Zhang, J.; Gao, Y.; Li, C. Evaluation of Beta-Cyfluthrin Biodegradation by a Novel Bacterial Consortium: Microbial Succession, Degradation Pathway and Toxicity Assessment. SSRN J. 2021, 1–34. [Google Scholar] [CrossRef]
- Bhatt, P.; Huang, Y.; Zhan, H.; Chen, S. Insight into Microbial Applications for the Biodegradation of Pyrethroid Insecticides. Front. Microbiol. 2019, 10, 1778. [Google Scholar] [CrossRef]
- Ghodke, V.M.; Punekar, N.S. Environmental Role of Aromatic Carboxylesterases. Environ. Microbiol. 2022, 24, 2657–2668. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, W.; Lin, L.; He, D.; Huang, G.; Shen, Y.; Wei, W.; Wei, D. Autotransporter Domain-Dependent Enzymatic Analysis of a Novel Extremely Thermostable Carboxylesterase with High Biodegradability towards Pyrethroid Pesticides. Sci. Rep. 2017, 7, 3461. [Google Scholar] [CrossRef] [PubMed]
- Schleier, J.J.; Peterson, R.K.D. The Joint Toxicity of Type I, II, and Nonester Pyrethroid Insecticides. JNL Econ. Entom. 2012, 105, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Ensley, S.M. Pyrethrins and Pyrethroids. In Veterinary Toxicology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 515–520. ISBN 978-0-12-811410-0. [Google Scholar]
- Sheng, Y.; Benmati, M.; Guendouzi, S.; Benmati, H.; Yuan, Y.; Song, J.; Xia, C.; Berkani, M. Latest Eco-Friendly Approaches for Pesticides Decontamination Using Microorganisms and Consortia Microalgae: A Comprehensive Insights, Challenges, and Perspectives. Chemosphere 2022, 308, 136183. [Google Scholar] [CrossRef] [PubMed]
- Gajendiran, A.; Abraham, J. An Overview of Pyrethroid Insecticides. Front. Biol. 2018, 13, 79–90. [Google Scholar] [CrossRef]
- Reddy, V.D.; Rao, P.N.; Rao, K.V. Pests and Pathogens: Management Strategies; BS Publish: Hyderabad, India, 2011; ISBN 978-0-415-66576-6. [Google Scholar]
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary Enzyme Based Technologies for Bioremediation: A Review. J. Environ. Manag. 2018, 210, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Zhang, W.; Lin, Z.; Pang, S.; Huang, Y.; Bhatt, P.; Chen, S. Carbofuran Toxicity and Its Microbial Degradation in Contaminated Environments. Chemosphere 2020, 259, 127419. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, B.; Dhamale, T.; Saha, B.K.; Phale, P.S. Microbial Degradation of Aromatic Pollutants: Metabolic Routes, Pathway Diversity, and Strategies for Bioremediation. In Microbial Biodegradation and Bioremediation; Elsevier: Amsterdam, The Netherlands, 2022; pp. 365–394. ISBN 978-0-323-85455-9. [Google Scholar]
- Shin, D.-H. Genetic and Phenotypic Diversity of Carbofuran-Degrading Bacteria Isolated from Agricultural Soils. J. Microbiol. Biotechnol. 2012, 22, 448–456. [Google Scholar] [CrossRef]
- Suhara, H.; Adachi, A.; Kamei, I.; Maekawa, N. Degradation of Chlorinated Pesticide DDT by Litter-Decomposing Basidiomycetes. Biodegradation 2011, 22, 1075–1086. [Google Scholar] [CrossRef]
- Ma, J.; Pan, L.; Yang, X.; Liu, X.; Tao, S.; Zhao, L.; Qin, X.; Sun, Z.; Hou, H.; Zhou, Y. DDT, DDD, and DDE in Soil of Xiangfen County, China: Residues, Sources, Spatial Distribution, and Health Risks. Chemosphere 2016, 163, 578–583. [Google Scholar] [CrossRef]
- Nikolaivits, E.; Dimarogona, M.; Fokialakis, N.; Topakas, E. Marine-Derived Biocatalysts: Importance, Accessing, and Application in Aromatic Pollutant Bioremediation. Front. Microbiol. 2017, 8, 265. [Google Scholar] [CrossRef] [PubMed]
- Cutright, T.J.; Erdem, Z. Overview of the bioremediation and the degradation pathways of ddt. J. Adnan Menderes Univ. Agric. Fac. 2012, 9, 39–45. [Google Scholar]
- Abbes, C.; Mansouri, A.; Werfelli, N.; Landoulsi, A. Aerobic Biodegradation of DDT by Advenella kashmirensis and Its Potential Use in Soil Bioremediation. Soil Sediment Contam. Int. J. 2018, 27, 455–468. [Google Scholar] [CrossRef]
- Xu, H.-J.; Bai, J.; Li, W.; Murrell, J.C.; Zhang, Y.; Wang, J.; Luo, C.; Li, Y. Mechanisms of the Enhanced DDT Removal from Soils by Earthworms: Identification of DDT Degraders in Drilosphere and Non-Drilosphere Matrices. J. Hazard. Mater. 2021, 404, 124006. [Google Scholar] [CrossRef] [PubMed]
- Colombo, R.; Yariwake, J.; Lanza, M. Degradation Products of Lambda-Cyhalothrin in Aqueous Solution as Determined by SBSE-GC-IT-MS. J. Braz. Chem. Soc. 2018, 29, 2207–2212. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Shen, C.; Dong, S.; Hu, X.; Lin, M.; Zhang, X.; Xu, C.; Zhong, J.; Xie, Y.; et al. Construction and Characterization of a Class-Specific Single-Chain Variable Fragment against Pyrethroid Metabolites. Appl. Microbiol. Biotechnol. 2020, 104, 7345–7354. [Google Scholar] [CrossRef] [PubMed]
- Cycoń, M.; Piotrowska-Seget, Z. Pyrethroid-Degrading Microorganisms and Their Potential for the Bioremediation of Contaminated Soils: A Review. Front. Microbiol. 2016, 7, 1463. [Google Scholar] [CrossRef]
- Wang, X.; Martínez, M.-A.; Dai, M.; Chen, D.; Ares, I.; Romero, A.; Castellano, V.; Martínez, M.; Rodríguez, J.L.; Martínez-Larrañaga, M.-R.; et al. Permethrin-Induced Oxidative Stress and Toxicity and Metabolism. A Review. Environ. Res. 2016, 149, 86–104. [Google Scholar] [CrossRef]
- Bhatt, P.; Gangola, S.; Chaudhary, P.; Khati, P.; Kumar, G.; Sharma, A.; Srivastava, A. Pesticide Induced Up-Regulation of Esterase and Aldehyde Dehydrogenase in Indigenous Bacillus spp. Bioremediation J. 2019, 23, 42–52. [Google Scholar] [CrossRef]
- Zhang, M.; Mei, J.; Lv, S.; Lai, J.; Zheng, X.; Yang, J.; Cui, S. Simultaneous Extraction of Permethrin Diastereomers and Deltamethrin in Environmental Water Samples Based on Aperture Regulated Magnetic Mesoporous Silica. New J. Chem. 2020, 44, 16152–16162. [Google Scholar] [CrossRef]
- Qin, S.; Gan, J. Enantiomeric Differences in Permethrin Degradation Pathways in Soil and Sediment. J. Agric. Food Chem. 2006, 54, 9145–9151. [Google Scholar] [CrossRef]
- Feng, X.; Liu, N. Functional Analyses of House Fly Carboxylesterases Involved in Insecticide Resistance. Front. Physiol. 2020, 11, 595009. [Google Scholar] [CrossRef]
- Tyler, C.R.; Beresford, N.; Van Der Woning, M.; Sumpter, J.P.; Tchorpe, K. Metabolism and Environmental Degradation of Pyrethroid Insecticides Produce Compounds with Endocrine Activities. Environ. Toxicol. Chem. 2000, 19, 801–809. [Google Scholar] [CrossRef]
- Liu, H.-F.; Ku, C.-H.; Chang, S.-S.; Chang, C.-M.; Wang, I.-K.; Yang, H.-Y.; Weng, C.-H.; Huang, W.-H.; Hsu, C.-W.; Yen, T.-H. Outcome of Patients with Chlorpyrifos Intoxication. Hum. Exp. Toxicol. 2020, 39, 1291–1300. [Google Scholar] [CrossRef]
- Ahirwar, U.; Kollah, B.; Dubey, G.; Mohanty, S.R. Chlorpyrifos Biodegradation in Relation to Metabolic Attributes and 16S rRNA Gene Phylogeny of Bacteria in a Tropical Vertisol. SN Appl. Sci. 2019, 1, 228. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, W.; Pang, S.; Chen, J.; Bhatt, P.; Mishra, S.; Chen, S. Insights into the Microbial Degradation and Catalytic Mechanisms of Chlorpyrifos. Environ. Res. 2021, 194, 110660. [Google Scholar] [CrossRef]
- McLachlan, M.S.; Undeman, E.; Zhao, F.; MacLeod, M. Predicting Global Scale Exposure of Humans to PCB 153 from Historical Emissions. Environ. Sci. Process. Impacts 2018, 20, 747–756. [Google Scholar] [CrossRef]
- Dar, M.A.; Kaushik, G.; Villarreal-Chiu, J.F. Pollution Status and Bioremediation of Chlorpyrifos in Environmental Matrices by the Application of Bacterial Communities: A Review. J. Environ. Manag. 2019, 239, 124–136. [Google Scholar] [CrossRef]
- Magnoli, K.; Carranza, C.S.; Aluffi, M.E.; Magnoli, C.E.; Barberis, C.L. Herbicides Based on 2,4-D: Its Behavior in Agricultural Environments and Microbial Biodegradation Aspects. A Review. Env. Sci. Pollut. Res. 2020, 27, 38501–38512. [Google Scholar] [CrossRef]
- Islam, F.; Wang, J.; Farooq, M.A.; Khan, M.S.S.; Xu, L.; Zhu, J.; Zhao, M.; Muños, S.; Li, Q.X.; Zhou, W. Potential Impact of the Herbicide 2,4-Dichlorophenoxyacetic Acid on Human and Ecosystems. Environ. Int. 2018, 111, 332–351. [Google Scholar] [CrossRef]
- Trefault, N.; De La Iglesia, R.; Molina, A.M.; Manzano, M.; Ledger, T.; Perez-Pantoja, D.; Sanchez, M.A.; Stuardo, M.; Gonzalez, B. Genetic Organization of the Catabolic Plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) Reveals Mechanisms of Adaptation to Chloroaromatic Pollutants and Evolution of Specialized Chloroaromatic Degradation Pathways. Env. Microbiol. 2004, 6, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, Y.-S.; Gao, J.-J.; Xu, J.; Li, Z.-J.; Fu, X.-Y.; Han, H.-J.; Wang, L.-J.; Zhang, W.-H.; Deng, Y.-D.; et al. Complete Biodegradation of the Oldest Organic Herbicide 2,4-Dichlorophenoxyacetic Acid by Engineering Escherichia coli. J. Hazard. Mater. 2023, 451, 131099. [Google Scholar] [CrossRef] [PubMed]
- Nykiel-Szymańska, J.; Stolarek, P.; Bernat, P. Elimination and Detoxification of 2,4-D by Umbelopsis isabellina with the Involvement of Cytochrome P450. Env. Sci. Pollut. Res. 2018, 25, 2738–2743. [Google Scholar] [CrossRef] [PubMed]
- Reregistration Eligibility Decision for Dicamba and Associated Salts. Available online: https://nepis.epa.gov/Exe/ZyPDF.cgi/P10049M3.PDF?Dockey=P10049M3.PDF (accessed on 27 June 2023).
- Lerro, C.C.; Hofmann, J.N.; Andreotti, G.; Koutros, S.; Parks, C.G.; Blair, A.; Albert, P.S.; Lubin, J.H.; Sandler, D.P.; Beane Freeman, L.E. Dicamba Use and Cancer Incidence in the Agricultural Health Study: An Updated Analysis. Int. J. Epidemiol. 2020, 49, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Riter, L.S.; Pai, N.; Vieira, B.C.; MacInnes, A.; Reiss, R.; Hapeman, C.J.; Kruger, G.R. Conversations about the Future of Dicamba: The Science Behind Off-Target Movement. J. Agric. Food Chem. 2021, 69, 14435–14444. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Peng, Q.; Yao, L.; He, Q.; Qiu, J.; Cao, H.; He, J.; Niu, Q.; Lu, Y.; Hui, F. Roles of the Gentisate 1,2-Dioxygenases DsmD and GtdA in the Catabolism of the Herbicide Dicamba in Rhizorhabdus dicambivorans Ndbn-20. J. Agric. Food Chem. 2020, 68, 9287–9298. [Google Scholar] [CrossRef]
- Kanissery, R.; Gairhe, B.; Kadyampakeni, D.; Batuman, O.; Alferez, F. Glyphosate: Its Environmental Persistence and Impact on Crop Health and Nutrition. Plants 2019, 8, 499. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Gill, J.P.K.; Datta, S.; Singh, S.; Dhaka, V.; Kapoor, D.; Wani, A.B.; Dhanjal, D.S.; Kumar, M.; et al. Herbicide Glyphosate: Toxicity and Microbial Degradation. Int. J. Environ. Res. Public Health 2020, 17, 7519. [Google Scholar] [CrossRef]
- Rajendran, K.; Pujari, L.; Ethiraj, K. Biodegradation and Bioremediation of S-Triazine Herbicides. In Environmental Biotechnology Vol. 3; Gothandam, K.M., Ranjan, S., Dasgupta, N., Lichtfouse, E., Eds.; Environmental Chemistry for a Sustainable World; Springer International Publishing: Cham, Switzerland, 2021; Volume 50, pp. 31–54. ISBN 978-3-030-48972-4. [Google Scholar]
- Billet, L.; Devers, M.; Rouard, N.; Martin-Laurent, F.; Spor, A. Labour Sharing Promotes Coexistence in Atrazine Degrading Bacterial Communities. Sci. Rep. 2019, 9, 18363. [Google Scholar] [CrossRef]
- Böger, P.; Matthes, B.; Schmalfuß, J. Towards the Primary Target of Chloroacetamides-New Findings Pave the Way. Pest Manag. Sci. 2000, 56, 497–508. [Google Scholar] [CrossRef]
- Liu, S.Y.; Freyer, A.J.; Bollag, J.M. Microbial Dechlorination of the Herbicide Metolachlor. J. Agric. Food Chem. 1991, 39, 631–636. [Google Scholar] [CrossRef]
- Desaint, S.; Arrault, S.; Siblot, S.; Fournier, J.-C. Genetic Transfer of the mcd Gene in Soil. J. Appl. Microbiol. 2003, 95, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Fukui, M.; Hayano, K.; Hayatsu, M. Nucleotide Sequence and Genetic Structure of a Novel Carbaryl Hydrolase Gene (cehA) from Rhizobium sp. Strain AC100. Appl. Env. Microbiol. 2002, 68, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Horne, I.; Sutherland, T.D.; Harcourt, R.L.; Russell, R.J.; Oakeshott, J.G. Identification of an opd (Organophosphate Degradation) Gene in an Agrobacterium Isolate. Appl. Env. Microbiol. 2002, 68, 3371–3376. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Iken, B.; Damania, A. A Comparison of Organophosphate Degradation Genes and Bioremediation Applications: Review of OP Degradation Genes and Their Applications. Environ. Microbiol. Rep. 2013, 5, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.-J.; Bartlam, M.; Sun, L.; Zhou, Y.-F.; Zhang, Z.-P.; Zhang, C.-G.; Rao, Z.; Zhang, X.-E. Crystal Structure of Methyl Parathion Hydrolase from Pseudomonas sp. WBC-3. J. Mol. Biol. 2005, 353, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Stosiek, N.; Talma, M.; Klimek-Ochab, M. Carbon-Phosphorus Lyase—The State of the Art. Appl. Biochem. Biotechnol. 2020, 190, 1525–1552. [Google Scholar] [CrossRef] [PubMed]
- Hove-Jensen, B.; Zechel, D.L.; Jochimsen, B. Utilization of Glyphosate as Phosphate Source: Biochemistry and Genetics of Bacterial Carbon-Phosphorus Lyase. Microbiol. Mol. Biol. Rev. 2014, 78, 176–197. [Google Scholar] [CrossRef]
- Bhatt, P.; Bhatt, K.; Huang, Y.; Lin, Z.; Chen, S. Esterase Is a Powerful Tool for the Biodegradation of Pyrethroid Insecticides. Chemosphere 2020, 244, 125507. [Google Scholar] [CrossRef]
- Li, N.; Chen, L.; Chen, E.; Yuan, C.; Zhang, H.; He, J. Cloning of a Novel Tetrahydrofolate-Dependent Dicamba Demethylase Gene from Dicamba-Degrading Consortium and Characterization of the Gene Product. Front. Microbiol. 2022, 13, 978577. [Google Scholar] [CrossRef]
- Kumar, A.; Trefault, N.; Olaniran, A.O. Microbial Degradation of 2,4-Dichlorophenoxyacetic Acid: Insight into the Enzymes and Catabolic Genes Involved, Their Regulation and Biotechnological Implications. Crit. Rev. Microbiol. 2014, 42, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; He, S.; Li, X.; Shi, L.; Wang, F.; Yu, S.; Xu, S.; Ju, C.; Fang, H.; Yu, Y. Characterization, Genome Functional Analysis, and Detoxification of Atrazine by Arthrobacter sp. C2. Chemosphere 2021, 264, 128514. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, D.; Rath, S.K.; Mohapatra, P.K. Soil Fungi for Bioremediation of Pesticide Toxicants: A Perspective. Geomicrobiol. J. 2022, 39, 352–372. [Google Scholar] [CrossRef]
- Pileggi, M.; Pileggi, S.A.V.; Sadowsky, M.J. Herbicide Bioremediation: From Strains to Bacterial Communities. Heliyon 2020, 6, e05767. [Google Scholar] [CrossRef] [PubMed]
- Mawang, C.-I.; Azman, A.-S.; Fuad, A.-S.M.; Ahamad, M. Actinobacteria: An Eco-Friendly and Promising Technology for the Bioaugmentation of Contaminants. Biotechnol. Rep. 2021, 32, e00679. [Google Scholar] [CrossRef] [PubMed]
- Rafeeq, H.; Afsheen, N.; Rafique, S.; Arshad, A.; Intisar, M.; Hussain, A.; Bilal, M.; Iqbal, H.M.N. Genetically Engineered Microorganisms for Environmental Remediation. Chemosphere 2023, 310, 136751. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Hashemi, S.A.; Iman Moezzi, S.M.; Ravan, N.; Gholami, A.; Lai, C.W.; Chiang, W.-H.; Omidifar, N.; Yousefi, K.; Behbudi, G. Recent Advances in Enzymes for the Bioremediation of Pollutants. Biochem. Res. Int. 2021, 2021, 1–12. [Google Scholar] [CrossRef]
- Parisi, C.; Vigani, M.; Rodríguez-Cerezo, E. Agricultural Nanotechnologies: What Are the Current Possibilities? Nano Today 2015, 10, 124–127. [Google Scholar] [CrossRef]
- Awad, M.; Ibrahim, E.-D.S.; Osman, E.I.; Elmenofy, W.H.; Mahmoud, A.W.M.; Atia, M.A.M.; Moustafa, M.A.M. Nano-Insecticides against the Black Cutworm Agrotis ipsilon (Lepidoptera: Noctuidae): Toxicity, Development, Enzyme Activity, and DNA Mutagenicity. PLoS ONE 2022, 17, e0254285. [Google Scholar] [CrossRef]
- Aliyari Rad, S.; Nobaharan, K.; Pashapoor, N.; Pandey, J.; Dehghanian, Z.; Senapathi, V.; Minkina, T.; Ren, W.; Rajput, V.D.; Asgari Lajayer, B. Nano-Microbial Remediation of Polluted Soil: A Brief Insight. Sustainability 2023, 15, 876. [Google Scholar] [CrossRef]
- Qin, X.; Xiang, X.; Sun, X.; Ni, H.; Li, L. Preparation of Nanoscale Bacillus thuringiensis Chitinases Using Silica Nanoparticles for Nematicide Delivery. Int. J. Biol. Macromol. 2016, 82, 13–21. [Google Scholar] [CrossRef]
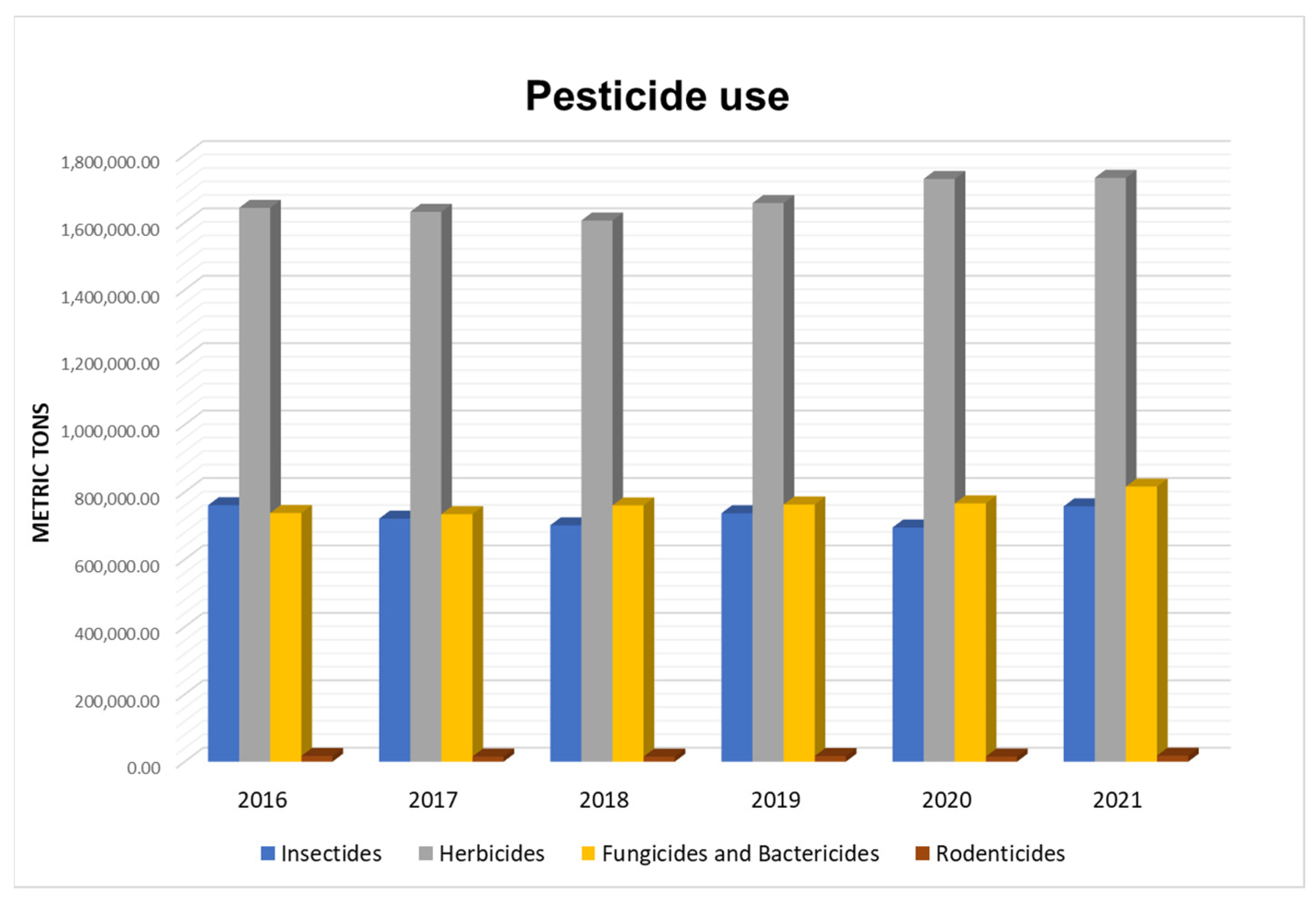

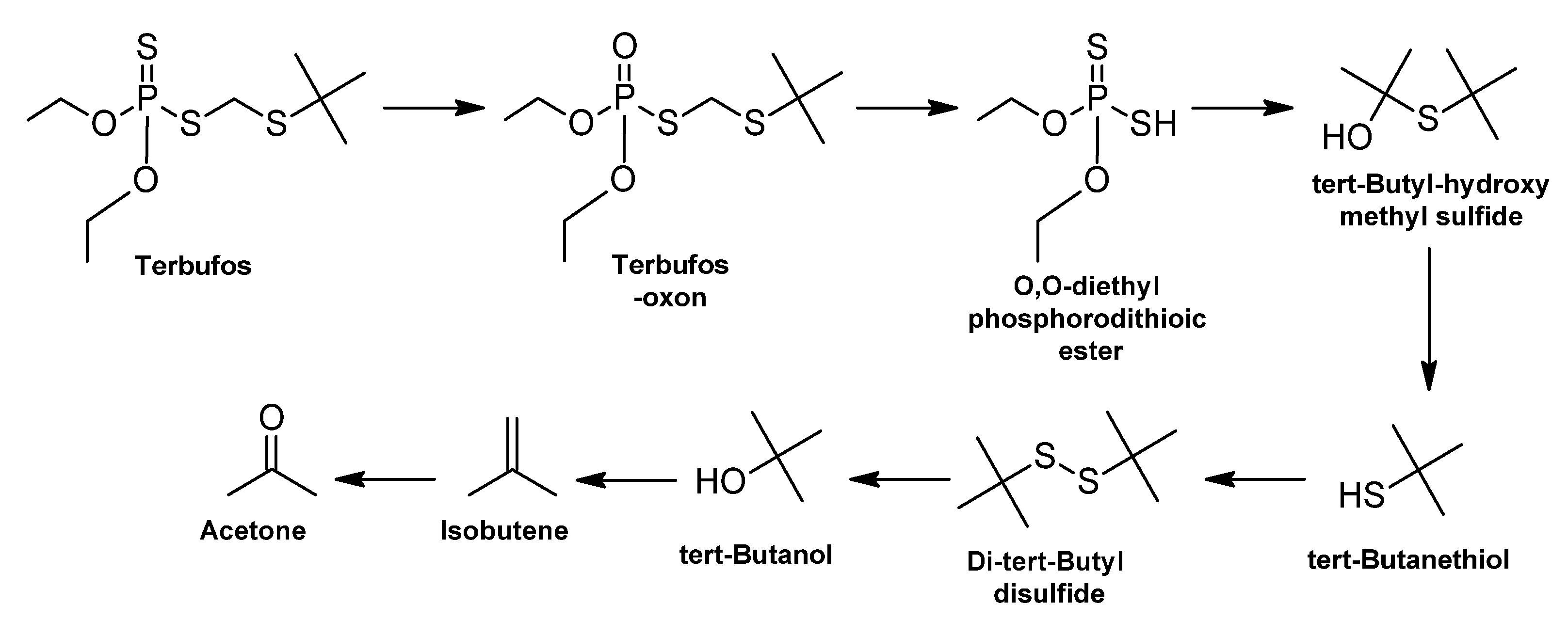
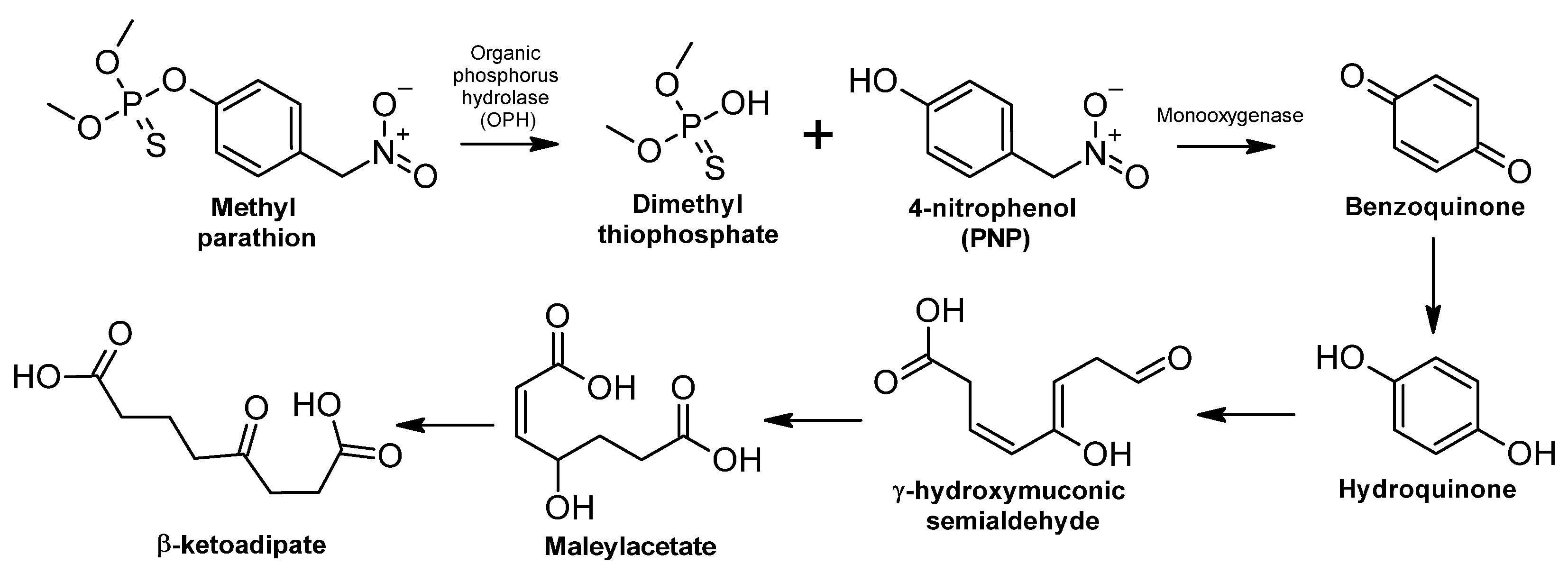
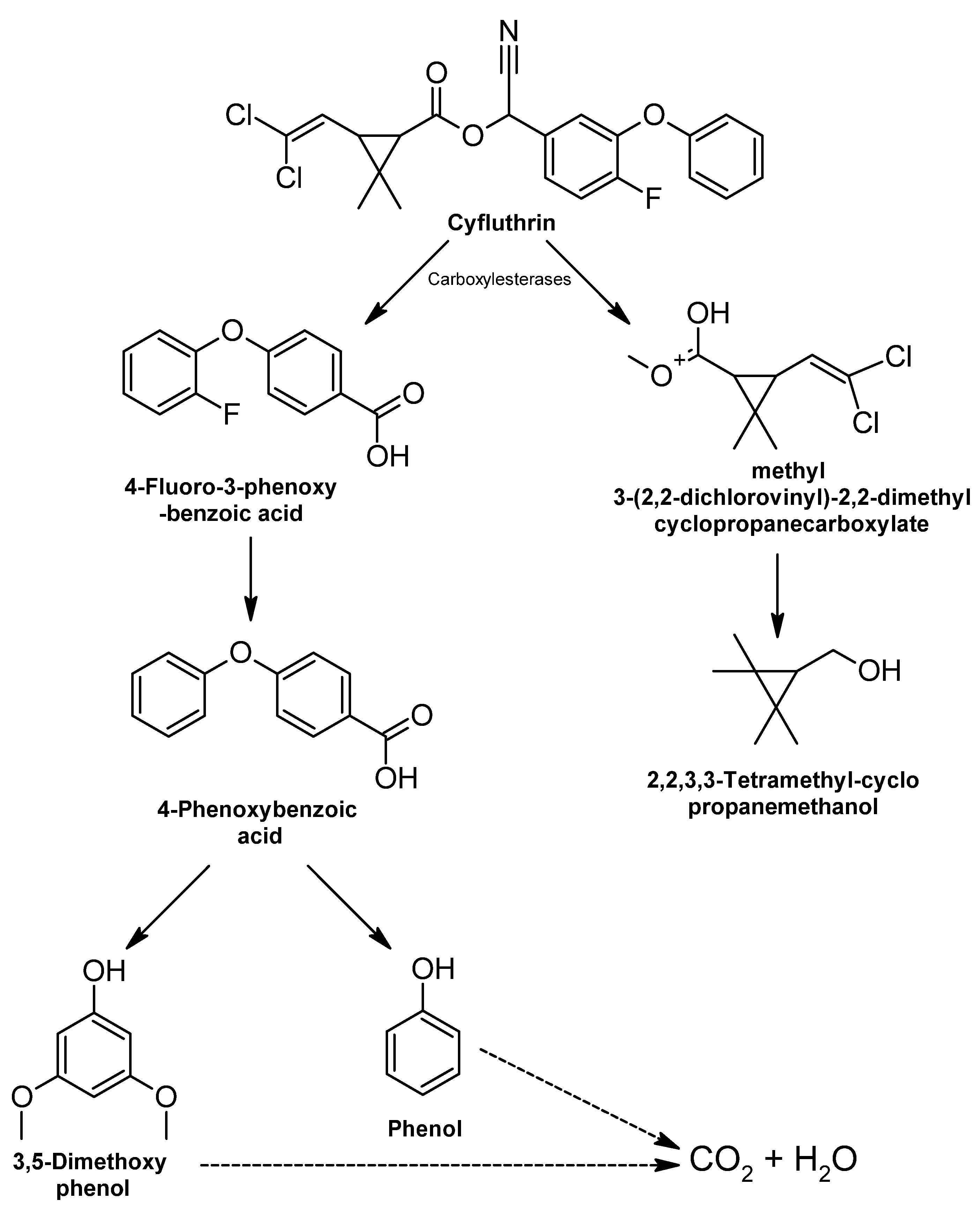


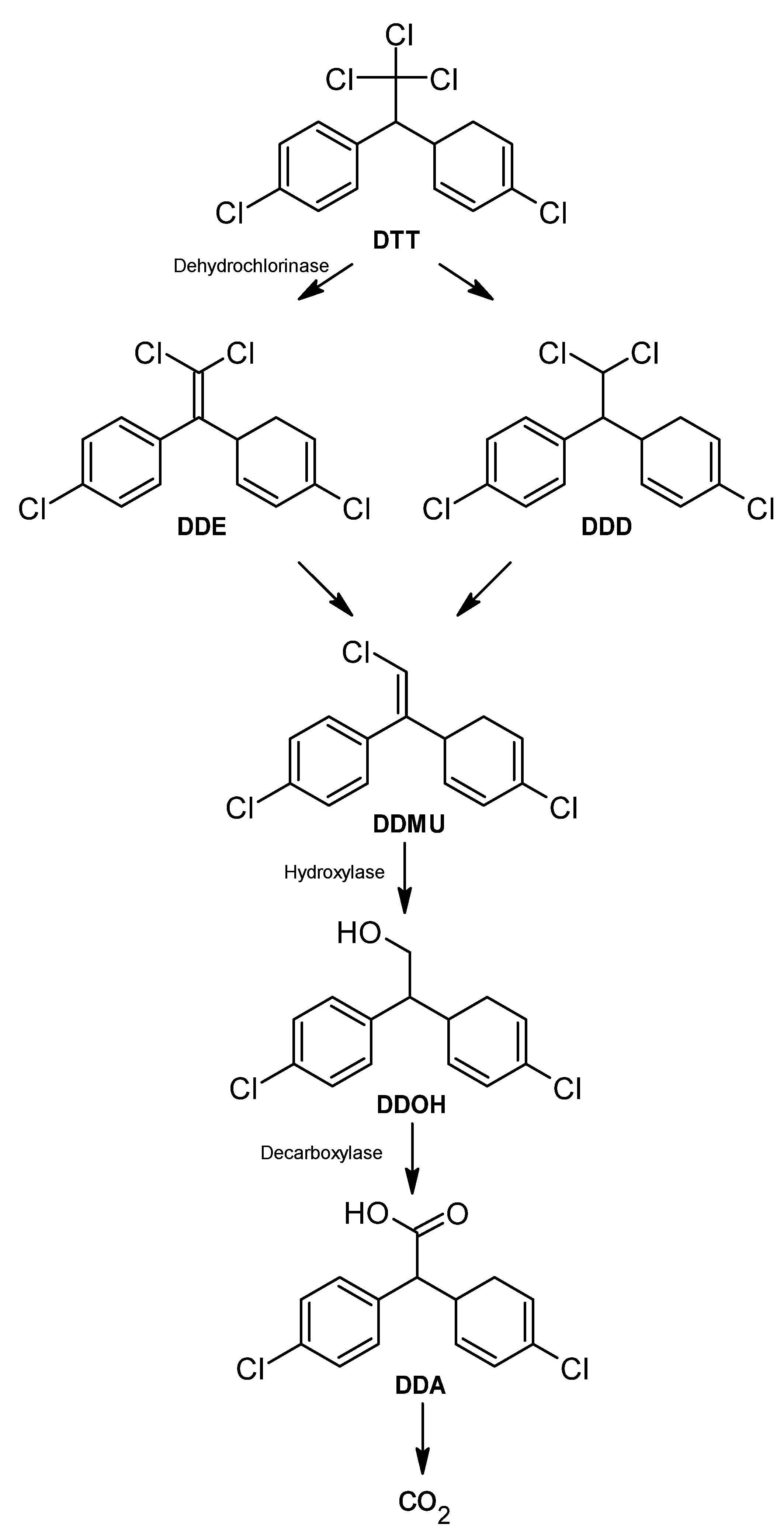
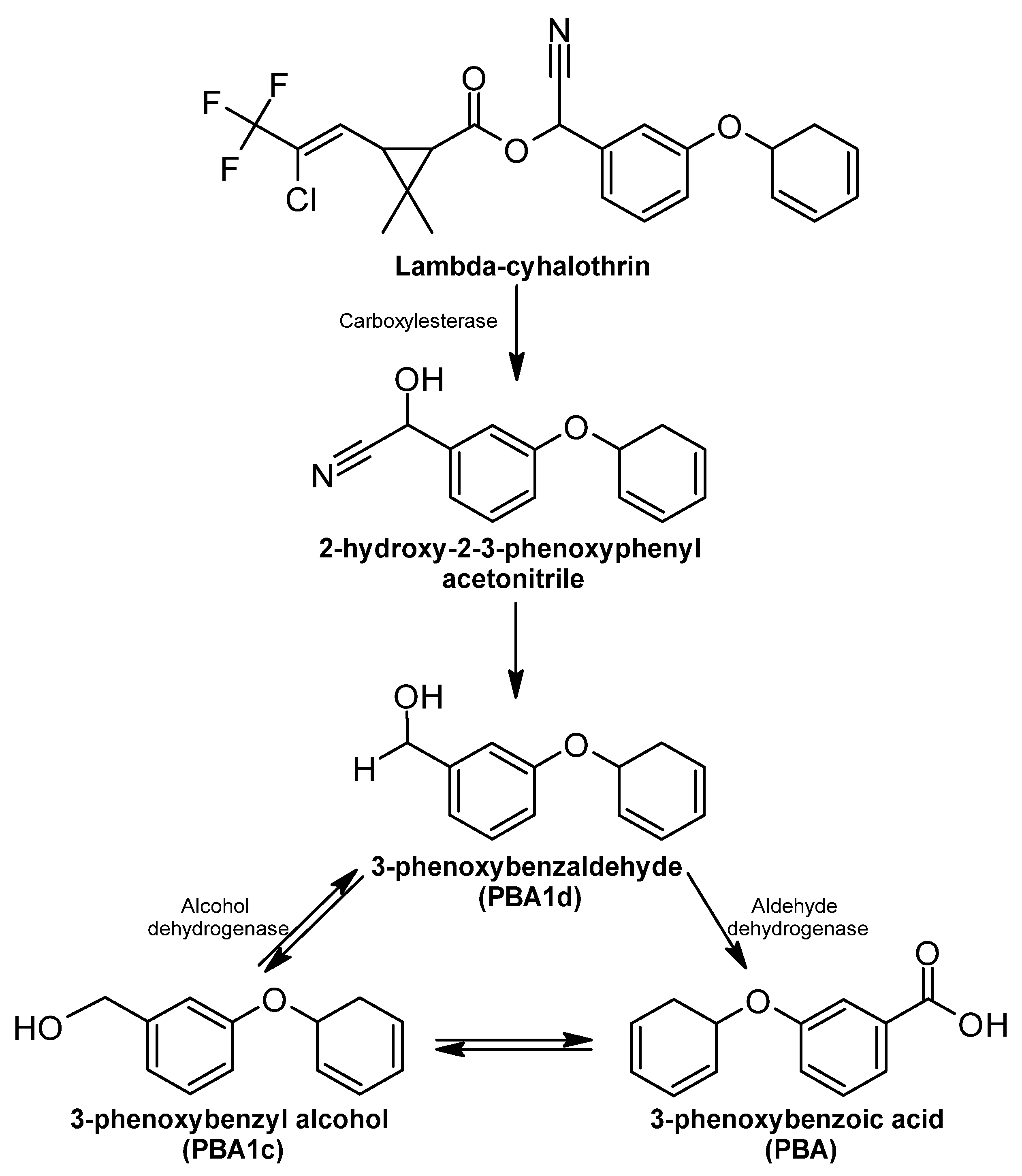
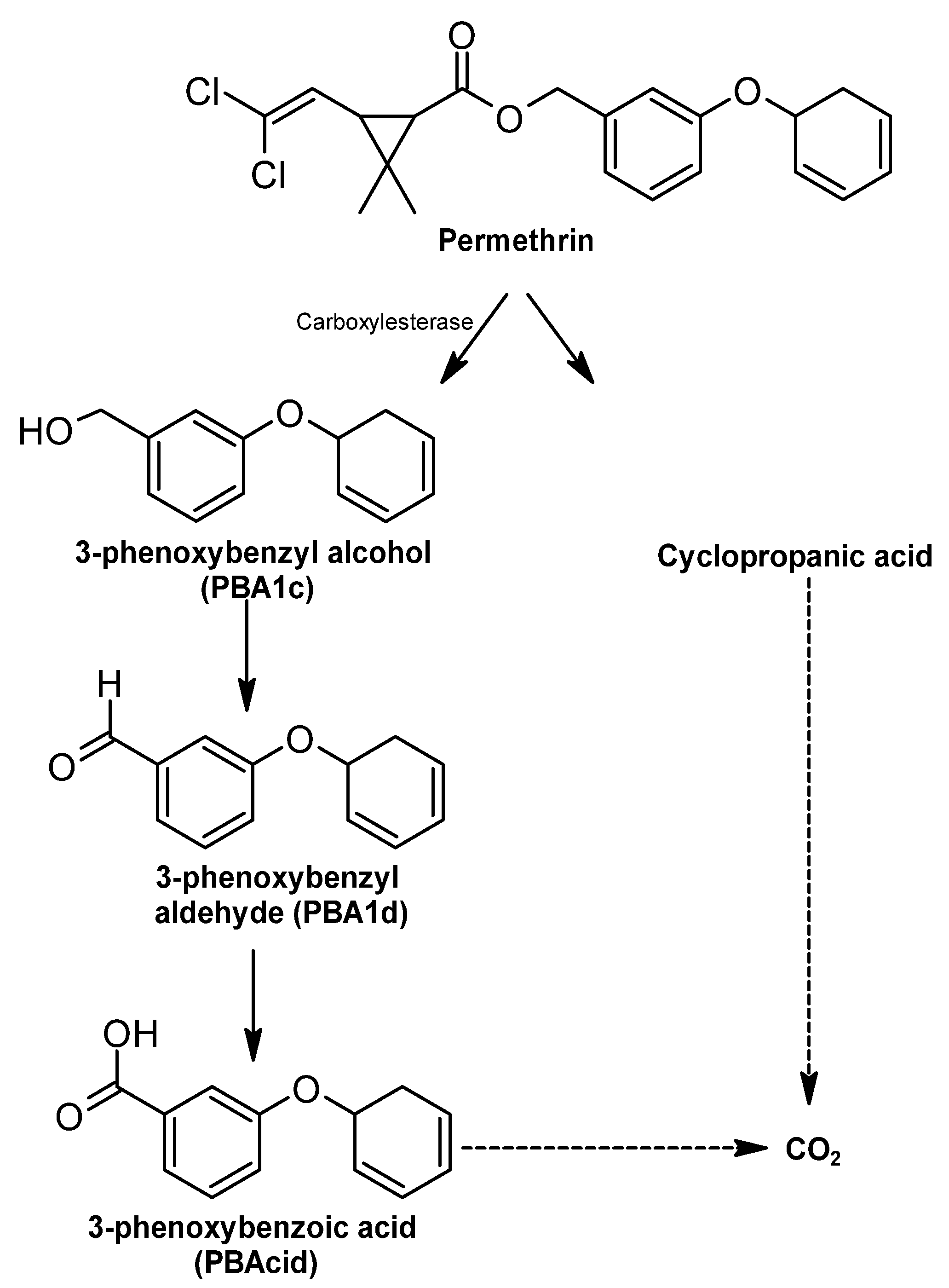
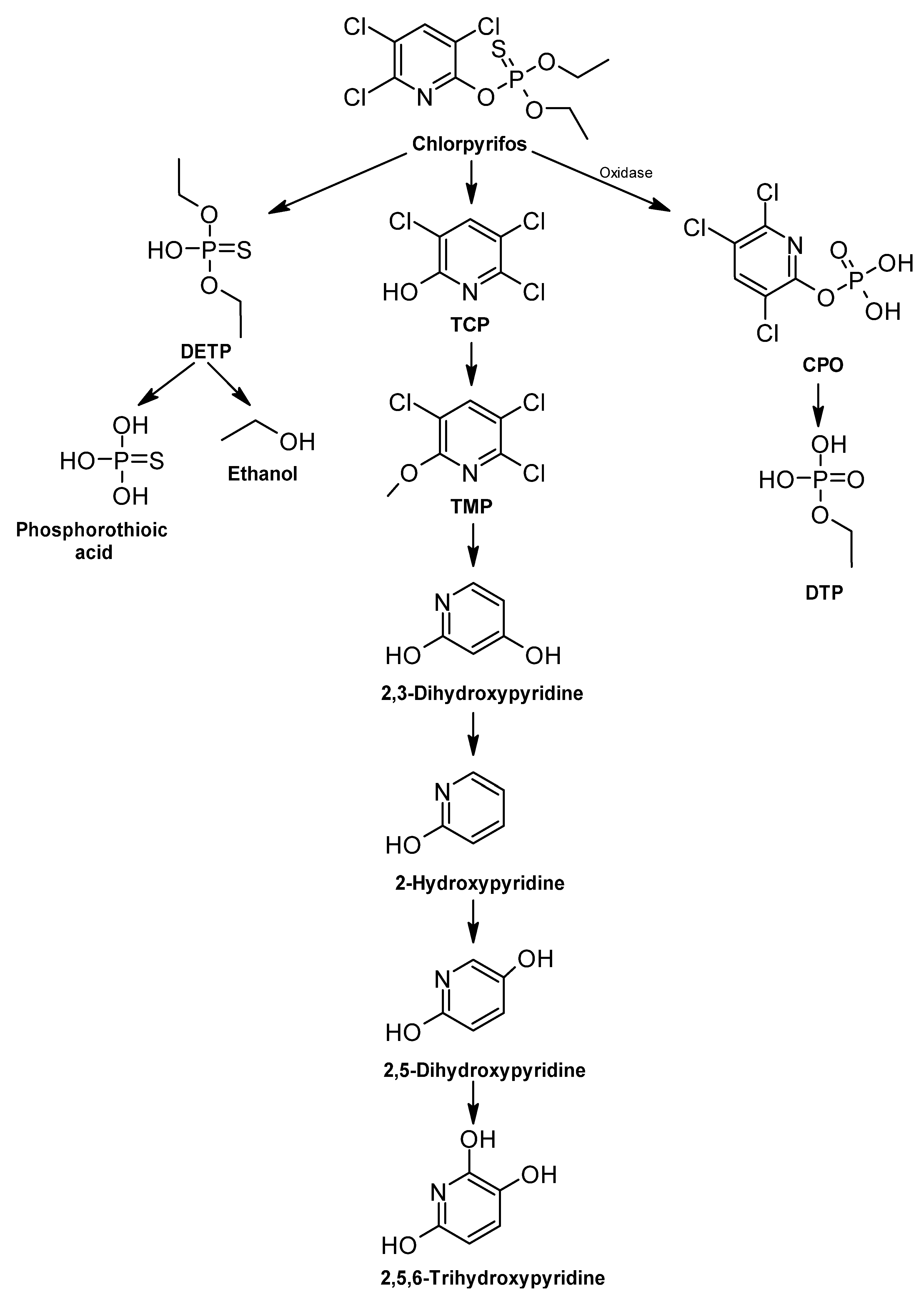
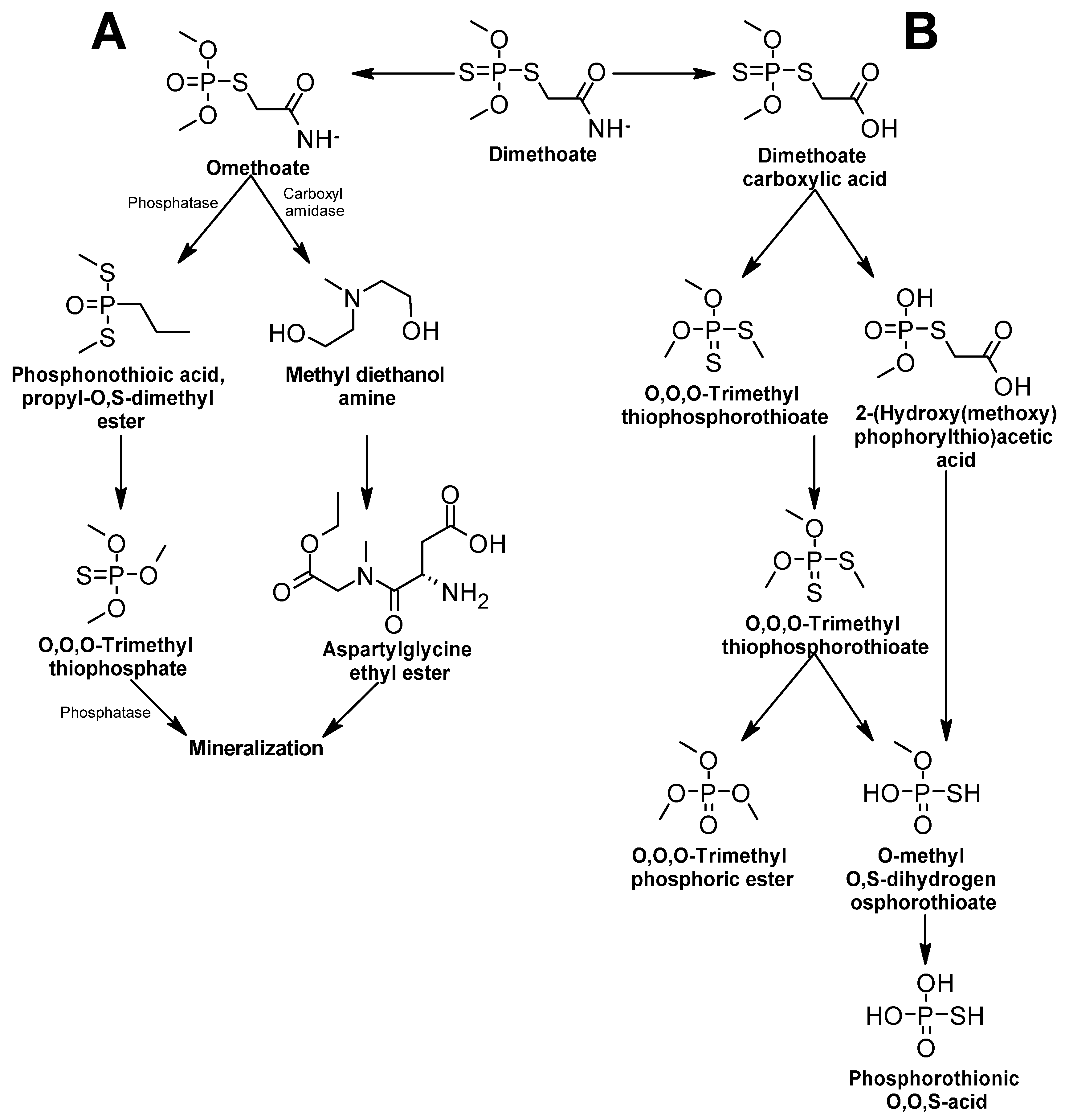
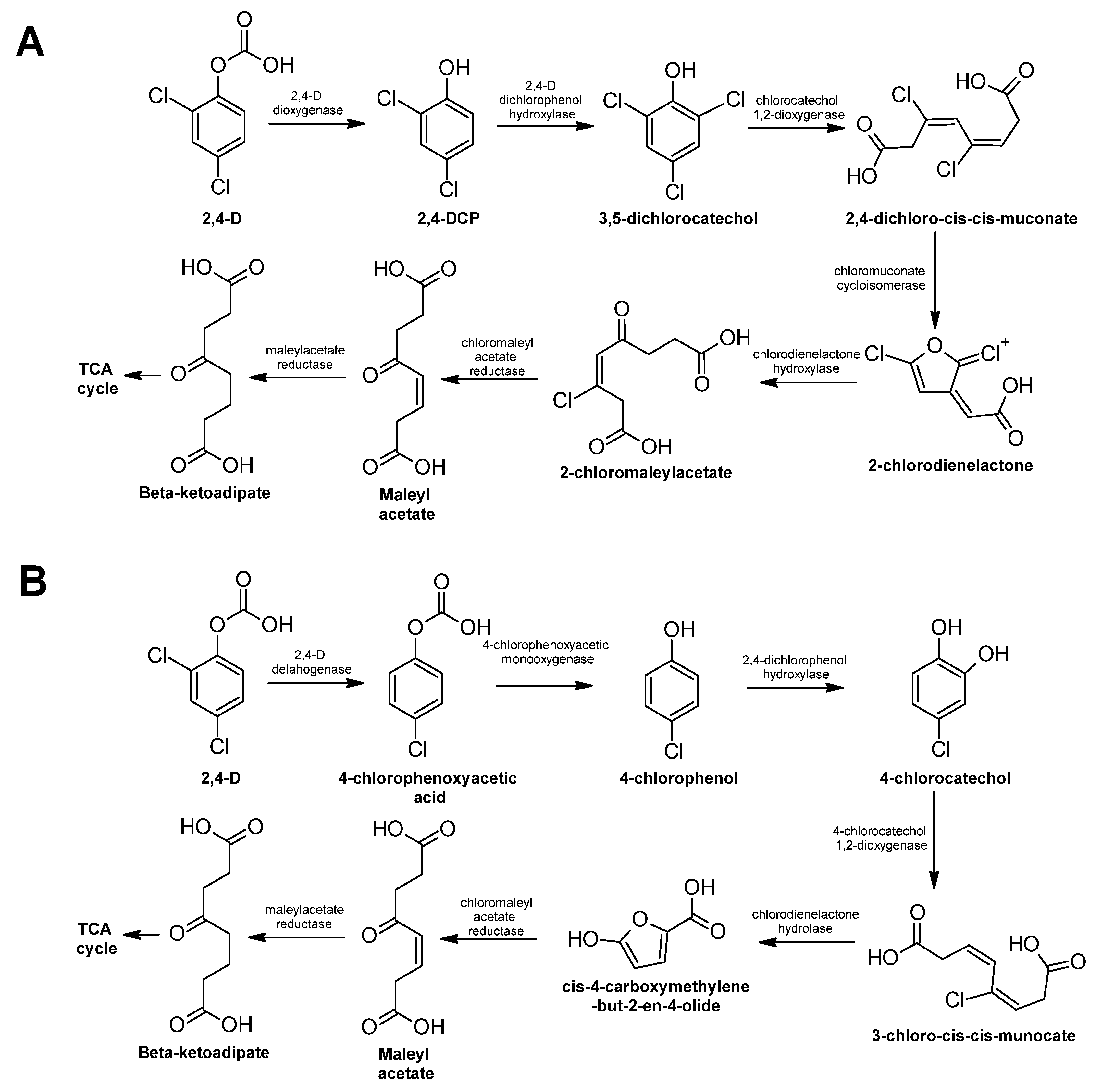

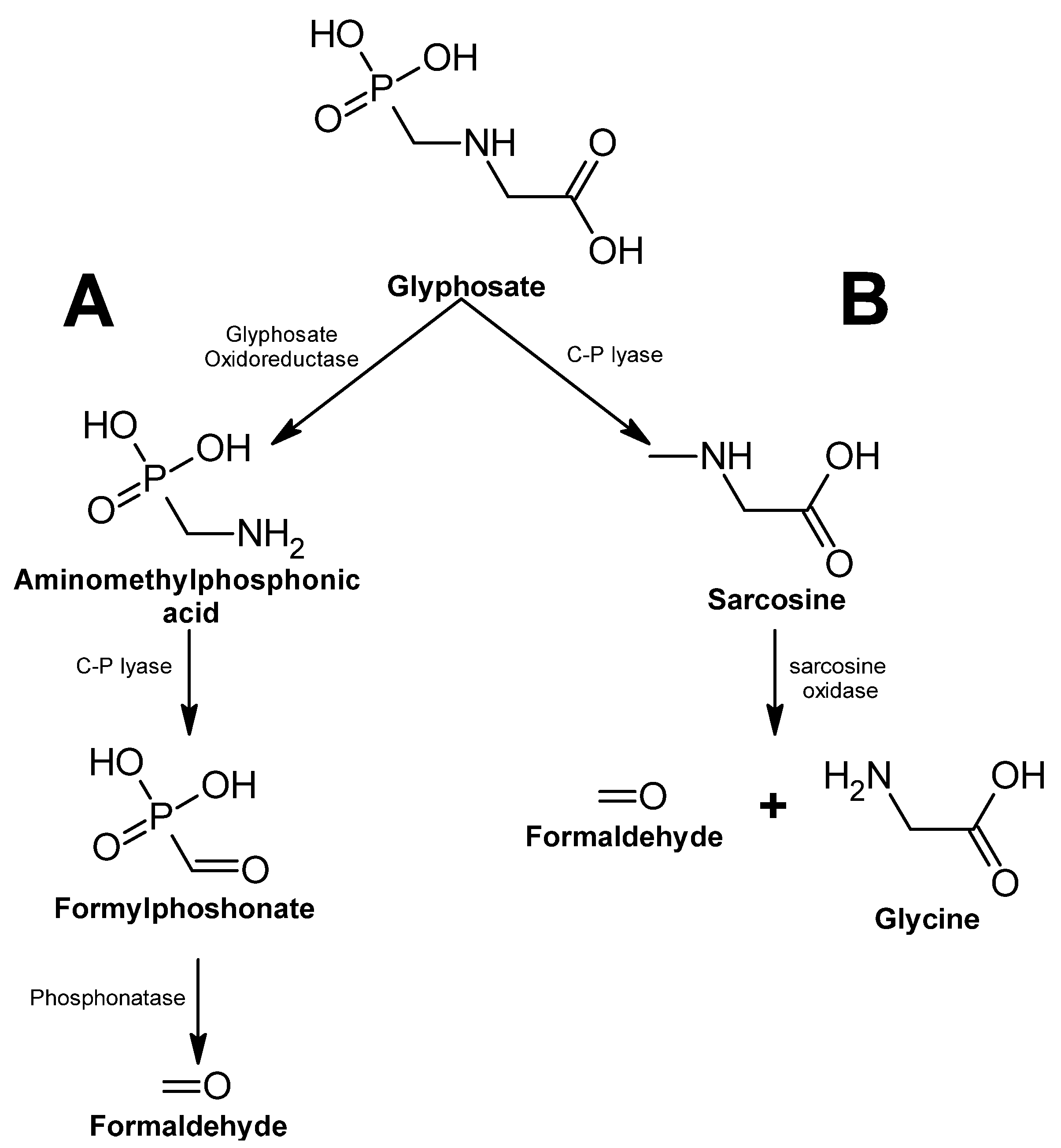
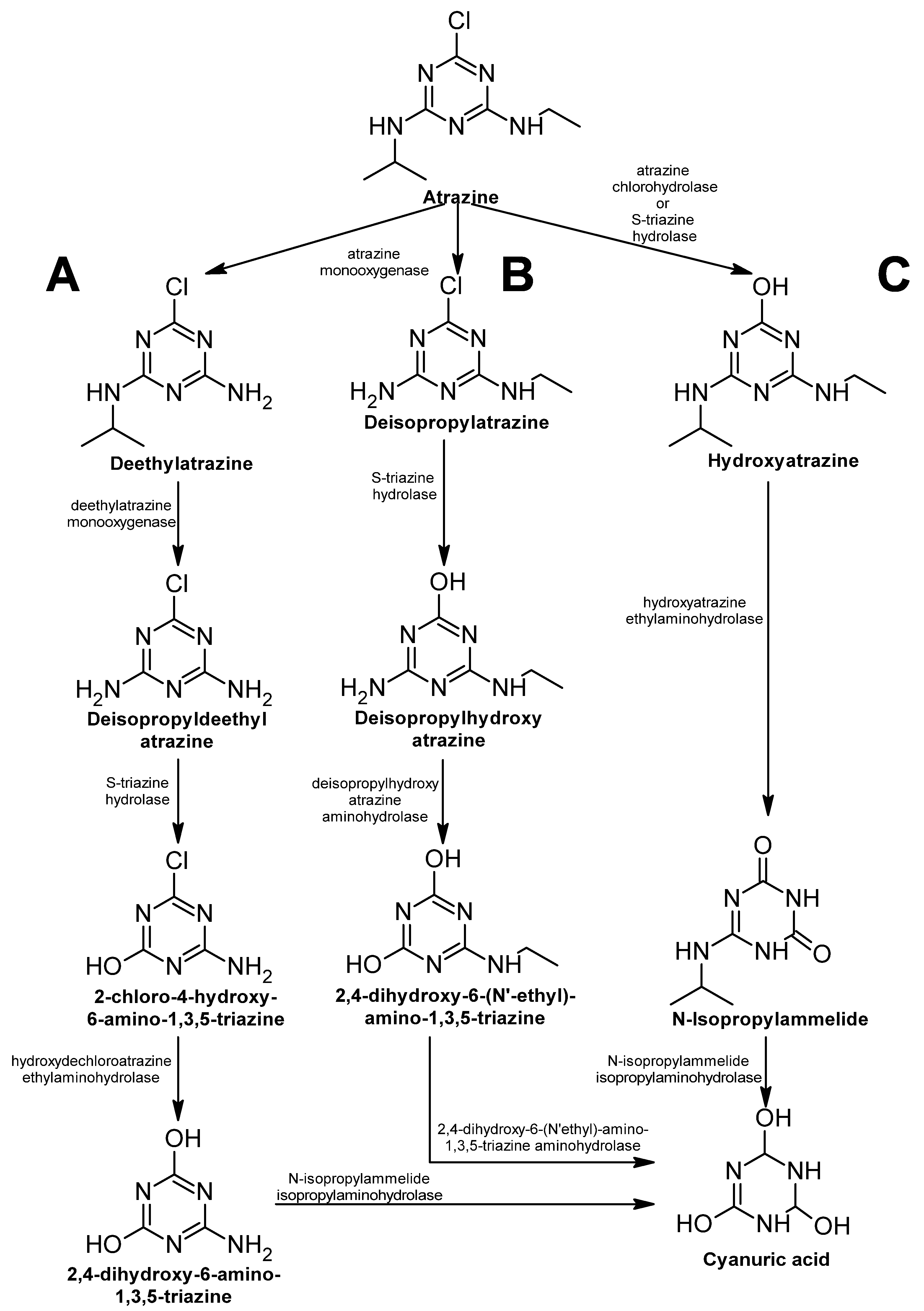
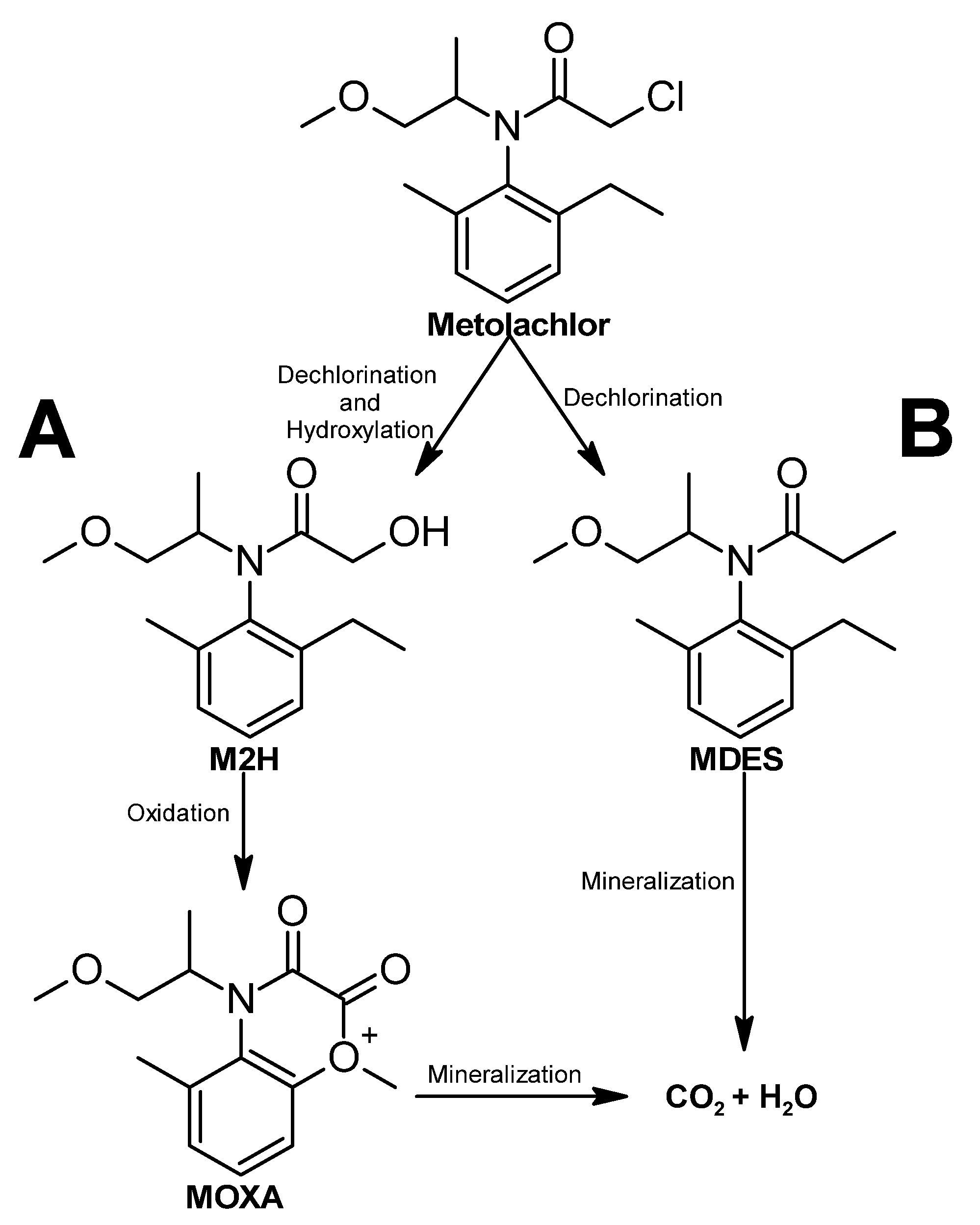
| Class | Oral LD50 for the Rat (mg/kg Body Weight) | Dermal LD50 for the Rat (mg/kg Body Weight) |
|---|---|---|
| Ia: Extremely hazardous | <5 | <50 |
| Ib: Highly hazardous | 5–50 | 50–200 |
| II: Moderately hazardous | 50–2000 | 200–2000 |
| III: Slightly hazardous | Over 2000 | |
| U: Unlikely to present acute hazard | 5000 or higher | |
| Substance | LD50 in Rats (mg/kg Body Weight) | Reference |
|---|---|---|
| Aldicarb (Carbamate) | 0.46–0.93 | [12] |
| Terbufos (Organophosphate) | 1.6–4.5 | [13] |
| Methyl Parathion (Organophosphate) | 6.9 | [14] |
| Substance | LD50 in Rats (mg/kg Body Weight) | Reference |
|---|---|---|
| Cyfluthrin (Pyrethroid) | 900 | [21] |
| Tefluthrin (Pyrethroid) | 21.8 | [22] |
| Carbofuran (Carbamate) | 7 | [22] |
| Substance | LD50 in Rats (mg/kg Body Weight) | Reference |
|---|---|---|
| DDT (Organochlorine) | 113–118 | [29] |
| λ-cyhalothrin (Pyrethroid) | 612 | [30] |
| Permethrin (Pyrethroid) | 430–4000 | [31] |
| Chlorpyrifos (Organophosphate) | 135 | [32] |
| Dimethoate (Organophosphate) | 245 | [33] |
| 2,4-D (Organochlorine) | 375 | [34] |
| Dicamba (Organochlorine) | 1581 | [35] |
| Cyanazine (Organochlorine) | 140–750 | [36] |
| Substance | LD50 in Rats (mg/kg Body Weight) | Reference |
|---|---|---|
| Glyphosate (Organophosphate) | 7203.58–7397.25 | [52] |
| Atrazine (Organochlorine) | 1869 | [53] |
| Metolachlor (Organochlorine) | 1500 | [54] |
| Substance | LD50 in Rats (mg/kg Body Weight) | Reference |
|---|---|---|
| Trifluralin (Organophosphate) | 10,000 | [59] |
| WHO Pesticide Classification | Pesticide (% of Biodegradation Rate) | Bacteria | Reference |
|---|---|---|---|
| Extremely Hazardous | Aldicarb (85%) Terbufos (42%) Methyl parathion (7–90%) | Enterobacter cloacae TA7 Micrococcus arborescens Pseudomonas aeruginosa Brachybacterium sp. Salsuginibacillus kocurii Stenotrophomonas sp. YC-1 Flavobacterium sp. Ochrobactrum sp. B2 Agrobacterium sp. YW12 Fischerella sp. Serratia sp. DS001 Bacillus sp. CBMAI 1833 Bacillus cereus P5CNB Pseudomonas sp. Z1 Burkholderia zhejiangensis CEIB S4–3 Nodularia linckia Nostoc muscorum Oscilatoria animalis Phormidium foveolarum Burkholderia cenocepacia CEIB S5-2 Acinetobacter sp. Pseudomonas putida Bacillus sp. Citrobacter freundii Stenotrophomonas sp. Flavobacterium sp. Proteus vulgaris Pseudomonas sp. Acinetobacter sp. Klebsiella sp. Proteus sp. Microcystis novacekii Alcaligenes sp. SRG Serratia marcescens MEW06 | [20,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83] |
| Highly Hazardous | Cyfluthrin (80%) Tefluthrin 1 Carbofuran (97.5%) | Photobacterium ganghwense T14 Novosphingobium sp. KN65.2 Pseudomonas stutzeri S1 Lysinibacillus sphaericus FLQ-11-1 Brevibacterium aureum Cupriavidus sp. ISTL7 Enterobacter cloacae TA7 Bacillus sp. Chryseobacterium sp. BSC2-3 Burkholderia cepacia PCL3 | [67,84,85,86,87,88,89,90,91,92] |
| Moderately Hazardous | DDT (5–98%) Lambda- Cyhalothrin (70–90%) Permethrin (80–100%) Chlorpyrifos (60–90%) Dimethoate (80–98%) 2,4-D (30–90%) Dicamba 1 Cyanazine 1 | Alcaligenes faecalis strain DSP3 Pseudomonas nitroreducens AR-3 Ralstonia pickettii Stenotrophomonas sp. Pseudomonas aeruginosa Ochrobactrum sp. DDT-2 Alcaligenes sp. KK Arthrobacter globiformis DC-1 Serratia marcescens NCIM 2919 Advenella kashmirensis Corynebacterium sp. Enterobacter cloacae Bacillus thuringiensis ZS-19 Bacillus velezensis sd Bacillus subtilis Paracoccus acridae SCU-M53 Brucella intermedia Halo1 Alcaligenes faecalis CH1S Aquamicrobium terrae CH1T Enterobacter ludwigii Bacillus thuringiensis Berliner Acinetobacter baumannii ZH-14 Hortaea sp. B15 Streptomyces sp. Enterobacter sp. SWLC2 Pseudomonas putida CBF10-2 Ochrobactrum anthropi FRAF13 Rhizobium radiobacter GHKF11 Methylobacterium extorquens Bacillus cereus Ct3 Stenotrophomonas maltophilia Acinetobacter calcoaceticus Bacillus amyloliquefaciens CP28 Pseudomonas putida T7 Pseudomonas aeruginosa M2 Klebsiella pneumoniae M6 Alcaligenes sp. Bacillus subtilis Enterobacter sp. Klebsiella sp. Micrococci sp. Cupriavidus nantongensis X1T Bacillus megaterium CCLP1 Bacillus safensis CCLP2 Shewanella sp. BT05 Pseudomonas fluorescens CD5 Achromobacter spanius C1 Pseudomonas rhodesiae C4 Weissella confusa Azotobacter vinelandii ATCC 12837 Coleofasciculus chthonoplastes Lysinibacillus sp. HBUM206408 Achromobacter insuavis Dyadobacter jiangsuensis 12851 Arthrobacter sp. HM01 Psychrobacter alimentarius T14 Streptomyces phaeochromogenes Streptomyces praecox SP1 Pseudomonas putida Xanthomonas campestris pv. Translucens Pseudomonas kilonensis MB490 Serratia sp. (100%) Sphingomonas sp. DC-6 Raoultella sp. X1 Lactobacillus plantarum Chryseobacterium Variovorax Aeromonas Xanthobacter Acidovorax Cupriavidus gilardii T-1 Enterobacter hormaechei subsp. xiangfangensis 19_357_F Cupriavidus campinensis Delftia sp. Cupriavidus necator Arthrobacter sp. SVMIICT25 Sphingomonas sp. SVMIICT11 Stenotrophomonas sp. SVMIICT13 Corynebacterium humireducens MFC-5 Cupriavidus oxalaticus X32 Thauera sp. DKT Pseudomonas simiae EGD-AQ6 Sphingobium sp. Ndbn-10 Sphingomonas sp. Ndbn-20 Pseudomonas maltophilia DI-6 Rhizorhabdus dicambivorans Ndbn-20 | [37,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155] |
| Slightly Hazardous | Glyphosate (30–90%) Atrazine (40–100%) Metolachlor (40–100%) | Rhodococcus soli G41 Stenotrophomonas maltophilia GP-1 Achromobacter sp. MPK7A Bacillus cereus Burkholderia vietnamiensis AO5–12 Burkholderia sp. AO5–13 Enterobacter cloacae K7 Ochrobacterium anthropic GPK3 Pseudomonas sp. Agrobacterium tumefaciens CNI28 Novosphingobium sp. CNI35 Ochrobactrum pituitosum CNI52 Gallinifaecis sp. CAS4 Chryseobacterium sp. Y16C Spirulina platensis Streptomyces lusitanus Lysinibacillus sphaericus Stenotrophomonas acidaminiphila Y4B Bradyrhizobium spp. Comamonas odontotermitis P2 Bacillus subtilis Rhizobium leguminosarum Serratia sp. Bacillus aryabhattai FACU Pseudomonas fluorescens Providencia rettgeri GDB 1 Bacillus cereus Pseudomonas alcaligenes Pseudomonas stutzeri Bacillus licheniformis Lactobacillus plantarum Lactobacillus rhamnosus Bacillus shackletonii Pseudomonas citronellolis ADA-23B Solicoccozyma terricola M 3.1.4. Achromobacter denitrificans Ochrobactrum haematophilum Pseudomonas putida Ch2 Ochrobactrum intermedium Sq20 Burkholderia cepacia PSBB1 Pseudomonas aeruginosa Ensifer adhaerens SZMC 25856 Pseudomonas resinovorans SZMC 25875 Burkholderia anthina Burkholderia cenocepacia Geobacillus caldoxylosilyticus T20 Bacillus licheniformis ATLJ-5. Bacillus megaterium ATLJ-11 Pseudomonas sp. Pseudaminobacter sp. Nocardioides sp. Klebsiella sp. A1 Comamonas sp. A2 Klebsiella variicola FH-1 Arthrobacter sp. NJ-1 Acinetobacter lwoffii DNS32 Agrobacterium rhizogenes AT13 Acetobacter Pseudomonas Clostridium-sensu-stricto Burkholderia Ensifer sp. Solibacillus sp. Bacillus sp. Arthrobacter sp. Bacillus velezensis MHNK1 Citricoccus sp. TT3 Paenarthrobacter sp. W11 Methylobacillus Enterobacter sp. P1 Arthrobacter sp. DNS10 Bradyrhizobium Rhodococcus Hydrogenophaga sp. Gsoil 1545 Sinorhizobium sp. TJ170 Rhizobium sp. Bacillus anthracis Pseudomonas balearica Pseudomonas otitidis Pseudomonas indica Providencia vermicola Pseudomonas spp. ACB Pseudomonas spp. TLB Methylobacterium Mycobacterium Bacillus atrophaeus Variovorax sp. 38R Arthrobacter sp. TES Chelatobacter sp. SR38 Bacillus megaterium Mes11 Ralstonia Phyllobacterium Stenotrophomonas Holophaga foetida | [55,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208] |
| Unlikely Hazardous | Trifuralin (30–95%) | Arthrobacter aurescens CTFL7 Herbaspirillum sp. Klebsiella sp. Pseudomonas fluorescens Bacillus simplex Bacillus muralis Micrococcus luteus Micrococcus yunnanensis Clostridium tetani Klebsiella oxytoca Herbaspirillum seropedicae Bacillus megaterium Brevundimonas diminuta Streptomyces PS1/5 | [123,209,210,211,212,213,214] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerrero Ramírez, J.R.; Ibarra Muñoz, L.A.; Balagurusamy, N.; Frías Ramírez, J.E.; Alfaro Hernández, L.; Carrillo Campos, J. Microbiology and Biochemistry of Pesticides Biodegradation. Int. J. Mol. Sci. 2023, 24, 15969. https://doi.org/10.3390/ijms242115969
Guerrero Ramírez JR, Ibarra Muñoz LA, Balagurusamy N, Frías Ramírez JE, Alfaro Hernández L, Carrillo Campos J. Microbiology and Biochemistry of Pesticides Biodegradation. International Journal of Molecular Sciences. 2023; 24(21):15969. https://doi.org/10.3390/ijms242115969
Chicago/Turabian StyleGuerrero Ramírez, José Roberto, Lizbeth Alejandra Ibarra Muñoz, Nagamani Balagurusamy, José Ernesto Frías Ramírez, Leticia Alfaro Hernández, and Javier Carrillo Campos. 2023. "Microbiology and Biochemistry of Pesticides Biodegradation" International Journal of Molecular Sciences 24, no. 21: 15969. https://doi.org/10.3390/ijms242115969
APA StyleGuerrero Ramírez, J. R., Ibarra Muñoz, L. A., Balagurusamy, N., Frías Ramírez, J. E., Alfaro Hernández, L., & Carrillo Campos, J. (2023). Microbiology and Biochemistry of Pesticides Biodegradation. International Journal of Molecular Sciences, 24(21), 15969. https://doi.org/10.3390/ijms242115969






