A Non-Canonical Pathway Induced by Externally Applied Virus-Specific dsRNA in Potato Plants
Abstract
:1. Introduction
2. Results
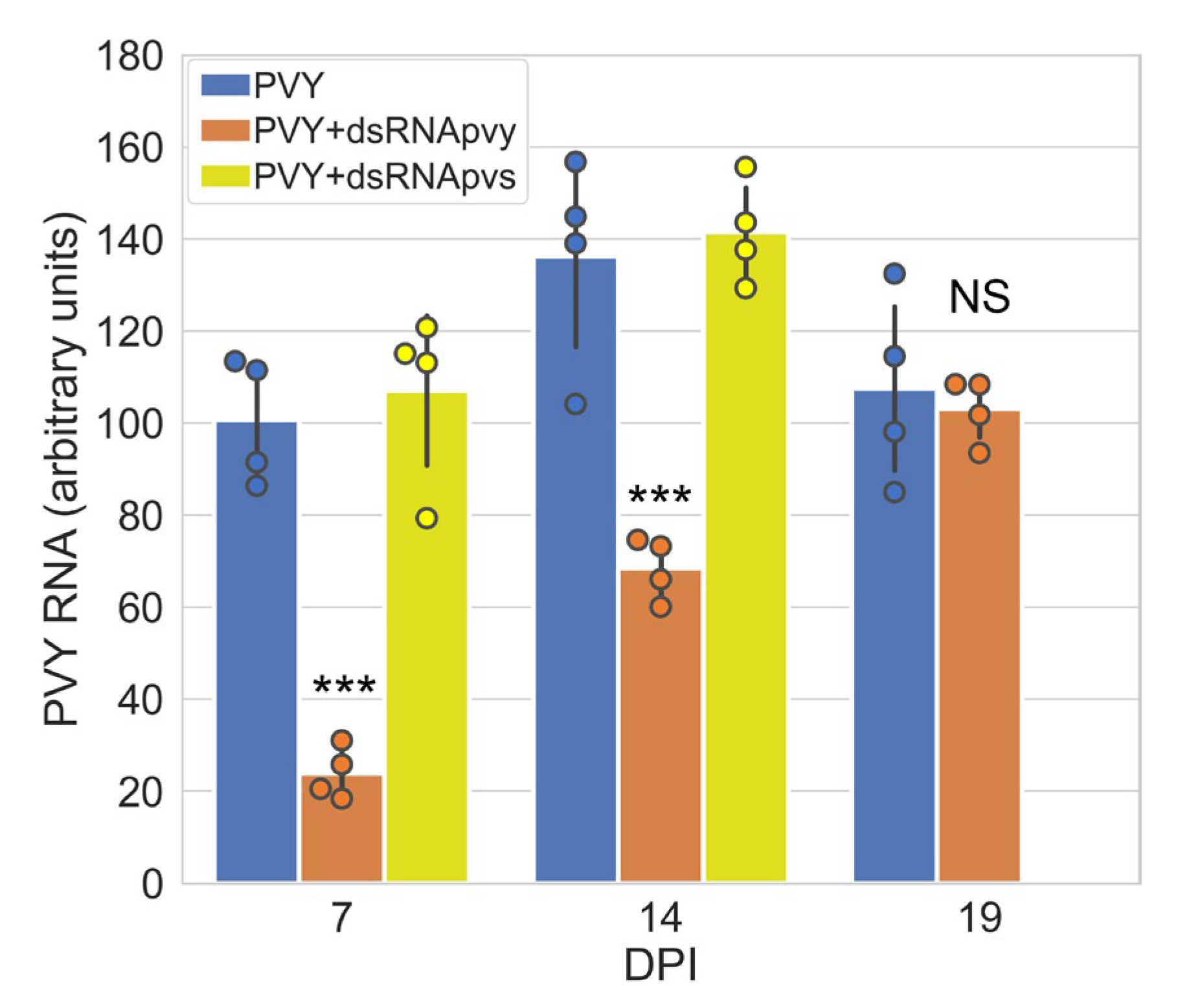
2.1. Impact of PVY-Specific dsRNA on PVY Accumulation in Potato Plants
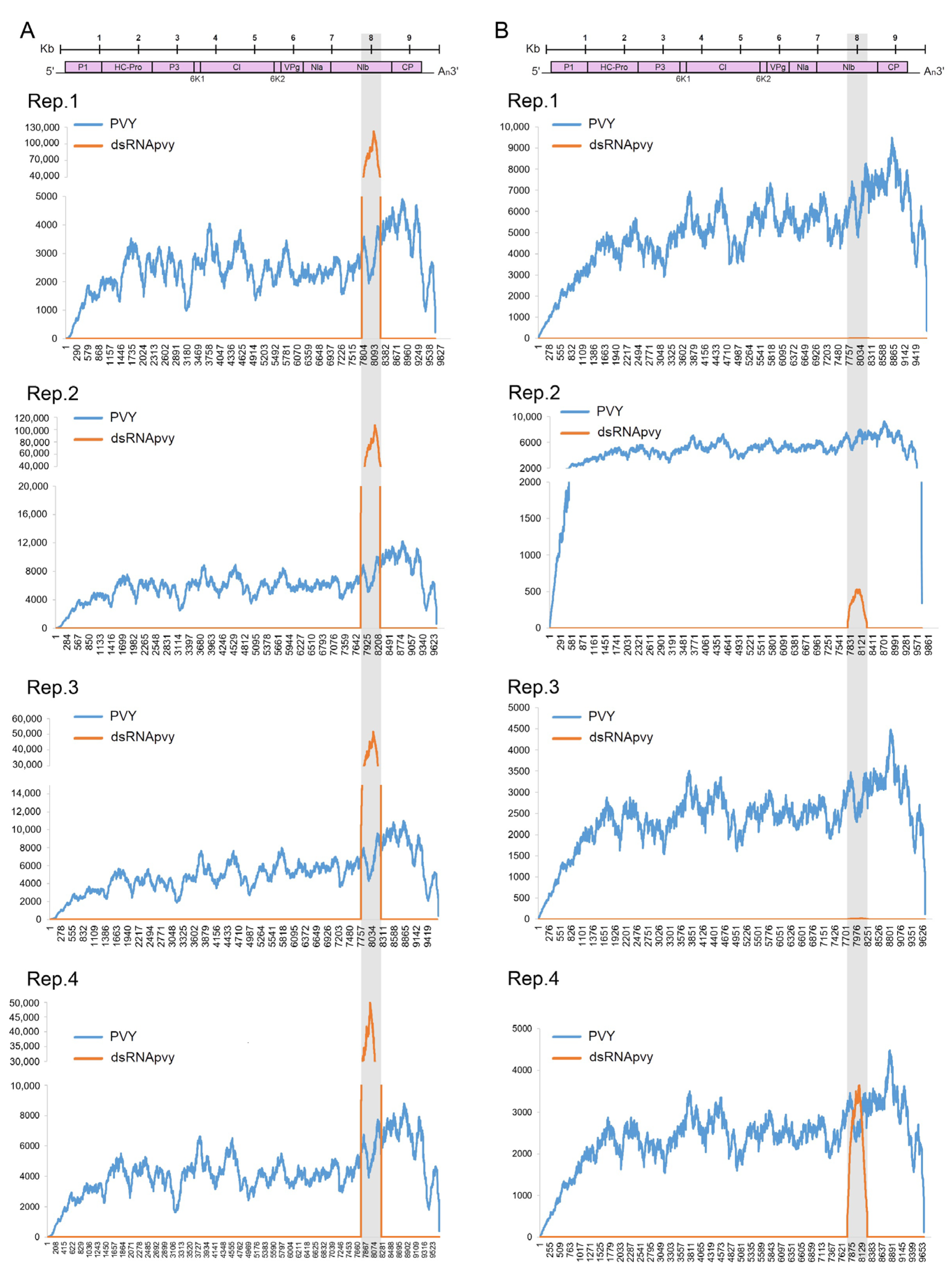
2.2. Persistence and Systemic Movement of PVY RNA-Targeting dsRNA
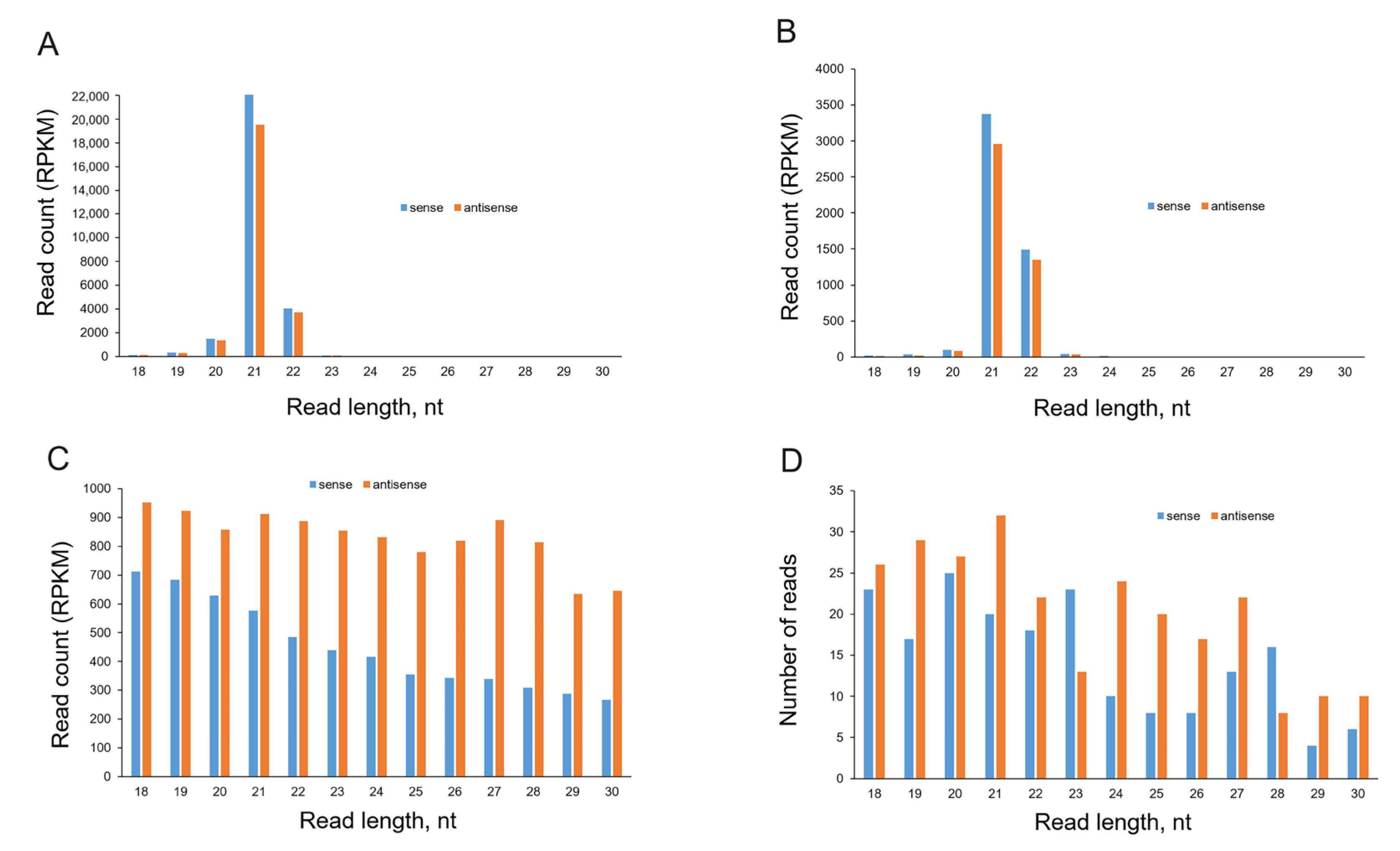
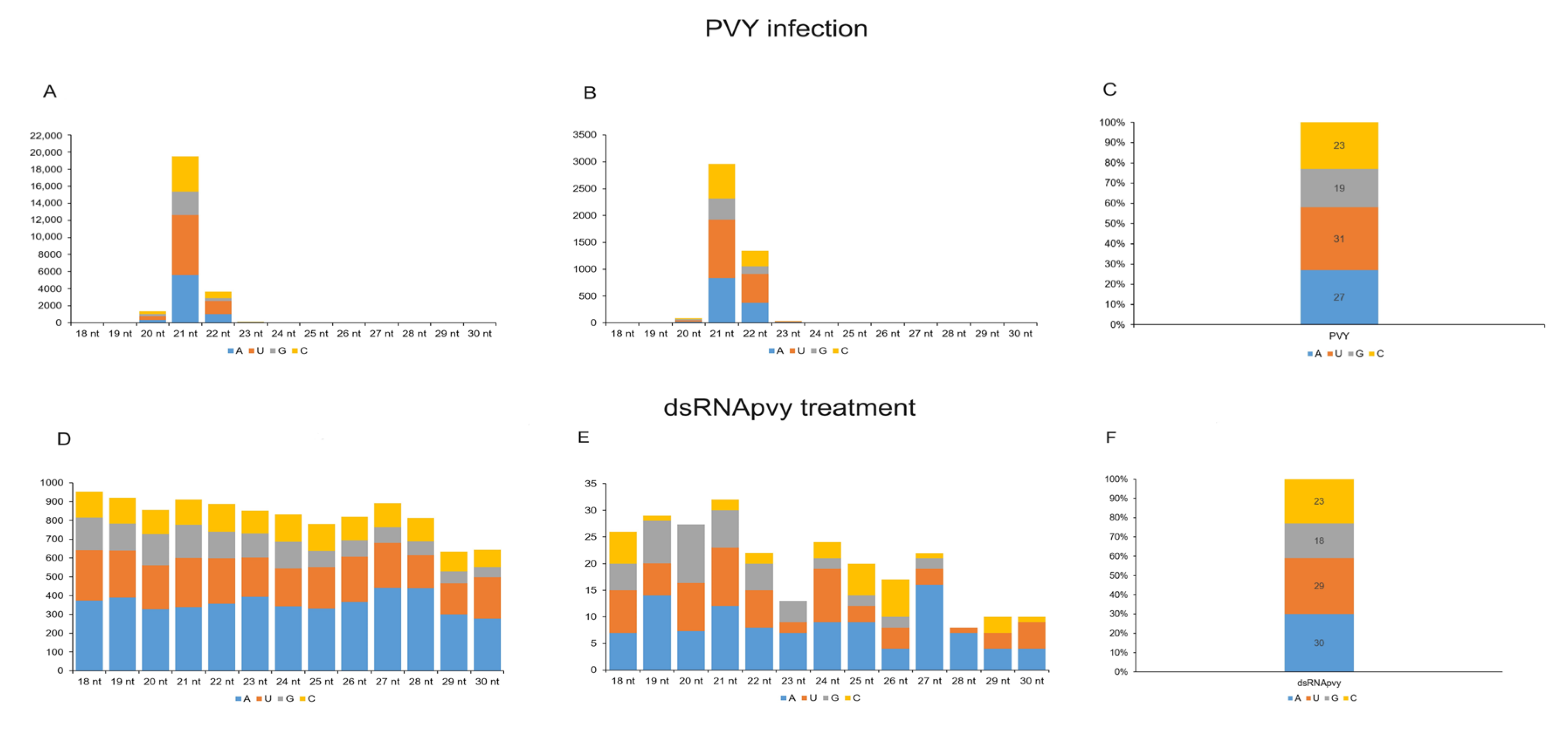
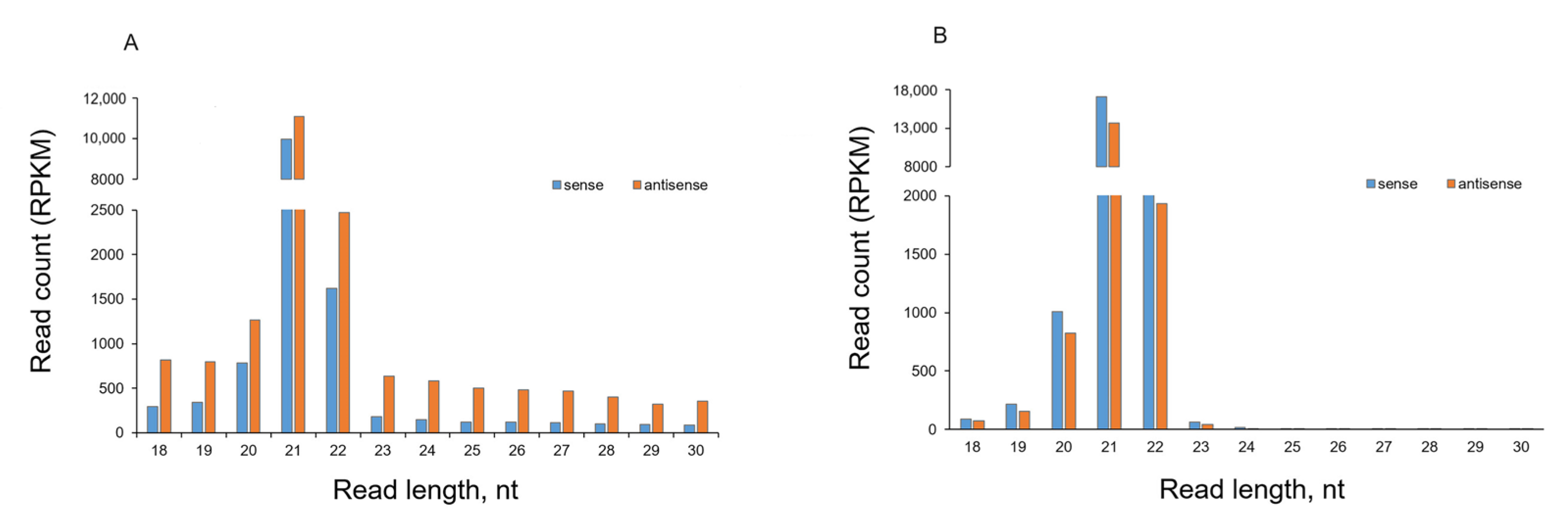
2.3. Analysis of sRNA Molecules
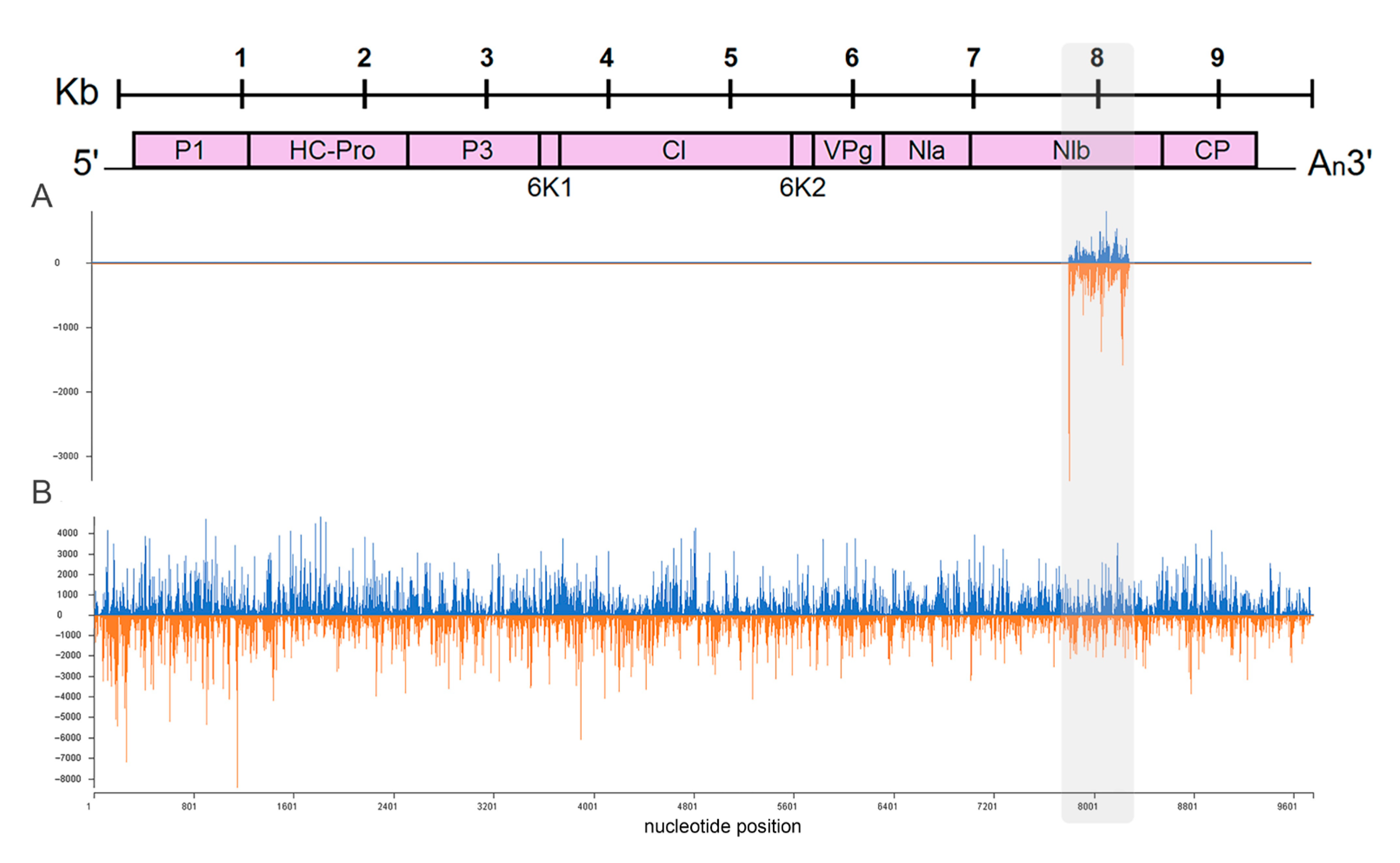
2.4. Effect of PVY Infection on Biogenesis of Non-Canonical sRNA Produced by PVY
3. Discussion
4. Materials and Methods
4.1. Virus, Plants and Growth Conditions
4.2. Production and Purification of dsRNA (dsRNApvy)
4.3. Exogenous dsRNA Application for Plant Protection against Virus Infection
4.4. Plant RNA Extraction and Real Time Quantitative RT-PCR (RT-qPCR)
4.5. RNA Sequencing and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect Vector-Mediated Transmission of Plant Viruses. Virology 2015, 479–480, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Harrison, B.D.; Baulcombe, D.C. A Similarity Between Viral Defense and Gene Silencing in Plants. Science 1997, 276, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Carrington, J.C. A Counterdefensive Strategy of Plant Viruses: Suppression of Posttranscriptional Gene Silencing. Cell 1998, 95, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C. The Role of Viruses in Identifying and Analyzing RNA Silencing. Annu. Rev. Virol. 2022, 9, 353–373. [Google Scholar] [CrossRef]
- Ding, S.-W. RNA-Based Antiviral Immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y. Dissection of RNAi-Based Antiviral Immunity in Plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA Genes Are Transcribed by RNA Polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Kørner, C.J.; Pitzalis, N.; Peña, E.J.; Erhardt, M.; Vazquez, F.; Heinlein, M. Crosstalk between PTGS and TGS Pathways in Natural Antiviral Immunity and Disease Recovery. Nat. Plants 2018, 4, 157–164. [Google Scholar] [CrossRef]
- Sanan-Mishra, N.; Abdul Kader Jailani, A.; Mandal, B.; Mukherjee, S.K. Secondary siRNAs in Plants: Biosynthesis, Various Functions, and Applications in Virology. Front. Plant Sci. 2021, 12, 610283. [Google Scholar] [CrossRef]
- Fang, X.; Qi, Y. RNAi in Plants: An Argonaute-Centered View. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef]
- Schröder, J.A.; Jullien, P.E. The Diversity of Plant Small RNAs Silencing Mechanisms. CHIMIA 2019, 73, 362. [Google Scholar] [CrossRef]
- Jin, L.; Chen, M.; Xiang, M.; Guo, Z. RNAi-Based Antiviral Innate Immunity in Plants. Viruses 2022, 14, 432. [Google Scholar] [CrossRef] [PubMed]
- Nagano, H.; Fukudome, A.; Hiraguri, A.; Moriyama, H.; Fukuhara, T. Distinct Substrate Specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res. 2014, 42, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shoji, K.; Naganuma, M.; Tomari, Y.; Iwakawa, H. The Mechanisms of siRNA Selection by Plant Argonaute Proteins Triggering DNA Methylation. Nucleic Acids Res. 2022, 50, 12997–13010. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Tomari, Y. Life of RISC: Formation, Action, and Degradation of RNA-Induced Silencing Complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef] [PubMed]
- De Felippes, F.F.; Waterhouse, P.M. The Whys and Wherefores of Transitivity in Plants. Front. Plant Sci. 2020, 11, 579376. [Google Scholar] [CrossRef] [PubMed]
- Uslu, V.V.; Dalakouras, A.; Steffens, V.A.; Krczal, G.; Wassenegger, M. High-Pressure Sprayed siRNAs Influence the Efficiency but Not the Profile of Transitive Silencing. Plant J. 2022, 109, 1199–1212. [Google Scholar] [CrossRef]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A Two-Hit Trigger for siRNA Biogenesis in Plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef]
- Lopez-Gomollon, S.; Baulcombe, D.C. Roles of RNA Silencing in Viral and Non-Viral Plant Immunity and in the Crosstalk between Disease Resistance Systems. Nat. Rev. Mol. Cell Biol. 2022, 23, 645–662. [Google Scholar] [CrossRef]
- Zand Karimi, H.; Innes, R.W. Molecular Mechanisms Underlying Host-Induced Gene Silencing. Plant Cell 2022, 34, 3183–3199. [Google Scholar] [CrossRef]
- Hernández-Soto, A.; Chacón-Cerdas, R. RNAi Crop Protection Advances. Int. J. Mol. Sci. 2021, 22, 12148. [Google Scholar] [CrossRef] [PubMed]
- Bramlett, M.; Plaetinck, G.; Maienfisch, P. RNA-Based Biocontrols—A New Paradigm in Crop Protection. Engineering 2020, 6, 522–527. [Google Scholar] [CrossRef]
- Akbar, S.; Wei, Y.; Zhang, M.-Q. RNA Interference: Promising Approach to Combat Plant Viruses. Int. J. Mol. Sci. 2022, 23, 5312. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-Based Technologies for Engineering Plant Virus Resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Sundaresha, S.; Sharma, S.; Bairwa, A.; Tomar, M.; Kumar, R.; Bhardwaj, V.; Jeevalatha, A.; Bakade, R.; Salaria, N.; Thakur, K.; et al. Spraying of dsRNA Molecules Derived from Phytophthora Infestans, along with Nanoclay Carriers as a Proof of Concept for Developing Novel Protection Strategy for Potato Late Blight. Pest Manag. Sci. 2022, 78, 3183–3192. [Google Scholar] [CrossRef]
- Necira, K.; Makki, M.; Sanz-García, E.; Canto, T.; Djilani-Khouadja, F.; Tenllado, F. Topical Application of Escherichia Coli-Encapsulated dsRNA Induces Resistance in Nicotiana Benthamiana to Potato Viruses and Involves RDR6 and Combined Activities of DCL2 and DCL4. Plants 2021, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Martín, J.; Ruiz, L.; Janssen, D.; Velasco, L. Exogenous Application of dsRNA for the Control of Viruses in Cucurbits. Front. Plant Sci. 2022, 13, 895953. [Google Scholar] [CrossRef]
- Nityagovsky, N.N.; Kiselev, K.V.; Suprun, A.R.; Dubrovina, A.S. Exogenous dsRNA Induces RNA Interference of a Chalcone Synthase Gene in Arabidopsis Thaliana. Int. J. Mol. Sci. 2022, 23, 5325. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay Nanosheets for Topical Delivery of RNAi for Sustained Protection against Plant Viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Tabein, S.; Jansen, M.; Noris, E.; Vaira, A.M.; Marian, D.; Behjatnia, S.A.A.; Accotto, G.P.; Miozzi, L. The Induction of an Effective dsRNA-Mediated Resistance Against Tomato Spotted Wilt Virus by Exogenous Application of Double-Stranded RNA Largely Depends on the Selection of the Viral RNA Target Region. Front. Plant Sci. 2020, 11, 533338. [Google Scholar] [CrossRef]
- Rego-Machado, C.M.; Nakasu, E.Y.T.; Silva, J.M.F.; Lucinda, N.; Nagata, T.; Inoue-Nagata, A.K. siRNA Biogenesis and Advances in Topically Applied dsRNA for Controlling Virus Infections in Tomato Plants. Sci. Rep. 2020, 10, 22277. [Google Scholar] [CrossRef] [PubMed]
- Konakalla, N.C.; Bag, S.; Deraniyagala, A.S.; Culbreath, A.K.; Pappu, H.R. Induction of Plant Resistance in Tobacco (Nicotiana tabacum) against Tomato Spotted Wilt Orthotospovirus through Foliar Application of dsRNA. Viruses 2021, 13, 662. [Google Scholar] [CrossRef] [PubMed]
- Samarskaya, V.O.; Spechenkova, N.; Markin, N.; Suprunova, T.P.; Zavriev, S.K.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Impact of Exogenous Application of Potato Virus Y-Specific dsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-Ribose) Metabolism. Int. J. Mol. Sci. 2022, 23, 7915. [Google Scholar] [CrossRef] [PubMed]
- Tenllado, F.; Díaz-Ruíz, J.R. Double-Stranded RNA-Mediated Interference with Plant Virus Infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Denli, A.M.; Hannon, G.J. Biochemical Specialization within Arabidopsis RNA Silencing Pathways. Mol. Cell 2005, 19, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Uslu, V.V.; Bassler, A.; Krczal, G.; Wassenegger, M. High-Pressure-Sprayed Double Stranded RNA Does Not Induce RNA Interference of a Reporter Gene. Front. Plant Sci. 2020, 11, 534391. [Google Scholar] [CrossRef]
- Donaire, L.; Barajas, D.; Martínez-García, B.; Martínez-Priego, L.; Pagán, I.; Llave, C. Structural and Genetic Requirements for the Biogenesis of Tobacco Rattle Virus-Derived Small Interfering RNAs. J. Virol. 2008, 82, 5167–5177. [Google Scholar] [CrossRef]
- Vetukuri, R.R.; Kalyandurg, P.B.; Saripella, G.V.; Sen, D.; Gil, J.F.; Lukhovitskaya, N.I.; Grenville-Briggs, L.J.; Savenkov, E.I. Effect of RNA Silencing Suppression Activity of Chrysanthemum Virus B P12 Protein on Small RNA Species. Arch. Virol. 2020, 165, 2953–2959. [Google Scholar] [CrossRef]
- Zhang, X.; Du, P.; Lu, L.; Xiao, Q.; Wang, W.; Cao, X.; Ren, B.; Wei, C.; Li, Y. Contrasting Effects of HC-Pro and 2b Viral Suppressors from Sugarcane Mosaic Virus and Tomato Aspermy Cucumovirus on the Accumulation of siRNAs. Virology 2008, 374, 351–360. [Google Scholar] [CrossRef]
- Alexandrova, A.; Karpova, O.; Kryldakov, R.; Golyaev, V.; Nargilova, R.; Iskakov, B.; Pooggin, M.M. Virus Elimination from Naturally Infected Field Cultivars of Potato (Solanum tuberosum) by Transgenic RNA Interference. Int. J. Mol. Sci. 2022, 23, 8020. [Google Scholar] [CrossRef]
- Hoang, B.T.L.; Fletcher, S.J.; Brosnan, C.A.; Ghodke, A.B.; Manzie, N.; Mitter, N. RNAi as a Foliar Spray: Efficiency and Challenges to Field Applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.; Deikman, J.; Hendrix, B.; Iandolino, A. Barriers to Efficient Foliar Uptake of dsRNA and Molecular Barriers to dsRNA Activity in Plant Cells. Front. Plant Sci. 2020, 11, 816. [Google Scholar] [CrossRef] [PubMed]
- Blevins, T.; Podicheti, R.; Mishra, V.; Marasco, M.; Wang, J.; Rusch, D.; Tang, H.; Pikaard, C.S. Identification of Pol IV and RDR2-Dependent Precursors of 24 Nt siRNAs Guiding de Novo DNA Methylation in Arabidopsis. eLife 2015, 4, e09591. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Chen, Z.; Lian, B.; Rowley, M.J.; Xia, N.; Chai, J.; Li, Y.; He, X.-J.; Wierzbicki, A.T.; Qi, Y. A Dicer-Independent Route for Biogenesis of siRNAs That Direct DNA Methylation in Arabidopsis. Mol. Cell 2016, 61, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-L.; Zhang, G.; Tang, K.; Li, J.; Yang, L.; Huang, H.; Zhang, H.; Zhu, J.-K. Dicer-Independent RNA-Directed DNA Methylation in Arabidopsis. Cell Res. 2016, 26, 66–82. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.-S.; Duan, C.-G.; Zhang, Z.-H.; Fang, Y.-Y.; Fang, R.-X.; Xie, Q.; Guo, H.-S. DCL4 Targets Cucumber Mosaic Virus Satellite RNA at Novel Secondary Structures. J. Virol. 2007, 81, 9142–9151. [Google Scholar] [CrossRef] [PubMed]
- Naveed, K.; Mitter, N.; Harper, A.; Dhingra, A.; Pappu, H.R. Comparative Analysis of Virus-Specific Small RNA Profiles of Three Biologically Distinct Strains of Potato Virus Y in Infected Potato (Solanum Tuberosum) Cv. Russet Burbank. Virus Res. 2014, 191, 153–160. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Donahue, K.; Koh, Y.; Martin, R.R.; Choi, M.-Y. Microbial-Based Double-Stranded RNA Production to Develop Cost-Effective RNA Interference Application for Insect Pest Management. Int. J. Insect Sci. 2019, 11, 1179543319840323. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping Gene Selection for Real-Time RT-PCR Normalization in Potato during Biotic and Abiotic Stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Baebler, Š.; Stare, K.; Kovač, M.; Blejec, A.; Prezelj, N.; Stare, T.; Kogovšek, P.; Pompe-Novak, M.; Rosahl, S.; Ravnikar, M.; et al. Dynamics of Responses in Compatible Potato—Potato Virus Y Interaction Are Modulated by Salicylic Acid. PLoS ONE 2011, 6, e29009. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Segonds-Pichon, A.; Biggins, L.; Krueger, C.; Wingett, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 September 2023).
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Seguin, J.; Otten, P.; Baerlocher, L.; Farinelli, L.; Pooggin, M.M. MISIS: A Bioinformatics Tool to View and Analyze Maps of Small RNAs Derived from Viruses and Genomic Loci Generating Multiple Small RNAs. J. Virol. Methods 2014, 195, 120–122. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Mezzetti, B.; Kleter, G.; Smagghe, G.; Baraldi, E. Does RNAi-Based Technology Fit within EU Sustainability Goals? Trends Biotechnol. 2021, 39, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Mezzetti, B.; Sabbadini, S.; Sorteberg, H.-G.; Sweet, J.; Ventura, V.; et al. RNA-Based Biocontrol Compounds: Current Status and Perspectives to Reach the Market. Pest Manag. Sci. 2020, 76, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Sundaresha, S.; Bairwa, A.; Tomar, M.; Kumar, R.; Venkatasalam, E.P.; Sagar, V.; Bhardwaj, V.; Sharma, S. In Vitro Method for Synthesis of Large-Scale dsRNA Molecule as a Novel Plant Protection Strategy. In Plant Gene Silencing: Methods and Protocols; Mysore, K.S., Senthil-Kumar, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; pp. 211–226. ISBN 978-1-07-161875-2. [Google Scholar]
- Thagun, C.; Horii, Y.; Mori, M.; Fujita, S.; Ohtani, M.; Tsuchiya, K.; Kodama, Y.; Odahara, M.; Numata, K. Non-Transgenic Gene Modulation via Spray Delivery of Nucleic Acid/Peptide Complexes into Plant Nuclei and Chloroplasts. ACS Nano 2022, 16, 3506–3521. [Google Scholar] [CrossRef] [PubMed]
- Samarskaya, V.O.; Ryabov, E.V.; Gryzunov, N.; Spechenkova, N.; Kuznetsova, M.; Ilina, I.; Suprunova, T.; Taliansky, M.E.; Ivanov, P.A.; Kalinina, N.O. The Temporal and Geographical Dynamics of Potato Virus Y Diversity in Russia. Int. J. Mol. Sci. 2023, 24, 14833. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarskaya, V.O.; Spechenkova, N.; Ilina, I.; Suprunova, T.P.; Kalinina, N.O.; Love, A.J.; Taliansky, M.E. A Non-Canonical Pathway Induced by Externally Applied Virus-Specific dsRNA in Potato Plants. Int. J. Mol. Sci. 2023, 24, 15769. https://doi.org/10.3390/ijms242115769
Samarskaya VO, Spechenkova N, Ilina I, Suprunova TP, Kalinina NO, Love AJ, Taliansky ME. A Non-Canonical Pathway Induced by Externally Applied Virus-Specific dsRNA in Potato Plants. International Journal of Molecular Sciences. 2023; 24(21):15769. https://doi.org/10.3390/ijms242115769
Chicago/Turabian StyleSamarskaya, Viktoriya O., Nadezhda Spechenkova, Irina Ilina, Tatiana P. Suprunova, Natalia O. Kalinina, Andrew J. Love, and Michael E. Taliansky. 2023. "A Non-Canonical Pathway Induced by Externally Applied Virus-Specific dsRNA in Potato Plants" International Journal of Molecular Sciences 24, no. 21: 15769. https://doi.org/10.3390/ijms242115769
APA StyleSamarskaya, V. O., Spechenkova, N., Ilina, I., Suprunova, T. P., Kalinina, N. O., Love, A. J., & Taliansky, M. E. (2023). A Non-Canonical Pathway Induced by Externally Applied Virus-Specific dsRNA in Potato Plants. International Journal of Molecular Sciences, 24(21), 15769. https://doi.org/10.3390/ijms242115769







