Heterogeneity of Phase II Enzyme Ligands on Controlling the Progression of Human Gastric Cancer Organoids as Stem Cell Therapy Model
Abstract
:1. Introduction
2. Results
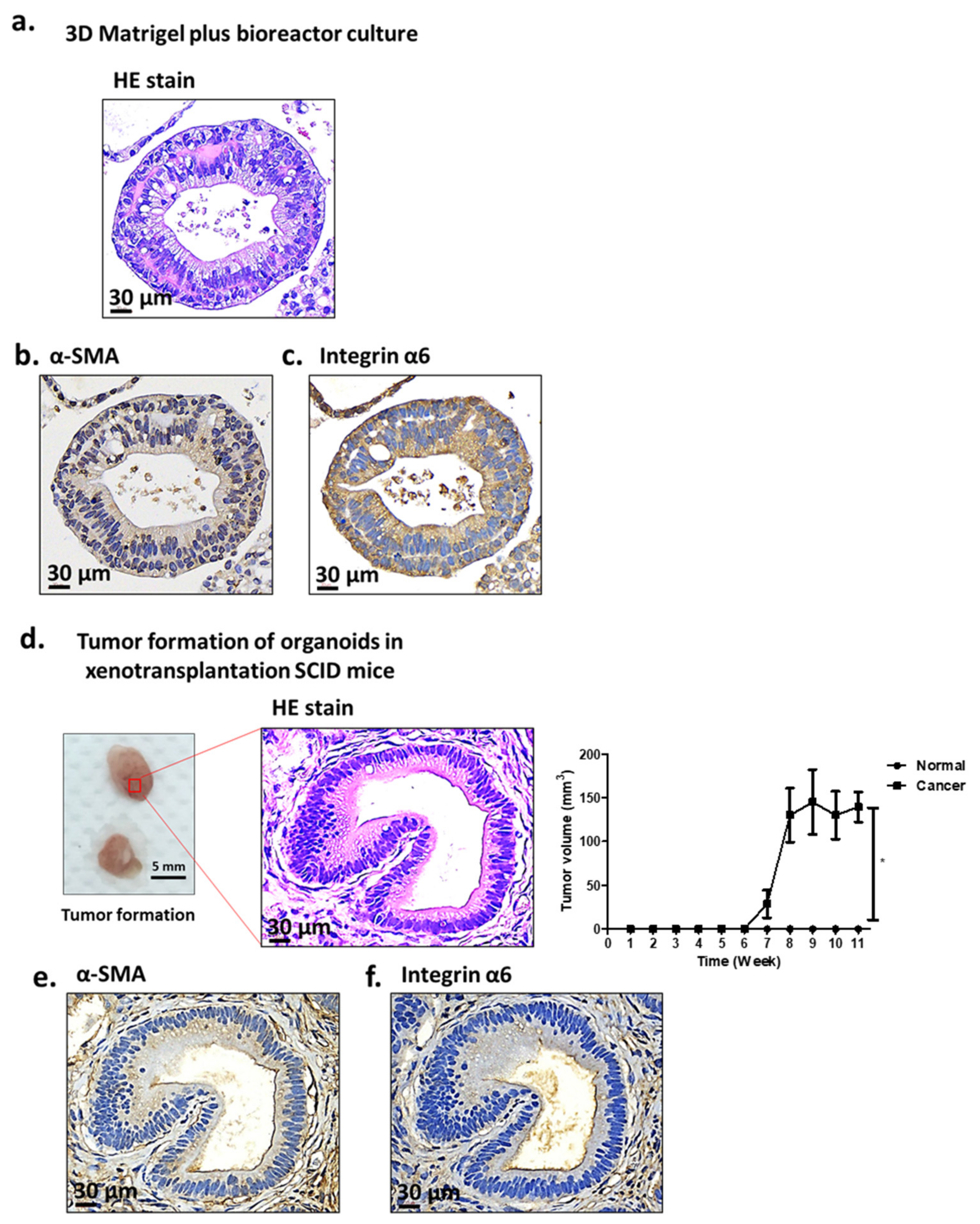
2.1. Characterization of Antrum Organoids of Patients with GC
2.2. Effect of Antioxidant Drugs on Organoid Growth
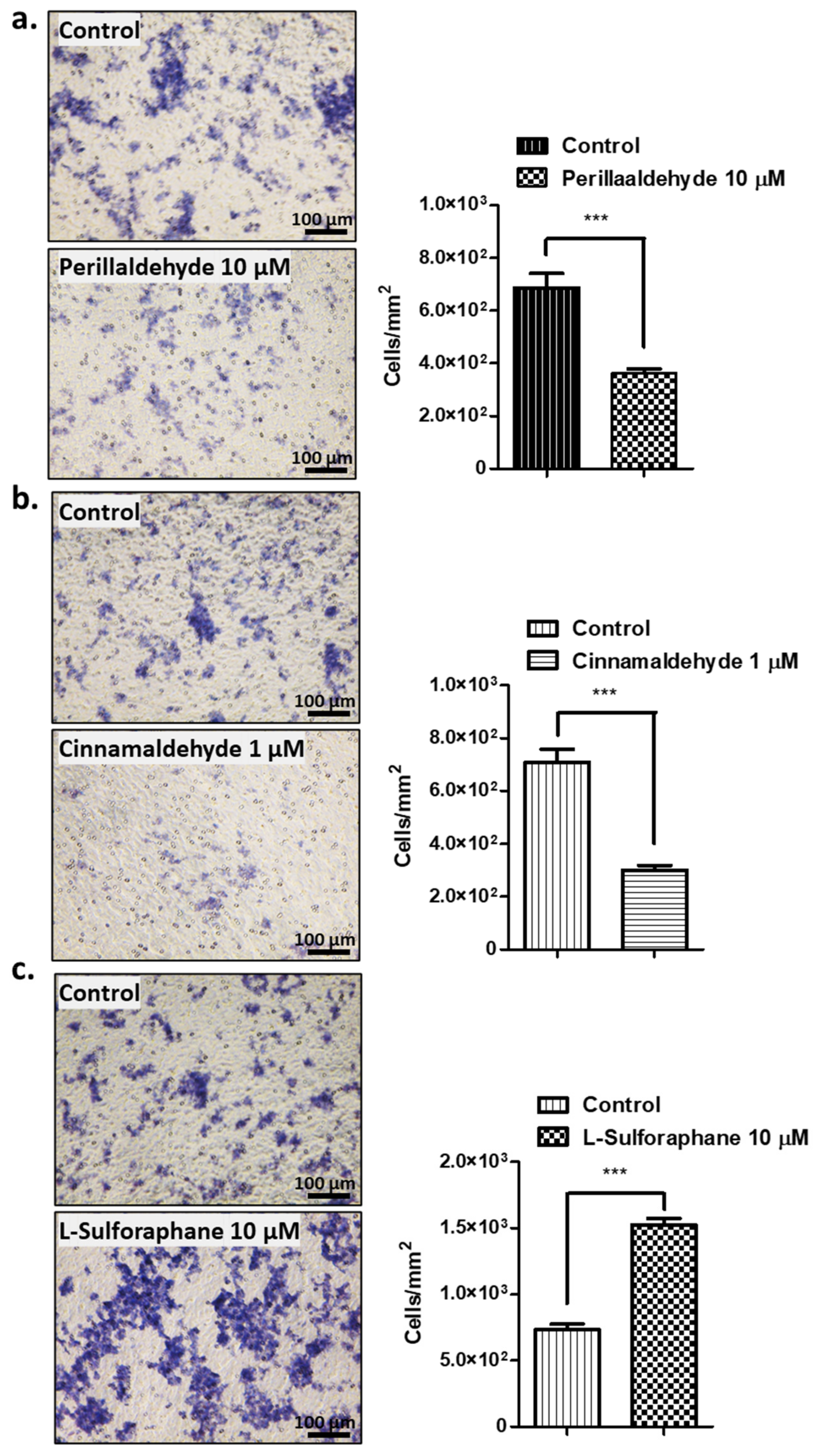
2.3. Effect of Antioxidant drugs on Invasive Activities
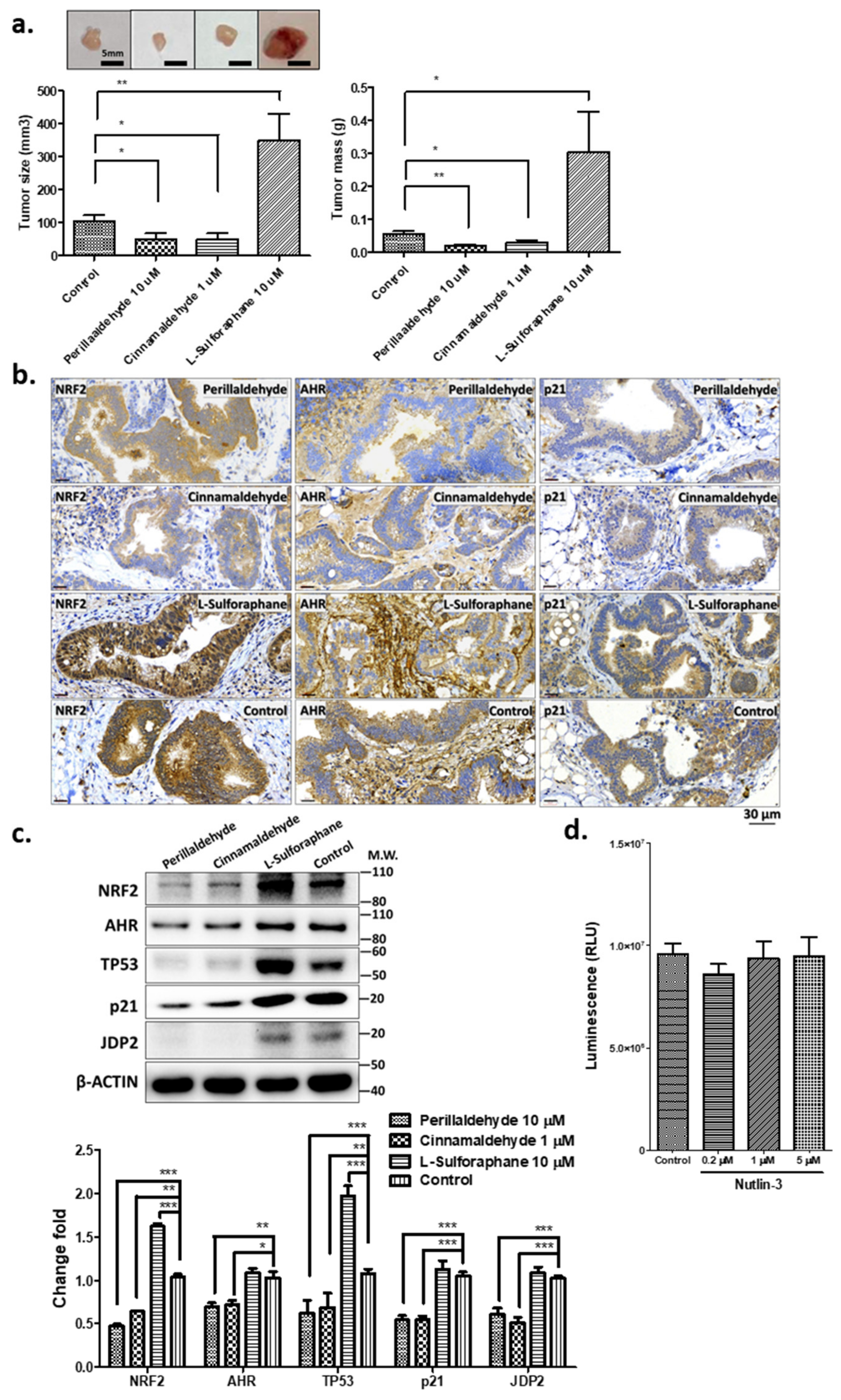
2.4. PEA and CA Repress the Organoid-Mediated Tumor Formation, but SFN Increases Tumor Development
2.5. Expression of Stress-Related Factors and TP53
2.6. PEA and CA Decrease ROS and Caspase 3/7 Activities, but SFN Increases Them
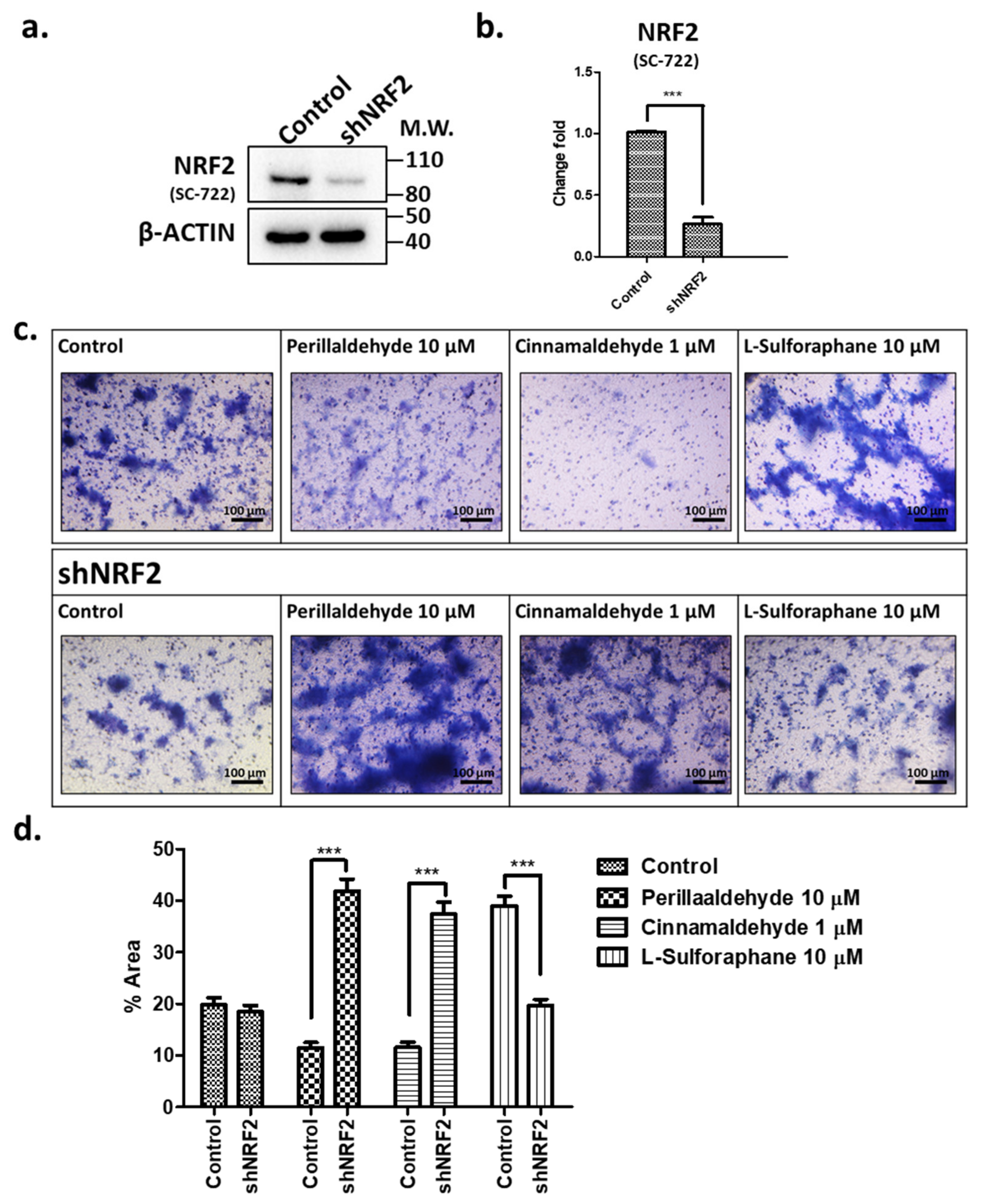
2.7. Effects of shRNA against NRF2 on Invasive Activities in Response to PEA, CA, and SFN
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Organoid Preparation and Chemicals
4.3. Bioreactor Culture
4.4. Immunohistochemistry
4.5. Preparation of shNRF2 and shRNA-Mediated Knockdown
4.6. Measurement of Cellular ROS Levels
4.7. Measurement of Viable Cells
4.8. Measurement of Caspase-Glo 3/7 Activities
4.9. Invasion Assay
4.10. Xenograft Injection
4.11. Western Blotting
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fitzmaurice, C.; Dicker, D.; Pain, A.; Hamavid, H.; Moradi-Lakeh, M.; MacIntyre, M.F.; Allen, C.; Hansen, G.; Woodbrook, R.; Wolfe, C.; et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015, 1, 505–527. [Google Scholar] [CrossRef] [PubMed]
- Cancer Registry Annual Report: 2021 Taiwan Health and Welfare Report; Department of Health, Ministry of Health and Welfare: Taipei, Taiwan, 2022.
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Hunt, R.H.; Camilleri, M.; Crowe, S.E.; El-Omar, E.M.; Fox, J.G.; Kuipers, E.J.; Malfertheiner, P.; McColl, K.E.; Pritchard, D.M.; Rugge, M.; et al. The stomach in health and disease. Gut 2015, 64, 1650–1668. [Google Scholar] [CrossRef] [PubMed]
- Ychou, M.; Boige, V.; Pignon, J.P.; Conroy, T.; Bouché, O.; Lebreton, G.; Ducourtieux, M.; Bedenne, L.; Fabre, J.M.; Saint-Aubert, B.; et al. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: An FNCLCC and FFCD multicenter phase III trial. J. Clin. Oncol. 2011, 29, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Al-Batran, S.E.; Hofheinz, R.D.; Pauligk, C.; Kopp, H.G.; Haag, G.M.; Luley, K.B.; Meiler, J.; Homann, N.; Lorenzen, S.; Schmalenberg, H.; et al. Histopathological regression after neoadjuvant docetaxel, oxaliplatin, fluorouracil, and leucovorin versus epirubicin, cisplatin, and fluorouracil or capecitabine in patients with resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4-AIO): Results from the phase 2 part of a multicentre, open-label, randomised phase 2/3 trial. Lancet Oncol. 2016, 17, 1697–1708. [Google Scholar] [CrossRef]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; Dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Barker, N.; Huch, M.; Kujala, P.; van de Wetering, M.; Snippert, H.J.; van Es, J.H.; Sato, T.; Stange, D.E.; Begthel, H.; van den Born, M.; et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010, 6, 25–36. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Stange, D.E.; Koo, B.K.; Huch, M.; Sibbel, G.; Basak, O.; Lyubimova, A.; Kujala, P.; Bartfeld, S.; Koster, J.; Geahlen, J.H.; et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 2013, 155, 357–368. [Google Scholar] [CrossRef]
- Koo, B.K.; Stange, D.E.; Sato, T.; Karthaus, W.; Farin, H.F.; Huch, M.; van Es, J.H.; Clevers, H. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods 2011, 9, 81–83. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Seidlitz, T.; Merker, S.R.; Rothe, A.; Zakrzewski, F.; von Neubeck, C.; Grützmann, K.; Sommer, U.; Schweitzer, C.; Schölch, S.; Uhlemann, H.; et al. Human gastric cancer modelling using organoids. Gut 2019, 68, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Ahmed, H.M. Ethnomedicinal, Phytochemical and Pharmacological Investigations of Perilla frutescens (L.) Britt. Molecules 2018, 24, 102. [Google Scholar] [CrossRef]
- Ji, W.W.; Wang, S.Y.; Ma, Z.Q.; Li, R.P.; Li, S.S.; Xue, J.S.; Li, W.; Niu, X.X.; Yan, L.; Zhang, X.; et al. Effects of perillaldehyde on alternations in serum cytokines and depressive-like behavior in mice after lipopolysaccharide administration. Pharmacol. Biochem. Behav. 2014, 116, 1–8. [Google Scholar] [CrossRef]
- Song, Y.; Sun, R.; Ji, Z.; Li, X.; Fu, Q.; Ma, S. Perilla aldehyde attenuates CUMS-induced depressive-like behaviors via regulating TXNIP/TRX/NLRP3 pathway in rats. Life Sci. 2018, 206, 117–124. [Google Scholar] [CrossRef]
- Uemura, T.; Yashiro, T.; Oda, R.; Shioya, N.; Nakajima, T.; Hachisu, M.; Kobayashi, S.; Nishiyama, C.; Arimura, G.I. Intestinal Anti-Inflammatory Activity of Perillaldehyde. J. Agric. Food Chem. 2018, 66, 3443–3448. [Google Scholar] [CrossRef]
- Fuyuno, Y.; Uchi, H.; Yasumatsu, M.; Morino-Koga, S.; Tanaka, Y.; Mitoma, C.; Furue, M. Perillaldehyde Inhibits AHR Signaling and Activates NRF2 Antioxidant Pathway in Human Keratinocytes. Oxid. Med. Cell. Longev. 2018, 2018, 9524657. [Google Scholar] [CrossRef]
- Cabello, C.M.; Bair, W.B., 3rd; Lamore, S.D.; Ley, S.; Bause, A.S.; Azimian, S.; Wondrak, G.T. The cinnamon-derived Michael acceptor cinnamic aldehyde impairs melanoma cell proliferation, invasiveness, and tumor growth. Free Radic. Biol. Med. 2009, 46, 220–231. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Dong, L.; Wen, Y.; Zheng, X.; Zhang, C.; Chen, R.; Zhang, Y.; Li, Y.; He, T.; et al. Cinnamaldehyde inhibits inflammation and brain damage in a mouse model of permanent cerebral ischaemia. Br. J. Pharmacol. 2015, 172, 5009–5023. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W. Cinnamaldehyde induces autophagy-mediated cell death through ER stress and epigenetic modification in gastric cancer cells. Acta Pharmacol. Sin. 2022, 43, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Uchi, H.; Yasumatsu, M.; Morino-Koga, S.; Mitoma, C.; Furue, M. Inhibition of aryl hydrocarbon receptor signaling and induction of NRF2-mediated antioxidant activity by cinnamaldehyde in human keratinocytes. J. Dermatol. Sci. 2017, 85, 36–43. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, H.; Dong, N.; Su, X.; Duan, M.; Wei, Y.; Wei, J.; Liu, G.; Peng, Q.; Zhao, Y. Sulforaphane induces S-phase arrest and apoptosis via p53-dependent manner in gastric cancer cells. Sci. Rep. 2021, 11, 2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Q.; Li, N.; Xu, M.; Miyamoto, T.; Liu, J. Sulforaphane suppresses metastasis of triple-negative breast cancer cells by targeting the RAF/MEK/ERK pathway. NPJ Breast Cancer 2022, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Silva-Palacios, A.; Ostolga-Chavarria, M.; Sanchez-Garibay, C.; Rojas-Marales, P.; Galvan-Arzate, S.; Buelna-Cotai, M.; Pavon, N.; Pedraza-Chaverri Konigsberg, M.; Zazueta, Z. Sulforaphane protects from myocardial ischemia-reperfusion damage through the balanced activation of Nrf2/AhR. Free. Radic. Biol. Med. 2019, 143, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Hu, S.; Jiang, P.; Zhang, Z.; Li, L.; Wu, Q. Effects of sulforaphane on breast cancer based on metabolome and microbiome. Food Sci. Nutr. 2023, 11, 2277–2287. [Google Scholar] [CrossRef]
- Broda, T.R.; McCracken, K.W.; Wells, J.M. Generation of human antral and fundic gastric organoids from pluripotent stem cells. Nat. Protoc. 2019, 14, 28–50. [Google Scholar] [CrossRef]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef]
- Wuputra, K.; Ku, C.C.; Pan, J.B.; Liu, C.J.; Kato, K.; Lin, Y.C.; Liu, Y.C.; Lin, C.S.; Hsiao, M.; Tai, M.H.; et al. Independent Signaling of Hepatoma Derived Growth Factor and Tumor Necrosis Factor-Alpha in Human Gastric Cancer Organoids Infected by Helicobacter pylori. Int. J. Mol. Sci. 2023, 24, 6567. [Google Scholar] [CrossRef]
- Przepiorski, A.; Sander, V.; Tran, T.; Hollywood, J.A.; Sorrenson, B.; Shih, J.H.; Wolvetang, E.J.; McMahon, A.P.; Holm, T.M.; Davidson, A.J. A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells. Stem Cell Rep. 2018, 11, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Nakamura, S.; Sugimoto, N.; Shigemori, T.; Kato, Y.; Ohno, M.; Sakuma, S.; Ito, K.; Kumon, H.; Hirose, H.; et al. Turbulence Activates Platelet Biogenesis to Enable Clinical Scale Ex Vivo Production. Cell 2018, 174, 636–648.e18. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Nanki, K.; Toshimitsu, K.; Takano, A.; Fujii, M.; Shimokawa, M.; Ohta, Y.; Matano, M.; Seino, T.; Nishikori, S.; Ishikawa, K.; et al. Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell 2018, 174, 856–869.e17. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef]
- Hira, V.V.; Breznik, B.; Van Noorden, C.J.; Lah, T.; Molenaar, R.J. 2D and 3D in vitro assays to quantify the invasive behavior of glioblastoma stem cells in response to SDF-1α. Biotechniques 2020, 69, 339–346. [Google Scholar] [CrossRef]
- Vinci, M.; Box, C.; Eccles, S.A. Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp. 2015, e52686. [Google Scholar] [CrossRef]
- Tovar, C.; Rosinski, J.; Filipovic, Z.; Higgins, B.; Kolinsky, K.; Hilton, H.; Zhao, X.; Vu, B.T.; Qing, W.; Packman, K.; et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc. Natl. Acad. Sci. USA 2006, 103, 1888–1893. [Google Scholar] [CrossRef]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, X.; Sun, K.H.; Zhou, L. α-smooth muscle actin is not a marker of fibrogenic cell activity in skeletal muscle fibrosis. PLoS ONE 2018, 13, e0191031. [Google Scholar] [CrossRef]
- Lipscomb, E.A.; Simpson, K.J.; Lyle, S.R.; Ring, J.E.; Dugan, A.S.; Mercurio, A.M. The alpha6beta4 integrin maintains the survival of human breast carcinoma cells in vivo. Cancer Res. 2005, 65, 10970–10976. [Google Scholar] [CrossRef] [PubMed]
- Faraonio, R.; Vergara, P.; Di Marzo, D.; Pierantoni, M.G.; Napolitano, M.; Russo, T.; Cimino, F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 2006, 281, 39776–39784. [Google Scholar] [CrossRef] [PubMed]
- Lisek, K.; Campaner, E.; Ciani, Y.; Walerych, D.; Del Sal, G. Mutant p53 tunes the NRF2-dependent antioxidant response to support survival of cancer cells. Oncotarget 2018, 9, 20508–20523. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, S.; Lee, C.H.; Lin, C.S.; Ku, C.C.; Hasegawa, H.; Qin, S.; Kawahara, A.; Korenori, Y.; Miyamori, K.; Noguchi, M.; et al. Jun dimerization protein 2 is a critical component of the Nrf2/MafK complex regulating the response to ROS homeostasis. Cell Death Dis. 2013, 4, e921. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jin, C.; Liu, Z.; Pan, J.; Li, H.; Zhang, Z.; Bi, S.; Yokoyama, K.K. Cloning and characterization of the mouse JDP2 gene promoter reveal negative regulation by p53. Biochem. Biophys. Res. Commun. 2014, 450, 1531–1536. [Google Scholar] [CrossRef]
- Ge, M.; Zhang, L.; Cao, L.; Xie, C.; Li, X.; Li, Y.; Meng, Y.; Chen, Y.; Wang, X.; Chen, J.; et al. Sulforaphane inhibits gastric cancer stem cells via suppressing sonic hedgehog pathway. Int. J. Food Sci. Nutr. 2019, 70, 570–578. [Google Scholar] [CrossRef]
- Li, S.; Khoi, P.N.; Yin, H.; Sah, D.K.; Kim, N.H.; Lian, S.; Jung, Y.D. Sulforaphane Suppresses the Nicotine-Induced Expression of the Matrix Metalloproteinase-9 via Inhibiting ROS-Mediated AP-1 and NF-κB Signaling in Human Gastric Cancer Cells. Int. J. Mol. Sci. 2022, 23, 5172. [Google Scholar] [CrossRef]
- Liang, J.; Hänsch, G.M.; Hübner, K.; Samstag, Y. Sulforaphane as anticancer agent: A double-edged sword? Tricky balance between effects on tumor cells and immune cells. Adv. Biol. Regul. 2019, 71, 79–87. [Google Scholar] [CrossRef]
- Masutani, H.; Otsuki, R.; Yamaguchi, Y.; Takenaka, M.; Kanoh, N.; Takatera, K.; Kunimoto, Y.; Yodoi, J. Fragrant unsaturated aldehydes elicit activation of the Keap1/Nrf2 system leading to the upregulation of thioredoxin expression and protection against oxidative stress. Antioxid. Redox Signal. 2009, 11, 949–962. [Google Scholar] [CrossRef]
- Shetzer, Y.; Solomon, H.; Koifman, G.; Molchadsky, A.; Horesh, S.; Rotter, V. The paradigm of mutant p53-expressing cancer stem cells and drug resistance. Carcinogenesis 2014, 35, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.V.; Srivastava, S.K.; Choi, S.; Lew, K.L.; Antosiewicz, J.; Xiao, D.; Zeng, Y.; Watkins, S.C.; Johnson, C.S.; Trump, D.L.; et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J. Biol. Chem. 2005, 280, 19911–19924. [Google Scholar] [CrossRef] [PubMed]
- Atwell, L.L.; Zhang, Z.; Mori, M.; Farris, P.; Vetto, J.T.; Naik, A.M.; Oh, K.Y.; Thuillier, P.; Ho, E.; Shannon, J. Sulforaphane Bioavailability and Chemopreventive Activity in Women Scheduled for Breast Biopsy. Cancer Prev. Res. 2015, 8, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Honma, M.; Yamada, M.; Yasui, M.; Horibata, K.; Sugiyama, K.I.; Masumura, K. In vivo and in vitro mutagenicity of perillaldehyde and cinnamaldehyde. Genes Environ. 2021, 43, 30. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Martinez-Silgado, A.; Akkerman, N.; Saftien, A.; Boot, C.; de Waal, A.; Beumer, J.; Dutta, D.; Heo, I.; et al. Intestinal organoid cocultures with microbes. Nat. Protoc. 2021, 16, 4633–4649. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Wuputra, K.; Tsai, M.H.; Kato, K.; Yang, Y.H.; Pan, J.B.; Ku, C.C.; Noguchi, M.; Kishikawa, S.; Nakade, K.; Chen, H.L.; et al. Dimethyl sulfoxide stimulates the AhR-Jdp2 axis to control ROS accumulation in mouse embryonic fibroblasts. Cell Biol. Toxicol. 2022, 38, 203–222. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.-C.; Ku, C.-C.; Pan, J.-B.; Wuputra, K.; Yang, Y.-H.; Liu, C.-J.; Liu, Y.-C.; Kato, K.; Saito, S.; Lin, Y.-C.; et al. Heterogeneity of Phase II Enzyme Ligands on Controlling the Progression of Human Gastric Cancer Organoids as Stem Cell Therapy Model. Int. J. Mol. Sci. 2023, 24, 15911. https://doi.org/10.3390/ijms242115911
Wu D-C, Ku C-C, Pan J-B, Wuputra K, Yang Y-H, Liu C-J, Liu Y-C, Kato K, Saito S, Lin Y-C, et al. Heterogeneity of Phase II Enzyme Ligands on Controlling the Progression of Human Gastric Cancer Organoids as Stem Cell Therapy Model. International Journal of Molecular Sciences. 2023; 24(21):15911. https://doi.org/10.3390/ijms242115911
Chicago/Turabian StyleWu, Deng-Chyang, Chia-Chen Ku, Jia-Bin Pan, Kenly Wuputra, Ya-Han Yang, Chung-Jung Liu, Yi-Chang Liu, Kohsuke Kato, Shigeo Saito, Ying-Chu Lin, and et al. 2023. "Heterogeneity of Phase II Enzyme Ligands on Controlling the Progression of Human Gastric Cancer Organoids as Stem Cell Therapy Model" International Journal of Molecular Sciences 24, no. 21: 15911. https://doi.org/10.3390/ijms242115911
APA StyleWu, D.-C., Ku, C.-C., Pan, J.-B., Wuputra, K., Yang, Y.-H., Liu, C.-J., Liu, Y.-C., Kato, K., Saito, S., Lin, Y.-C., Chong, I.-W., Hsiao, M., Hu, H.-M., Kuo, C.-H., Kuo, K.-K., Lin, C.-S., & Yokoyama, K. K. (2023). Heterogeneity of Phase II Enzyme Ligands on Controlling the Progression of Human Gastric Cancer Organoids as Stem Cell Therapy Model. International Journal of Molecular Sciences, 24(21), 15911. https://doi.org/10.3390/ijms242115911








