Advances and Prospects of Virus-Resistant Breeding in Tomatoes
Abstract
1. Introduction
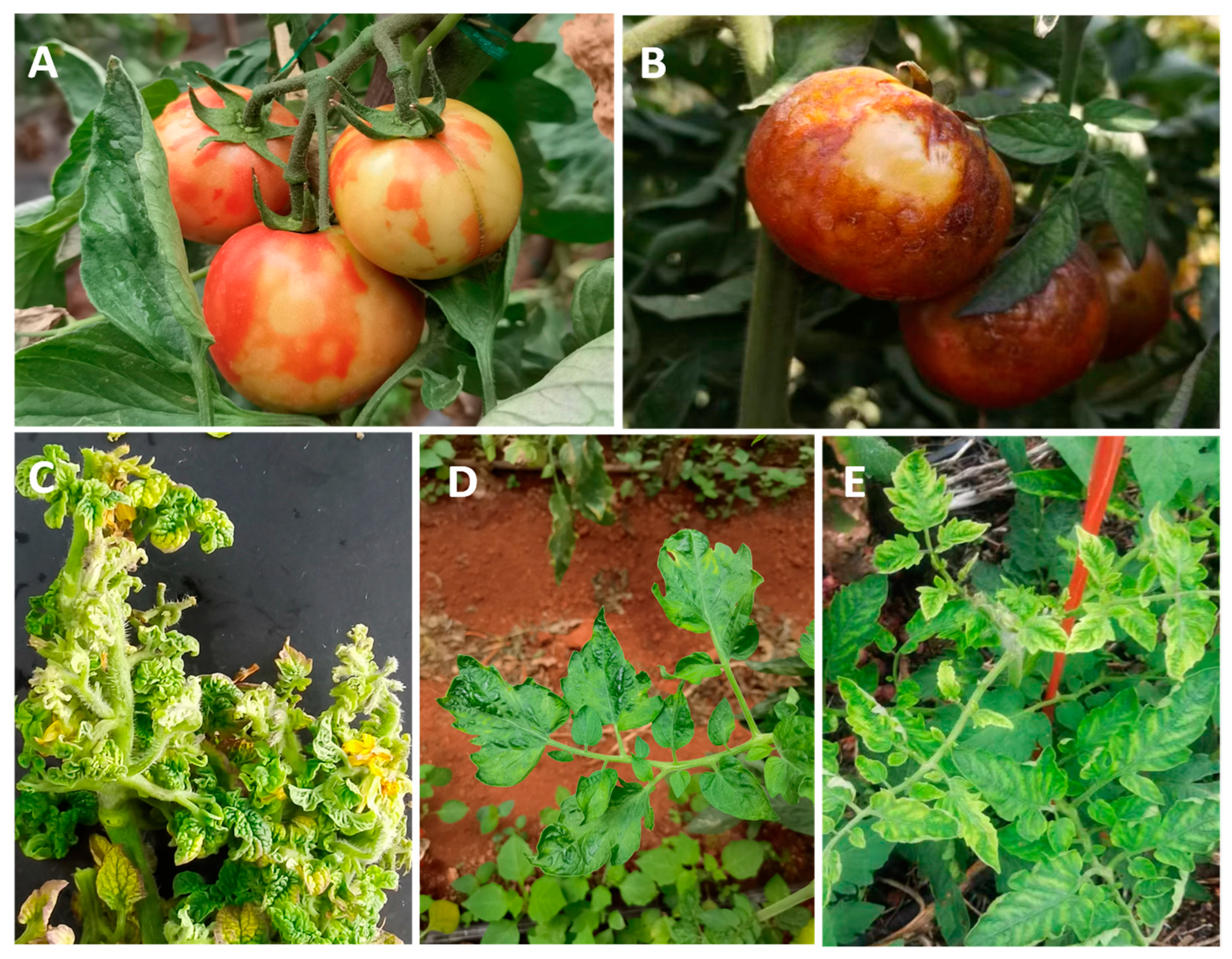
2. Tomato Viruses and Their Epidemic Characteristics
| Genus | Epidemic Species Worldwide | Genome | Transmission | Symptoms | References |
|---|---|---|---|---|---|
| Begomovirus | Tomato yellow leaf curl virus (TYLCV)) and tomato leaf curl virus (ToLCV) | Single-stranded DNA (ssDNA) | Whitefly, seed | Yellowing, curling, and a significant loss in apical leaf. Early-infected plants are frequently infertile. Since most blooms (>90%) droop after infection, there is almost no or fewer small fruit. | [23,24,25,26] |
| Orthotospovirus | Tomato spotted wilt virus (TSWV) | Negative-sense single-stranded ambisense (-ssRNA) RNA | Thrips, seed | Stunting, necrosis, bronzing, chlorosis, ring spots, and ring patterns on the leaves, stems, and fruits. | [27,28,29] |
| Tobamovirus | Tobacco mosaic virus (TMV), tomato mosaic virus (ToMV), and tomato brown rugose fruit virus (ToBRFV) | Single-stranded positive-sense RNA (+ssRNA) | Seed, mechanical transmission such as by hand, pruning tools, soil, etc. | Yellow–green mottling on the leaves; stunted growth; flowers and leaflets may be curled, distorted, and smaller than normal in size. | [15,30,31,32,33,34] |
| Potyvirus | Potato virus Y (PVY), and, chilli veinal mottle virus (ChiVMV) | Single-stranded positive-sense RNA (+ssRNA) viruses | Aphid, seed | Leaf mosaic, mottle and crinkling, vein necrosis and necrotic spots, stem and petiole necrosis, leaf drop, and yield reduction. | [2,21,35,36] |
| Crinivirus | Tomato chlorosis virus (ToCV) | Single-stranded positive-sense RNA (+ssRNA) viruses | Whitefly | Leaf chlorosis, chlorotic flecking, and bronzing. Fruits are symptomless but with reduced yield. | [37,38] |
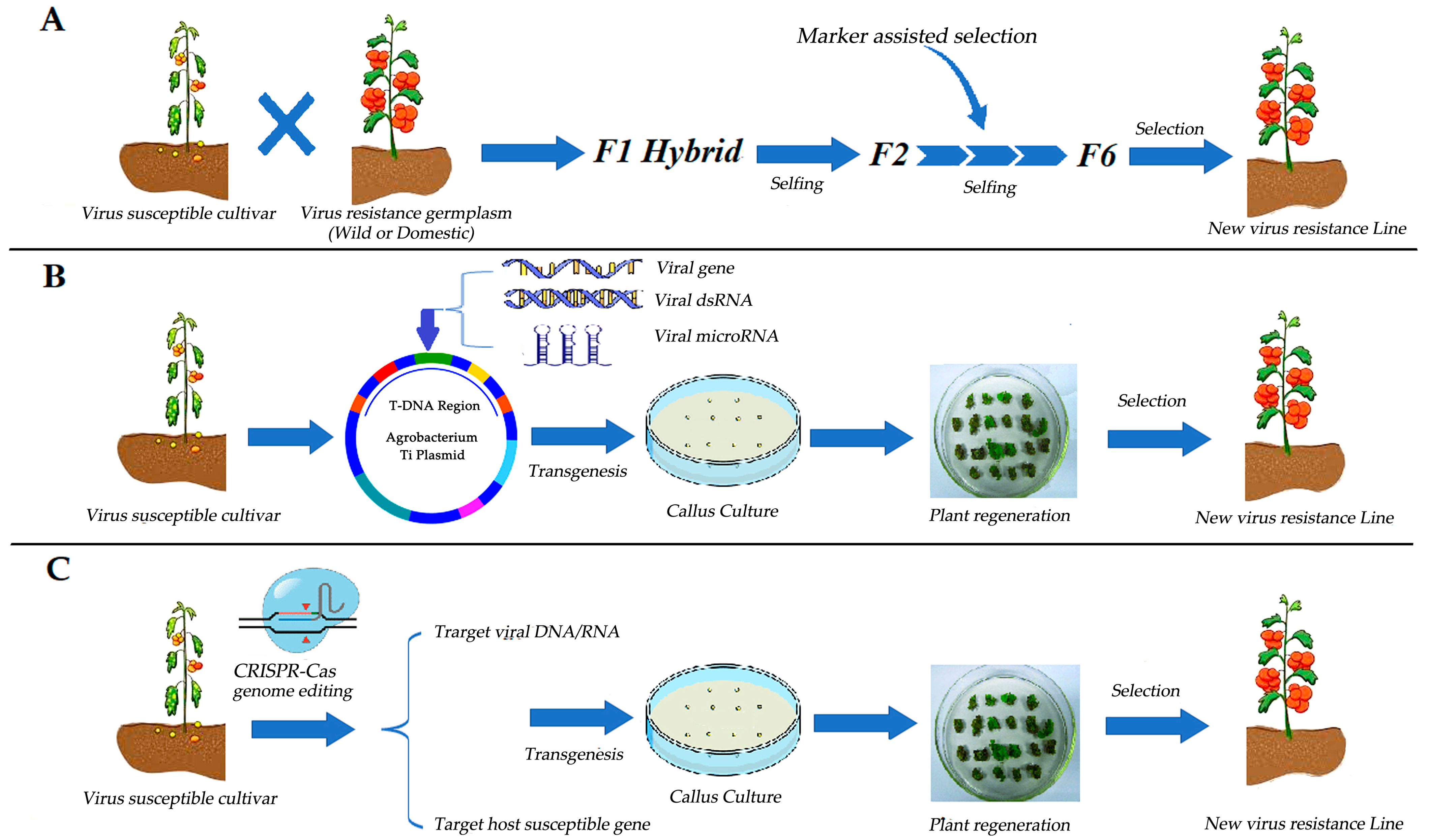
3. Strategies of Tomato Resistance Breeding against Viruses
4. Virus Resistance Gene and MAS-Based Hybrid Breeding in Tomatoes
4.1. The Tomato Ty Gene Family Encoding for Resistance against Begomoviruses
4.2. The Tomato Sw Gene Family Conferring Resistance against Orthotospoviruses
4.3. The Tomato Tm Gene Family Conferring Resistance against Tobamoviruses
4.4. The Tomato Pot 1 Gene Conferring Resistance against Potyviruses
4.5. The Challenges in Breeding Tomato Hybrid Cultivars against Viruses
5. RNAi-Based Transgenic Breeding
| Strategy | Target Virus | Genus | RNAi Induction Method | Targeted Region | Precursor(s) | Efficiency | Reference |
|---|---|---|---|---|---|---|---|
| First-generation antiviral transgenic strategy | TMV | Tobamovirus | ssRNA | CP | cDNA | Delayed symptom development; 10 to 60 percent of the transgenic plants failed to develop symptoms. | [96] |
| TMV | Tobamovirus | ssRNA | CP | cDNA | The resistance level of expression TMV CP from the pal2 promoter is less than that of the 35S promoter. | [97] | |
| TSWV | Orthotospovirus | ssRNA | N | cDNA | Lack of systemic symptoms and little or no systemic accumulation of virus. | [98] | |
| ToLCV | Begomovirus | ssRNA | Rep | cDNA | A high level of resistance and inheritability of the transgene was observed up to T2. | [99] | |
| TLCV | Begomovirus | ssRNA | CP | cDNA | T1-generation transgenic plants were showed variable degrees of disease resistance/tolerance compared to the untransformed control. | [100] | |
| ToLCNDV | Begomovirus | ssRNA | AV2 | cDNA | Transgenic plants showed symptomless, although viral DNA could be detected in some plants by PCR. | [101] | |
| PRSV | Potyvirus | ssRNA | CP | cDNA | PRSV infection was not observed on any of the transgenic resistance (TR) plants. TR plant yields were at least three times higher than the industry average. | [102] | |
| Second-generation antiviral transgenic strategy | TMV | Tobamovirus | dsRNA | CP, p126 | dsRNA | The application of TMV p126 dsRNA onto tobacco plants induced greater resistance against TMV infection as compared to CP dsRNA (65 vs. 50%). | [103] |
| ToLCV | Begomovirus | hpRNA | AC1, AC4 | hpRNA | Provides a promising approach to suppress a wide spectrum of ToLCV infection in the tomato. | [104] | |
| ToLCV | Begomovirus | dsRNA | AC4 | dsRNA | Absolute absence of leaf curl virus disease symptoms and reduction in nematode symptoms. | [105] | |
| ToCMoV | Begomovirus | hpRNA | AC1, AC4, AV1, AC5 | hpRNA | Most transgenic lines showed significant delays in symptom development, and two lines had immune plants. | [106] | |
| PVY | Potyvirus | dsRNA | CP | dsRNA | Highly resistant to three strains of PVY. | [107] | |
| PVY | Potyvirus | hpRNA | CP | hpRNA | Nine of the ten transgenic lines showed no infection by PVYO, and six of the nine showed no infection by PVYNTN. | [108] | |
| CaCV GBNV CMV ChiVMV | Orthotospovirus Cucumovirus Potyvirus | hpRNA | viral silencing suppressors gene | hpRNA | Efficiently controls multiple viruses | [109] | |
| Third-generation antiviral transgenic strategy | ToLCNDV | Begomovirus | amiRNA | AV1, AV1 + AV2 | Ath-miR319a | High tolerance when targeting AV1 + AV2. Moderate tolerance when targeting AV1. | [87] |
| TYLCV | Begomovirus | amiRNA | AC1+Rep | Ath-miR159a | Confer resistance to TYLCV. | [110] | |
| TSWV | Orthotospovirus | syn-tasiRNA | NSm+RdRP | TAS1c | 100% of the plants were resistant. | [111] | |
| TSWV | Orthotospovirus | syn-tasiRNA | RdRP | TAS1c | Delay of viroid accumulation | [112] | |
| PhCMoV, ToBRFV | Alphanucleorhabdovirus Tobamovirus | amiRNA | L, M, G | To-miR6026 | Bioinformatic assay showed successful results in controlling both viruses. | [93] | |
| PVY | Potyvirus | amiRNA | CI, NIa, NIb, CP | Ath-miR319a | Higher protection when targeting NIb or CP. | [113] | |
| PVY, PVX, PSTVd | Potyvirus Potexvirus Pospiviroid | amiRNA | P25, HC-Pro, Brp1 | Ath-miR159a | Resistance against PVX, PVY, and PSTVd coinfection simultaneously, whereas the untransformed controls developed severe symptoms. | [92] |
6. Virus Resistance Breeding Based on the CRISPR/Cas Genome Editing
6.1. CRISPR/Cas Genome Editing Targeting DNA Viruses
6.2. CRISPR/Cas Genome Editing Targeting RNA Viruses
| Target Virus | Genus | Plant | Targeted Genome | CRISPR/Cas | Targeted Region | Efficiency | Reference |
|---|---|---|---|---|---|---|---|
| TYLCV | Begomovirus | Tomato | Viral DNA | CRISPR/Cas9 | CP, IR | Significant reduction or delayed accumulation of viral DNA compared to the control plants. | [132] |
| TYLCV | Begomovirus | Tomato | Viral DNA | CRISPR/Cas9 | CP, Rep | Low accumulation of the viral DNA genome compared to the control plants. | [114] |
| TYLCV | Begomovirus | Tomato | Viral DNA | CRISPR/Cas9 | IR, CP, RCRII | Reduction or delayed accumulation of viral DNA, abolishing or significantly attenuating symptoms of infection. | [125] |
| CLCuKoV | Begomovirus | Tomato | Viral DNA | CRISPR/Cas9 | IR, CP, RCRII | Significantly limits CLCuKoV and MeMV replication and systemic infection. | [126] |
| MeMV | Begomovirus | Tomato | Viral DNA | CRISPR/Cas9 | IR, CP, RCRII | ||
| CMV | Cucumovirus | Arabidopsis | Viral RNA | CRISPR/FnCas9 | ORF1a, ORF CP, 3′UTR | Significantly attenuated infection symptoms and reduced viral RNA accumulation. The resistance was inheritable, and the progenies showed significantly low virus accumulation. | [133] |
| TMV | Tobamovirus | Tobacco | Viral RNA | CRISPR/FnCas9 | 3′ORFs | ||
| TuMV | Potyvirus | Tobacco | Viral RNA | CRISPR/LshCas13a | HC-Pro, CP | Targeting the HC-Pro rather than those targeting the coat protein (CP) sequence significantly inhibits TuMV-GFP accumulation and systematic movement. | [134] |
| TuMV | Potyvirus | Arabidopsis | Viral RNA | CRISPR/LshCas13a | HC-Pro, CP | Significant inhibition of TuMV-GFP accumulation level and systematic movement in T1 and T2 plants. | [135] |
| PVY | Potyvirus | Potato | Viral RNA | CRISPR/LshCas13a | P3, CI, NIb, CP | Specifically resistant to multiple PVY strains while having no effect on unrelated viruses such as PVA or Potato virus S. | [136] |
| TMV | Tobamovirus | Tobacco | Viral RNA | CRISPR/LshCas13a | RdRp, MP, CP | Significant reduction or delayed accumulation of viral RNA compared to the control plants. | [137] |
| SRBSDV | Fijivirus | Rice | Viral RNA | CRISPR/LshCas13a | ORF | Abolishing or significantly attenuating symptoms of infection. T3 transgenic plants we tested showed stable resistance to SRBSDV. | [137] |
| RSMV | Cytorhabdovirus | Rice | Viral RNA | CRISPR/LshCas13a | ORF | Abolishing or significantly attenuating symptoms of infection. T3 transgenic plants we tested showed stable resistance to RSMV. | [137] |
| SPCSV | Crinivirus | Sweet potato | Viral RNA | CRISPR/LwaCas13a CRISPR/13d | RNase3 | Transgenic plants and their grafted plants showed a significant reduction in virus accumulation and were asymptomatic. | [138] |
| TYLCV | Begomovirus | Tomato | Tomato genome | CRISPR/Cas9 | SlPelo | Knocking out the bialleles of SlPelo proved to suppress systematic infection of TYLCV. | [117] |
| PVY | Potyvirus | Tomato | Tomato genome | CRISPR/Cas9 | eIF4E1 | Significant reduction in susceptibility to the N strain (PVY-N) but not to the ordinary strain (PVY-O). | [139] |
| CMV | Cucumovirus | Tomato | Tomato genome | CRISPR/Cas9 | eIF4E1 | Viral aphid transmission from an infected susceptible plant to gene-edited plants was reduced compared with the parental control. | [139] |
| TEV | Potyvirus | Tomato | Tomato genome | CRISPR/Cas9 | eIF4E1 | A combination of mutations in regions I and II of eIF4E1 associates with resistance to several isolates of potyviruses. | [140] |
| PVY | Potyvirus | Tomato | Tomato genome | CRISPR/Cas9 | eIF4E1 | Differences in silent targets showed differences in resistance levels. | [140] |
| PepMoV | Potyvirus | Tomato | Tomato genome | CRISPR/Cas9 | eIF4E1 | Knocking out eIF4E1 exhibited a significant reduction accumulation of PepMoV but not TEV. | [141] |
| PVMV | Potyvirus | Tomato | Tomato genome | CRISPR/Cas9 | 4E2 (eIF4E2) | Knocking out eIF4E2 exhibited resistance to six of the eight PVMV isolates but not to other potyviruses. | [142] |
| TuMV | Potyvirus | Tomato | Tomato genome | CRISPR/Cas9 | eIF(iso)4E | Homozygous mutations and transgene-free T2 and T3 generation in self-pollinating species showed no differences in dry weights and flowering times with wild-type plants under standard growth conditions. | [143] |
| ToBRFV | Tobamovirus | Tomato | Tomato genome | CRISPR/Cas9 | SlTOM1a-e | Quadruple-mutant plants did not show detectable ToBRFV CP accumulation or obvious defects in growth or fruit production. The quadruple-mutant plants also showed resistance to three other tobamoviruses. | [31] |
| ToBRFV | Tobamovirus | Tomato | Tomato genome | CRISPR/Cas9 | SlTOM1 SlTOM3 | SlTOM1a and SlTOM3 are essential for the replication of ToBRFV but not for ToMV and TMV. | [144] |
6.3. CRISPR/Cas Genome Editing Targeting Host Susceptible Genes
7. Future Prospects
7.1. CRISPR/Cas-Mediated Tomato Breeding for Resistance against Orthotospoviruses
7.2. Challenges of the CRISPR/Cas-Mediated Antiviral Breeding in Tomato
7.3. Suggestions for Improving Tomato Antiviral Breeding in the Future
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P. Emerging viral diseases of tomato crops. Mol. Plant-Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.X.; Sun, X.P.; Taylor, A.; Jiao, C.; Xu, Y.M.; Cai, X.F.; Wang, X.L.; Ge, C.H.; Pan, G.H.; Wang, Q.X.; et al. Diversity, Distribution, and Evolution of Tomato Viruses in China Uncovered by Small RNA Sequencing. J. Virol. 2017, 91, e00173–17. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, G.Q.; Silva, J.O.; Copati, M.G.F.; Dias, F.O.; Santos, M.C. Tomato breeding for disease resistance. Multi-Sci. J. 2020, 3, 8–16. [Google Scholar] [CrossRef]
- García-Estrada, R.S.; Diaz-Lara, A.; Aguilar-Molina, V.H.; Tovar-Pedraza, J.M. Viruses of Economic Impact on Tomato Crops in Mexico: From Diagnosis to Management-A Review. Viruses 2022, 14, 1251. [Google Scholar] [CrossRef]
- Rodríguez, E.; Téllez, M.M.; Janssen, D. Whitefly Control Strategies against Tomato Leaf Curl New Delhi Virus in Greenhouse Zucchini. Int. J. Environ. Res. Public Health 2019, 16, 2673. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef]
- De Oliveira, A.S.; Boiteux, L.S.; Kormelink, R.; Resende, R.O. The Sw-5 gene cluster: Tomato breeding and research toward Orthotospovirus disease control. Front. Plant Sci. 2018, 9, 1055. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.X.; Tao, X.R.; Li, D.W.; Yang, X.L.; Zhou, X.P. ty-5 Confers Broad-Spectrum Resistance to Geminiviruses. Viruses 2022, 14, 1804. [Google Scholar] [CrossRef]
- Nevame, A.Y.M.; Xia, L.; Nchongboh, C.G.; Hasan, M.M.; Alam, M.A.; Yongbo, L.; Wenting, Z.; Yafei, H.; Emon, R.M.; Ismail, M.R.; et al. Development of a New Molecular Marker for the Resistance to Tomato Yellow Leaf Curl Virus. BioMed Res. Int. 2018, 8120281. [Google Scholar] [CrossRef]
- Qi, S.M.; Zhang, S.J.; Islam, M.M.; El-Sappah, A.H.; Zhang, F.; Liang, Y. Natural Resources Resistance to Tomato Spotted Wilt Virus (TSWV) in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2021, 22, 10978. [Google Scholar] [CrossRef]
- Gautam, S.; Chinnaiah, S.; Workneh, F.; Crosby, K.; Rush, C.; Gadhave, K.R. First report of a resistance-breaking strain of tomato spotted wilt orthotospovirus infecting Capsicum annuum with the Tsw resistance gene in Texas. Plant Dis. 2022, 107, PDIS-09. [Google Scholar] [CrossRef] [PubMed]
- Lahre, K.A.; Shekasteband, R.; Meadows, I.; Whitfield, A.E.; Rotenberg, D. First Report of Resistance-Breaking Variants of Tomato Spotted Wilt Virus (TSWV) Infecting Tomatoes with the Sw-5 Tospovirus-Resistance Gene in North Carolina. Plant Dis. 2023, 107. [Google Scholar] [CrossRef]
- García-Cano, E.; Resende, R.O.; Fernández-Muñoz, R.; Moriones, E. Synergistic Interaction Between Tomato chlorosis virus and Tomato spotted wilt virus Results in Breakdown of Resistance in Tomato. Phytopathology 2006, 96, 1263–1269. [Google Scholar] [CrossRef]
- Jewehan, A.; Kiemo, F.W.; Salem, N.; Tóth, Z.; Salamon, P.; Szabó, Z. Isolation and molecular characterization of a tomato brown rugose fruit virus mutant breaking the tobamovirus resistance found in wild Solanum species. Arch. Virol. 2022, 167, 1559–1563. [Google Scholar] [CrossRef]
- Tettey, C.K.; Yan, Z.Y.; Ma, H.Y.; Zhao, M.S.; Geng, C.; Tian, Y.P.; Li, X.D. Tomato mottle mosaic virus: Characterization, resistance gene effectiveness, and quintuplex RT-PCR detection system. J. Integr. Agric. 2022, 21, 2641–2651. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Navas-Castillo, J. Tomato chlorosis virus, an emergent plant virus still expanding its geographical and host ranges. Mol. Plant Pathol. 2019, 20, 1307–1320. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef]
- Elena, S.F.; García-Arenal, F. Plant Virus Adaptation to New Hosts: A Multi-scale Approach. Curr. Top. Microbiol. Immunol. 2023, 439, 167–196. [Google Scholar] [PubMed]
- Díaz-Pendón, J.A.; Sánchez-Campos, S.; Fortes, I.M.; Moriones, E. Tomato Yellow Leaf Curl Sardinia Virus, a Begomovirus Species Evolving by Mutation and Recombination: A Challenge for Virus Control. Viruses 2019, 11, 45. [Google Scholar] [CrossRef]
- Rivarez, M.P.S.; Vučurović, A.; Mehle, N.; Ravnikar, M.; Kutnjak, D. Global Advances in Tomato Virome Research: Current Status and the Impact of High-Throughput Sequencing. Front. Microbiol. 2021, 12, 671925. [Google Scholar] [CrossRef]
- Choi, H.; Jo, Y.; Cho, W.K.; Yu, J.; Tran, P.T.; Salaipeth, L.; Kwak, H.R.; Choi, H.S.; Kim, K.H. Identification of Viruses and Viroids Infecting Tomato and Pepper Plants in Vietnam by Metatranscriptomics. Int. J. Mol. Sci. 2020, 21, 7565. [Google Scholar] [CrossRef]
- Ontiveros, I.; López-Moya, J.J.; Díaz-Pendón, J.A. Coinfection of Tomato Plants with Tomato yellow leaf curl virus and Tomato chlorosis virus Affects the Interaction with Host and Whiteflies. Phytopathology 2022, 112, 944–952. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Lett, J.M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Virus Taxonomy Profile: Geminiviridae 2021. J. Gen. Virol. 2021, 102, 001696. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, L.; Tsai, W.S.; Shih, S.L.; Lee, L.M. Emergence and diversity of begomoviruses infecting solanaceous crops in East and Southeast Asia. Virus Res. 2014, 186, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato Yellow Leaf Curl Virus: Impact, Challenges, and Management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Kil, E.J.; Kim, S.; Lee, Y.J.; Byun, H.S.; Park, J.; Seo, H.; Kim, C.S.; Shim, J.K.; Lee, J.H.; Kim, J.K.; et al. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016, 6, 19013. [Google Scholar] [CrossRef]
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avšič-Županc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, É.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Zheng, K.Y.; Zhao, L.H.; Su, X.X.; Zheng, X.; Wang, T.T. Occurrence, Distribution, Evolutionary Relationships, Epidemiology, and Management of Orthotospoviruses in China. Front. Microbiol. 2021, 12, 686025. [Google Scholar] [CrossRef]
- Wang, H.; Wu, X.; Huang, X.; Wei, S.; Lu, Z.; Ye, J. Seed Transmission of Tomato Spotted Wilt Orthotospovirus in Peppers. Viruses 2022, 14, 1873. [Google Scholar] [CrossRef]
- Adams, M.J.; Adkins, S.; Bragard, C.; Gilmer, D.; Li, D.; MacFarlane, S.A.; Wong, S.M.; Melcher, U.; Ratti, C.; Ryu, K.H.; et al. ICTV Virus Taxonomy Profile: Virgaviridae. J. Gen. Virol. 2017, 98, 1999–2000. [Google Scholar] [CrossRef]
- Ishikawa, M.; Yoshida, T.; Matsuyama, M.; Kouzai, Y.; Kano, A.; Ishibashi, K. Tomato brown rugose fruit virus resistance generated by quadruple knockout of homologs of TOBAMOVIRUS MULTIPLICATION1 in tomato. Plant Physiol. 2022, 189, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.K.; Griffiths, J.S.; Marchand, G.; Bernards, M.A.; Wang, A.M. Tomato brown rugose fruit virus: An emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 2022, 23, 1262–1277. [Google Scholar] [CrossRef] [PubMed]
- Sakem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar]
- Xu, Y.T.; Zhang, S.L.; Shen, J.G.; Wu, Z.J.; Du, Z.G.; Gao, F.L. The phylogeographic history of tomato mosaic virus in Eurasia. Virology 2021, 554, 42–47. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Jordan, R.; Kreuze, J.; Li, F.; López-Moya, J.J.; Mäkinen, K.; Ohshima, K.; Wylie, S.J.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Potyviridae 2022. J. Gen. Virol. 2022, 103, 001738. [Google Scholar] [CrossRef]
- Quenouille, J.; Vassilakos, N.; Moury, B. Potato virus Y: A major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol. Plant Pathol. 2013, 14, 439–452. [Google Scholar] [CrossRef]
- Orfanidou, C.G.; Pappi, P.G.; Efthimiou, K.E.; Katis, N.I.; Maliogka, V.I. Transmission of Tomato chlorosis virus (ToCV) by Bemisia tabaci Biotype Q and Evaluation of Four Weed Species as Viral Sources. Plant Dis. 2016, 100, 2043–2049. [Google Scholar] [CrossRef]
- Moodley, V.; Gubba, A.; Mafongoya, P.L. Prevalence, epidemiology and molecular studies of Tomato chlorosis virus (ToCV) in South Africa. PLoS ONE 2019, 14, e0220298. [Google Scholar] [CrossRef]
- Maurya, D.; Mukherjee, A.; Bhagyashree; Sangam, S.; Kumar, R.; Akhtar, S.; Chattopadhyay, T. Marker assisted stacking of Ty3, Mi1.2 and Ph3 resistance alleles for leaf curl, root knot and late blight diseases in tomato. Physiol. Mol. Biol. Plants 2023, 29, 121–129. [Google Scholar] [CrossRef]
- Akbar, S.; Wei, Y.; Zhang, M.Q. RNA Interference: Promising Approach to Combat Plant Viruses. Int. J. Mol. Sci. 2022, 23, 5312. [Google Scholar] [CrossRef]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef]
- HEl-Sappah, A.H.; Qi, S.M.; A Soaud, S.; Huang, Q.L.; M Saleh, A.; A S Abourehab, M.; Wan, L.Y.; Cheng, G.T.; Liu, J.Y.; Ihtisham, M.; et al. Natural resistance of tomato plants to Tomato yellow leaf curl virus. Front. Plant Sci. 2022, 13, 1081549. [Google Scholar] [CrossRef]
- Mangal, M.; Srivastava, A.; Sharma, R.; Kalia, P. Conservation and Dispersion of Genes Conferring Resistance to Tomato Begomoviruses between Tomato and Pepper Genomes. Front. Plant Sci. 2017, 8, 1803. [Google Scholar] [CrossRef] [PubMed]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.; Scott, J.W.; D Edwards, J.; Bai, Y.L. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA–dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [PubMed]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.; Bai, Y.L.; Kormelink, R. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef] [PubMed]
- Voorburg, C.M.; Yan, Z.; Bergua-Vidal, M.; Wolters, A.M.A.; Bai, Y.; Kormelink, R. Ty-1, a universal resistance gene against geminiviruses that is compromised by co-replication of a betasatellite. Mol. Plant Pathol. 2020, 21, 160–172. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Gill, U.; Scott, J.W.; Shekasteband, R.; Ogundiwin, E.; Schuit, C.; Francis, D.M.; Sim, S.C.; Smith, H.; Hutton, S.F. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. TAG. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar] [CrossRef]
- Arens, P.; Mansilla, C.; Deinum, D.; Cavellini, L.; Moretti, A.; Rolland, S.; van der Schoot, H.; Calvache, D.; Ponz, F.; Collonnier, C. Development and evaluation of robust molecular markers linked to disease resistance in tomato for distinctness, uniformity and stability testing. Theor. Appl. Genet. 2010, 120, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Osei, M.K.; Prempeh, R.; Adjebeng-Danquah, J.; Opoku, J.A.; Danquah, A.; Danquah, A.; Danquah, E.Y.; Blay, E.; Dapaah, H.A. Marker-Assisted Selection (MAS): A Fast-Track Tool in Tomato Breeding. In Recent Advances in Tomato Breeding and Production; IntechOpen: London, UK, 2018; pp. 93–113. [Google Scholar]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Bai, Y.; Wolters, A.A. The NLR Protein Encoded by the Resistance Gene Ty-2 Is Triggered by the Replication-Associated Protein Rep/C1 of Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2020, 11, 545306. [Google Scholar] [CrossRef]
- Hallwass, M.; de Oliveira, A.S.; de Campos Dianese, E.; Lohuis, D.; Boiteux, L.S.; Inoue-Nagata, A.K.; Resende, R.O.; Kormel-ink, R. The Tomato spotted wilt virus cell-to-cell movement protein (NSM) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 2014, 15, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, C.; Ma, Q.; Shekasteband, R.; Stewart, K.S.; Hutton, S.F.; Scott, J.W.; Fei, Z.; Ling, K.S. Comprehensive transcriptome analysis and functional characterization of PR-5 for its involvement in tomato Sw-7 resistance to tomato spotted wilt tospovirus. Sci. Rep. 2019, 9, 7673. [Google Scholar] [CrossRef]
- Loon, L.C.V.; Strien, E.A.V. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathology 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.; Haan, P.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1039. [Google Scholar] [CrossRef][Green Version]
- Ishibashi, K.; Kezuka, Y.; Kobayashi, C.; Kato, M.; Inoue, T.; Nonaka, T.; Ishikawa, M.; Matsumura, H.; Katoh, E. Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1. Proc. Natl. Acad. Sci. USA 2014, 111, E3486–E3495. [Google Scholar] [CrossRef]
- Hak, H.; Spiegelman, Z. The Tomato Brown Rugose Fruit Virus Movement Protein Overcomes Tm-22 Resistance in Tomato While Attenuating Viral Transport. Mol. Plant-Microbe Interact. 2021, 34, 1024–1032. [Google Scholar] [CrossRef]
- Ishibashi, K.; Ishikawa, M. Mechanisms of tomato mosaic virus RNA replication and its inhibition by the host resistance factor Tm-1. Curr. Opin. Virol. 2014, 9, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Hak, H.; Raanan, H.; Schwarz, S.; Sherman, Y.; Dinesh-Kumar, S.P.; Spiegelman, Z. Activation of Tm-22 resistance is mediated by a conserved cysteine essential for tobacco mosaic virus movement. Mol. Plant Pathol. 2023, 24, 838–848. [Google Scholar] [CrossRef]
- Weber, H.; Schultze, S.; Pfitzner, A.J. Two amino acid substitutions in the tomato mosaic virus 30-kilodalton movement protein confer the ability to overcome the Tm-2(2) resistance gene in the tomato. J. Virol. 1993, 67, 6432–6438. [Google Scholar] [CrossRef]
- Parrella, G.; Ruffel, S.; Moretti, A.; Morel, C.; Palloix, A.; Caranta, C. Recessive resistance genes against potyviruses are localized in colinear genomic regions of the tomato (Lycopersicon spp.) and pepper (Capsicum spp.) Genomes. Theor. Appl. Genet. 2002, 105, 855–861. [Google Scholar] [CrossRef]
- Diaz-Pendon, J.A.; Truniger, V.; Nieto, C.; Garcia-Mas, J.; Bendahmane, A.; Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004, 5, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Gallois, J.L.; Lesage, M.L.; Caranta, C. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genom. 2005, 274, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Scott, S.; Gergerich, R. Inheritance of a gene for resistance to Tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica 1992, 59, 9–17. [Google Scholar] [CrossRef]
- Boiteux, L.S.; Giordano, L.D. Genetic basis of resistance against two Tospovirus species in tomato (Lycopersicon-esculentum). Euphytica 1993, 71, 151–154. [Google Scholar] [CrossRef]
- Dianese, E.C.; Fonseca, M.E.N.; Inoue-Nagata, A.K.; Resende, R.O.; Boiteux, L.S. Search in Solanum (section Lycopersicon) germplasm for sources of broad-spectrum resistance to four Tospovirus species. Euphytica 2011, 180, 307–319. [Google Scholar] [CrossRef]
- Stevens, M.R.; Lamb, E.M.; Rhoads, D.D. Mapping the Sw-5 locus for Tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor. Appl. Genet. 1995, 90, 451–456. [Google Scholar] [CrossRef]
- Spassova, M.I.; Prins, T.W.; Folkertsma, R.T.; Klein-Lankhorst, R.M.; Hille, J.; Goldbach, R.W.; Prins, M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 2001, 7, 151–161. [Google Scholar] [CrossRef]
- Hanson, F.S. Viral Diseases of Tomato—Origins, Impact, and Future Prospects with a Focus on Tomato Spotted Wilt Virus and Tomato Yellow Leaf Curl Virus; IntechOpen: London, UK, 2022. [Google Scholar]
- Laterrot, H.; Pecaut, P. Gene Tm-2: New source. Rep. Tomato Genet. Coop. 1969, 19, 13–14. [Google Scholar]
- Lanfermeijer, F.C.; Jiang, G.Y.; Ferwerda, M.A.; Dijkhuis, J.; Haan, P.D.; Yang, R.; Hille, J. The durable resistance gene Tm-22 from tomato confers resistance against ToMV in tobacco and preserves its viral specificity. Plant Sci. 2004, 167, 687–692. [Google Scholar] [CrossRef]
- Ohmori, T.; Murata, M.; Motoyoshi, F. Molecular characterization of RAPD and SCAR markers linked to the Tm-1 locus in tomato. Theor. Appl. Genet. 1996, 92, 151–156. [Google Scholar] [CrossRef]
- Ohmori, T.; Murata, M.; Motoyoshi, F. Identification of RAPD markers linked to the Tm-2 locus in tomato. Theor. Appl. Genet. 1995, 90, 307–311. [Google Scholar] [CrossRef]
- Kuo, Y.W.; Falk, B.W. RNA interference approaches for plant disease control. BioTechniques 2020, 69, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pérez, M.G.; García-Luque, I.; Fraile, A.; García-Arenal, F. Mutations That Determine Resistance Breaking in a Plant RNA Virus Have Pleiotropic Effects on Its Fitness That Depend on the Host Environment and on the Type, Single or Mixed, of Infection. J. Virol. 2016, 90, 9128–9137. [Google Scholar] [CrossRef]
- Kumar, A.; Tiwari, K.L.; Datta, D.; Singh, M. Marker assisted gene pyramiding for enhanced Tomato leaf curl virus disease resistance in tomato cultivars. Biol. Plant 2014, 58, 792–797. [Google Scholar] [CrossRef]
- Kumar, A.; Jindal, S.K.; Dhaliwal, M.S.; Sharma, A.; Kaur, S.; Jain, S. Gene pyramiding for elite tomato genotypes against ToLCV (Begomovirus spp.), late blight (Phytophthora infestans) and RKN (Meloidogyne spp.) for northern India farmers. Physiol. Mol. Biol. Plants 2019, 25, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, S.; Grau, A.; Alonso, A.; Rubio, F.; Valero, M.; Ruiz, J.J. UMH 1422 and UMH 1415: Two Fresh-market Tomato Breeding Lines Resistant to Tomato Mosaic Virus and Tomato Spotted Wilt Virus. Hortscience A Publ. Am. Soc. Hortic. Sci. 2014, 49, 1465–1466. [Google Scholar] [CrossRef]
- Dormatey, R.; Sun, C.; Ali, K.; Coulter, J.A.; Bai, J. Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses. Agronomy 2020, 10, 1255. [Google Scholar] [CrossRef]
- García-Martínez, S.; Grau, A.; Alonso, A.; Rubio, F.; Valero, M.; Ruiz, J.J. UMH 1200, a breeding line within the Muchamiel tomato type resistant to three viruses. HortScience 2011, 46, 1054–1055. [Google Scholar] [CrossRef]
- Rubio, F.; García-Martínez, S.; Alonso, A.; Grau, A.; Valero, M.; Ruiz, J.J. Introgressing resistance genes into traditional tomato varie-ties: Effects on yield and quality. Acta Hortic. 2012, 935, 29–33. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Svoboda, P. Key Mechanistic Principles and Considerations Concerning RNA Interference. Front. Plant Sci. 2020, 11, 1237. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Prasad, M. Silencing AC1 of Tomato leaf curl virus using artificial microRNA confers resistance to leaf curl disease in transgenic tomato. Plant Cell Rep. 2020, 39, 1565–1579. [Google Scholar] [CrossRef]
- Singh, A.; Mohorianu, I.; Green, D.; Dalmay, T.; Dasgupta, I.; Mukherjee, S.K. Artificially induced phased siRNAs promote virus resistance in transgenic plants. Virology 2019, 537, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.V.; Choudhury, N.R.; Mukherjee, S.K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 2013, 172, 35–45. [Google Scholar] [CrossRef]
- Cisneros, A.E.; Carbonell, A. Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants. Plants 2020, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A. Secondary Small Interfering RNA-Based Silencing Tools in Plants: An Update. Front. Plant Sci. 2019, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Lison, P.; Daros, J.A. Multi-targeting of viral RNAs with synthetic trans-acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019, 100, 720–737. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Daròs, J.A. Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 2017, 18, 746–753. [Google Scholar] [CrossRef]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 2016, 228, 16–20. [Google Scholar] [CrossRef]
- Jiang, L.; Mu, R.; Wang, Z.; Liu, S.; Lu, D. Silencing P25, HC-Pro and Brp1 of Potato Virus (Viroid) Using Artificial microRNA Confers Resistance to PVX, PVY and PSTVd in Transgenic Potato. Potato Res. 2023, 66, 231–244. [Google Scholar] [CrossRef]
- Gaafar, Y.Z.A.; Ziebell, H. Novel Targets for Engineering Physostegia Chlorotic Mottle and Tomato Brown Rugose Fruit Virus-resistant Tomatoes: In Silico Prediction of Tomato microRNA Targets. PeerJ 2020, 8, e10096. [Google Scholar]
- Cisneros, A.E.; Martín-García, T.; Primc, A.; Kuziuta, W.; Sánchez-Vicente, J.; Aragonés, V.; Daròs, J.A.; Carbonell, A. Transgene-free, virus-based gene silencing in plants by artificial microRNAs derived from minimal precursors. Nucleic Acids Res. 2023, gkad747. [Google Scholar] [CrossRef]
- Halder, K.; Chaudhuri, A.; Abdin, M.Z.; Majee, M.; Datta, A. RNA Interference for Improving Disease Resistance in Plants and Its Relevance in This Clustered Regularly Interspaced Short Palindromic Repeats-Dominated Era in Terms of dsRNA-Based Biopesticides. Front. Plant Sci. 2022, 13, 885128. [Google Scholar] [CrossRef]
- Abel, P.P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of Disease Development in Transgenic Plants that Express the Tobacco Mosaic Virus Coat Protein Gene. Science 1986, 232, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Reimann-Philipp, U.; Beachy, R.N. Coat protein-mediated resistance in transgenic tobacco expressing the tobacco mosaic virus coat protein from tissue-specific promoters. Mol. Plant-Microbe Interact. 1993, 6, 323–330. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.J.; Ellis, P.J. Resistance to tomato spotted wilt virus infection in transgenic tobacco expressing the viral nucleocapsid gene. Mol. Plant-Microbe Interact. 1992, 5, 34–40. [Google Scholar] [CrossRef]
- Praveen, S.; Mishra, A.K.; Dasgupta, A. Antisense Suppression of Replicase Gene Expression Recovers Tomato Plants from Leaf Curl Infection. Plant Sci. 2005, 168, 1011–1014. [Google Scholar] [CrossRef]
- Raj, S.K.; Singh, R.; Pandey, S.K.; Singh, B.P. Agrobacterium-mediated Tomato Transformation and Regeneration of Transgenic Lines Expressing Tomato Leaf Curl Virus Coat Protein Gene for Resistance Against TLCV Infection. Curr. Sci. 2005, 88, 1674–1679. [Google Scholar]
- Mubin, M.; Mansoor, S.; Hussain, M.; Zafar, Y. Silencing of the AV2 Gene by Antisense RNA Protects Transgenic Plants Against a Bipartite Begomovirus. Virol. J. 2007, 4, 10. [Google Scholar] [CrossRef]
- Ferreira, S.A.; Pitz, K.Y.; Manshardt, R.; Zee, F.; Fitch, M.; Gonsalves, D. Virus Coat Protein Transgenic Papaya Provides Practical Control of Papaya Ringspot Virus in Hawaii. Plant Dis. 2002, 86, 101–105. [Google Scholar] [CrossRef]
- Konakalla, N.C.; Kaldis, A.; Berbati, M.; Masarapu, H.; Voloudakis, A.E. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 2016, 244, 961–969. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Mishra, A.K.; Praveen, S. Hairpin RNA-mediated Strategies for Silencing of Tomato Leaf Curl Virus AC1 and AC4 Genes for Effective Resistance in Plants. Oligonucleotide 2007, 17, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Koulagi, R.; Banerjee, S.; Gawade, B.H.; Singh, A.K.; Jain, P.K.; Praveen, S.; Subramaniam, K.; Sirohi, A. Host-delivered RNA Interference in Tomato for Mediating Resistance Against Meloidogyne Incognita and Tomato Leaf Curl Virus. Plant Cell Tiss. Organ Cult. 2020, 143, 345–361. [Google Scholar] [CrossRef]
- Ribeiro, S.G.; Lohuis, H.; Goldbach, R.; Prins, M. Tomato Chlorotic Mottle Virus is a Target of RNA Silencing but the Presence of Specific Short Interfering RNAs does not Guarantee Resistance in Transgenic Plants. J. Virol. 2007, 81, 1563–1573. [Google Scholar] [CrossRef]
- Missiou, A.; Kalantidis, K.; Boutla, A.; Tzortzakaki, S.; Tabler, M.; Tsagris, M. Generation of Transgenic Potato Plants Highly Resistant to Potato Virus Y (PVY) through RNA Silencing. Mol. Breed. 2004, 14, 185–197. [Google Scholar] [CrossRef]
- McCue, K.F.; Ponciano, G.; Rockhold, D.R.; Whitworth, J.L.; Gray, S.M.; Fofanov, Y.; Belknap, W.R. Generation of PVY Coat Protein siRNAs in Transgenic Potatoes Resistant to PVY. Am. J. Pot. Res. 2012, 89, 374–383. [Google Scholar] [CrossRef]
- Prasad Babu, K.; Maligeppagol, M.; Asokan, R.; Krishna Reddy, M. Screening of a Multi-virus Resistant RNAi Construct in Cowpea through Transient Vacuum Infiltration Method. Virus Dis. 2019, 30, 269–278. [Google Scholar] [CrossRef]
- Al-Roshdi, M.R.; Ammara, U.; Khan, J.; Al-Sadi, A.M.; Shahid, M.S. Artificial microRNA-mediated Resistance Against Oman strain of Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2023, 30, 14:1164921. [Google Scholar] [CrossRef]
- Carbonell, A.; Lopez, C.; Daròs, J.-A. Fast-Forward Identification of Highly Effective Artificial Small RNAs Against Different Tomato spotted wilt virus Isolates. Mol. Plant Microbe Interact. 2019, 32, 142–156. [Google Scholar] [CrossRef] [PubMed]
- López-Dolz, L.; Spada, M.; Daròs, J.-A.; Carbonell, A. Fine-Tune Control of Targeted RNAi Efficacy by Plant Artificial Small RNAs. Nucleic Acids Res. 2020, in press. [Google Scholar] [CrossRef]
- Song, Y.Z.; Han, Q.J.; Jiang, F.; Sun, R.Z.; Fan, Z.H.; Zhu, C.X.; Wen, F.J. Effects of the Sequence Characteristics of miRNAs on Multi-viral Resistance Mediated by Single amiRNAs in Transgenic Tobacco. Plant Physiol. Biochem. 2014, 77, 90–98. [Google Scholar] [CrossRef]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M.M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 2018, 13, e1525996. [Google Scholar] [CrossRef]
- Wang, N.; Tang, C.L.; Fan, X.; He, M.Y.; Gan, P.F.; Zhang, S.; Hu, Z.Y.; Wang, X.D.; Yan, T.; Shu, W.X.; et al. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rsust fungi. Cell 2022, 185, 2961–2974. [Google Scholar] [CrossRef]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.-Y. CRISPR/Cas9-mediated generation of pathogen resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 2021, 22, 1878. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.-Y. CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant. 2021, 14, 127–150. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Zhang, F.; Koonin, E.V. SnapShot: Class 1 CRISPRCas systems. Cell 2017, 168, 946–946.e1. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Zhang, F.; Koonin, E.V. SnapShot: Class 2 CRISPRCas systems. Cell 2017, 168, 328–328.e1. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, H.W.; Zhang, Y.; Wang, Y.P.; Gao, C.X. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef]
- van Beljouw, S.P.B.; Sanders, J.; Rodríguez-Molina, A.; Brouns, S.J.J. RNA-targeting CRISPR-Cas systems. Nat. Rev. Microbiol. 2023, 21, 21–34. [Google Scholar] [CrossRef]
- Lebedeva, M.V.; Nikonova, E.Y.; Terentiev, A.A.; Taranov, V.V.; Babakov, A.V.; Nikonov, O.S. VPg of Potato Virus Y and Potato Cap-Binding eIF4E Factors: Selective Interaction and Its Supposed Mechanism. Biochem 2021, 86, 1128–1138. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Shakila, A.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef]
- Ali, Z.; Shakila, A.; Tashkandi, M.; Zaidi, S.S.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: Differential interference and evasion. Sci. Rep. 2016, 6, 26912. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessoryproteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Smargon, A.A.; Cox, D.B.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 2018, 70, 327–339. [Google Scholar]
- Yan, Z.; Pérez-de-Castro, A.; Díez, M.J.; Hutton, S.F.; Visser, R.G.F.; Wolters, A.-M.A.; Bai, Y.L.; Li, J.M. Resistance to Tomato Yellow Leaf Curl Virus in Tomato Germplasm. Front. Plant Sci. 2018, 9, 1198. [Google Scholar] [CrossRef]
- Ghorbani Faal, P.; Farsi, M.; Seifi, A.; Mirshamsi Kakhki, A. Virus-induced CRISPR-Cas9 system improved resistance against tomato yellow leaf curl virus. Mol. Biol. Rep. 2020, 47, 3369–3376. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Q.F.; Yi, X.; An, H.; Zhao, Y.L.; Ma, S.Q.; Zhou, G.H. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Mahas, A.; Butt, H.; Aljedaani, F.; Mahfouz, M. Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses 2018, 10, 732. [Google Scholar] [CrossRef]
- Zhan, X.H.; Zhang, F.J.; Zhong, Z.Y.; Chen, R.H.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.H.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.L.; Ye, J.J.; Cao, X.; Xu, C.H.; Chen, B.; An, H.; Jiao, Y.T.; Zhang, F.S.; Yang, X.; et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185–1187. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.C.; Pan, Z.Y.; Wang, X.; Bian, X.F.; Wang, W.C.; Liang, Q.; Kou, M.; Ji, H.T.; Li, Y.J.; Ma, D.F.; et al. Targeting of SPCSV-RNase3 via CRISPR-Cas13 confers resistance against sweet potato virus disease. Mol. Plant Pathol. 2022, 23, 104–117. [Google Scholar] [CrossRef]
- Atarashi, H.; Jayasinghe, W.H.; Kwon, J.; Kim, H.; Taninaka, Y.; Igarashi, M.; Ito, K.; Yamada, T.; Masuta, C.; Nakahara, K.S. Artificially edited alleles of the eukaryotic translation initiation factor 4E1 gene differentially reduce susceptibility to cucumber mosaicvirus and potato virus Y in tomato. Front. Microbiol. 2020, 11, 564310. [Google Scholar] [CrossRef]
- Kuroiwa, K.; Danilo, B.; Perrot, L.; Thenault, C.; Veillet, F.; Delacote, F.; Duchateau, P.; Nogué, F.; Mazier, M.; Gallois, J.L. An iterative gene-editing strategy broadens eIF4E1 genetic diversity in Solanum lycopersicum and generates resistance to multiple potyvirus isolates. Plant Biotechnol. J. 2023, 21, 918–930. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Venkatesh, J.; Lee, J.H.; Kim, J.; Lee, H.E.; Kim, D.S.; Kang, B.C. Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus. Front. Plant Sci. 2020, 11, 1098. [Google Scholar] [CrossRef]
- Moury, B.; Lebaron, C.; Szadkowski, M.; Ben Khalifa, M.; Girardot, G.; Bolou Bi, B.A.; Koné, D.; Nitiema, L.W.; Fakhfakh, H.; Gallois, J.L. Knock-out mutation of eukaryotic initiation factor 4E2 (eIF4E2) confers resistance to pepper veinal mottle virus in tomato. Virology 2020, 539, 11–17. [Google Scholar] [CrossRef]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef]
- Kravchik, M.; Shnaider, Y.; Abebie, B.; Shtarkman, M.; Kumari, R.; Kumar, S.; Leibman, D.; Spiegelman, Z.; Gal-On, A. Knockout of SlTOM1 and SlTOM3 results in differential resistance to tobamovirus in tomato. Mol. Plant Pathol. 2022, 23, 1278–1289. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H. Susceptibility Genes to Plant Viruses. Viruses 2018, 10, 484. [Google Scholar] [CrossRef]
- Kan, J.; Cai, Y.; Cheng, C.; Jiang, C.; Jin, Y.; Yang, P. Simultaneous editing of host factor gene TaPDIL5-1 homoeoalleles confers wheat yellow mosaic virus resistance in hexaploid wheat. New Phytol. 2022, 234, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Eskelin, K.; Lohmus, A.; Makinen, K. Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 2014, 95, 1415–1429. [Google Scholar] [CrossRef]
- Yamanaka, T.; Ohta, T.; Takahashi, M.; Meshi, T.; Schmidt, R.; Dean, C.; Naito, S.; Ishikawa, M. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 2000, 97, 10107–10112. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L.; et al. A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota. PLoS Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef] [PubMed]
- Turina, M.; Kormelink, R.; Resende, R.O. Resistance to Tospoviruses in Vegetable Crops: Epidemiological and Molecular Aspects. Annu. Rev. Phytopathol. 2016, 54, 347–371. [Google Scholar] [CrossRef]
- Chinnaiah, S.; Gautam, S.; Workneh, F.; Crosby, K.; Rush, C.; Gadhave, K.R. First report of Sw-5 resistance-breaking strain of tomato spotted wilt orthotospovirus infecting tomato in Texas. Plant Dis. 2023, 107. [Google Scholar] [CrossRef]
- Zheng, K.Y.; Chen, T.C.; Wu, K.; Kang, Y.C.; Yeh, S.D.; Zhang, Z.K.; Dong, J.H. Characterization of a New Orthotospovirus from Chilli Pepper in Yunnan Province, China. Plant Dis. 2020, 104, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Helderman, T.A.; Deurhof, L.; Bertran, A.; Boeren, S.; Fokkens, L.; Kormelink, R.; Joosten, M.H.A.J.; Prins, M.; van den Burg, H.A. An Isoform of the Eukaryotic Translation Elongation Factor 1A (eEF1a) Acts as a Pro-Viral Factor Required for Tomato Spotted Wilt Virus Disease in Nicotiana benthamiana. Viruses 2021, 13, 2190. [Google Scholar] [CrossRef]
- Komoda, K.; Ishibashi, K.; Kawamura-Nagaya, K.; Ishikawa, M. Possible involvement of eEF1A in Tomato spotted wilt virus RNA synthesis. Virology 2014, 468–470, 81–87. [Google Scholar] [CrossRef]
- Helderman, T.A.; Deurhof, L.; Bertran, A.; Richard, M.M.S.; Kormelink, R.; Prins, M.; Joosten, M.H.A.J.; van den Burg, H.A. Members of the ribosomal protein S6 (RPS6) family act as pro-viral factor for tomato spotted wilt orthotospovirus infectivity in Nicotiana benthamiana. Mol. Plant Pathol. 2022, 23, 431–446. [Google Scholar] [CrossRef]
- Spiechowicz, M.; Zylicz, A.; Bieganowski, P.; Kuznicki, J.; Filipek, A. Hsp70 is a new target of Sgt1--an interaction modulated by S100A6. Biochem. Biophys. Res. Commun. 2007, 357, 1148–1153. [Google Scholar] [CrossRef]
- Qian, X.; Xiang, Q.; Yang, T.Q.; Ma, H.Y.; Ding, X.S.; Tao, X.R. Molecular Co-Chaperone SGT1 Is Critical for Cell-to-Cell Movement and Systemic Infection of Tomato Spotted Wild Virus in Nicotiana benthamiana. Viruses 2018, 10, 647. [Google Scholar] [CrossRef]
- Gao, X.H.; Chen, J.L.; Dai, X.H.; Zhang, D.D.; Zha, Y.D. An effective strategy for reliably isolating heritable and Cas9-free arabidopsis mutants generated by CRISPR/Cas9 mediated genome editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef]
- Lin, C.S.; Hsu, C.T.; Yuan, Y.H.; Zheng, P.X.; Wu, F.H.; Cheng, Q.W.; Wu, Y.L.; Wu, T.L.; Lin, S.; Yue, J.J.; et al. DNA-free CRISPR-Cas9 gene editing of wild tetraploid tomato Solanum peruvianum using protoplast regeneration. Plant Physiol. 2022, 188, 1917–1930. [Google Scholar] [CrossRef]
- Č ermák, T.; Baltes, N.J.; Č egan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.L.; Liu, Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, C.L.; Sun, K.; Deng, Y.L.; Li, Z.H. Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol. Plant 2023, 16, 616–631. [Google Scholar] [CrossRef]
- Li, T.D.; Hu, J.C.; Sun, Y.; Li, B.S.; Zhang, D.L.; Li, W.L.; Liu, J.X.; Li, D.W.; Gao, C.X.; Zhang, Y.L.; et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef]
- Ma, X.N.; Zhang, X.Y.; Liu, H.M.; Li, Z.H. Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef]
- Varanda, C.M.; Félix, M.D.R.; Campos, M.D.; Patanita, M.; Materatski, P. Plant Viruses: From Targets to Tools for CRISPR. Viruses 2021, 13, 141. [Google Scholar] [CrossRef]
- Zhang, W.N.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell 2016, 28, 1237–1249. [Google Scholar] [CrossRef]
- Yang, L.; Machin, F.; Wang, S.F.; Saplaoura, E.; Kragler, F. Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat. Biotechnol. 2023, 41, 958–967. [Google Scholar] [CrossRef]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.E.A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 80. [Google Scholar] [CrossRef]
- Mubarik, M.S.; Wang, X.; Khan, S.H.; Ahmad, A.; Khan, Z.; Amjid, M.W.; Razzaq, M.K.; Ali, Z.; Azhar, M.T. Engineering broad-spectrum resistance to cotton leaf curl disease by CRISPR-Cas9 based multiplex editing in plants. GM Crops Food 2021, 12, 647–658. [Google Scholar] [CrossRef]
- Xiao, N.; Pan, C.; Li, Y.; Wu, Y.; Cai, Y.; Lu, Y.; Wang, R.; Yu, L.; Shi, W.; Kang, H.; et al. Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 2021, 22, 283. [Google Scholar] [CrossRef]
- Khera, P.; Pandey, M.K.; Mallikarjuna, N.; Sriswathi, M.; Roorkiwal, M.; Janila, P.; Sharma, S.; Shilpa, K.; Sudini, H.; Guo, B.; et al. Genetic imprints of domestication for disease resistance, oil quality, and yield component traits in groundnut (Arachis hy-pogaea L.). Mol. Genet. Genom. 2019, 294, 365–378. [Google Scholar] [CrossRef]
- Hua, K.; Zhang, J.; Botella, J.R.; Ma, C.; Kong, F.; Liu, B.; Zhu, J.K. Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant. 2019, 12, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Salava, H.; Thula, S.; Mohan, V.; Kumar, R.; Maghuly, F. Application of Genome Editing in Tomato Breeding: Mechanisms, Advances, and Prospects. Int. J. Mol. Sci. 2021, 22, 682. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Escudero, M.; Osorio, A.N.; Cortés, A.J. Integrative Pre-Breeding for Biotic Resistance in Forest Trees. Plants 2021, 10, 2022. [Google Scholar] [CrossRef]
- Qi, S.; Shen, Y.; Wang, X.; Zhang, S.; Li, Y.; Islam, M.M.; Wang, J.; Zhao, P.; Zhan, X.; Zhang, F.; et al. A new NLR gene for resistance to Tomato spotted wilt virus in tomato (Solanum lycopersicum). Theor. Appl. Genet. 2022, 135, 1493–1509. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Deng, M.; Jiang, S.; Zhu, H.; Li, Z.; Wang, Z.; Li, J.; Yang, Z.; Yue, Y.; Xu, J.; et al. Mapping and functional characterization of the tomato spotted wilt virus resistance gene SlCHS3 in Solanum lycopersicum. Mol. Breed. 2022, 42, 55. [Google Scholar] [CrossRef] [PubMed]
- Sáez, C.; Martínez, C.; Montero-Pau, J.; Esteras, C.; Sifres, A.; Blanca, J.; Ferriol, M.; López, C.; Picó, B. A Major QTL Located in Chromosome 8 of Cucurbita moschata Is Responsible for Resistance to Tomato Leaf Curl New Delhi Virus. Front. Plant Sci. 2020, 11, 207. [Google Scholar] [CrossRef]
| Resistance Gene Family | Resistance Genes | Source of Resistance Genes | Location on Chromosome | Gene Action | Efficiency | Resistance Mechanism | References |
|---|---|---|---|---|---|---|---|
| Ty gene family (against begomoviruses) | Ty-1 | Solanum chilense | 6 | Dominant | Broad-spectrum begomoviruses resistance | Ty-1 encodes an RNA-dependent RNA polymerase (RDR) involved in the RNA silencing pathway, increasing antiviral RNAi responses and the viral genome’s cytosine methylation. | [8,42,44,45,46,47,51,52,53,54] |
| Ty-2 | S. habrochaites | 11 (Long arm) | Dominant | TYLCV resistance | Ty-2 encodes a nucleotide-binding leucine-rich repeat (NLR) protein. The Ty-2 could recognize TYLCV Rep/C1 protein and induce hypersensitive responses (HR) in host plant. | ||
| Ty-3 | S. chilense | 6 | Dominant | Complementary resistance | Ty-3 encodes an RNA-dependent RNA polymerase (RDR) involved in the RNA silencing pathway. | ||
| Ty-4 | S. chilense | 3 | Dominant | Increase virus resistance in combination with Ty-3 | Not reported. | ||
| Ty-5 | Tyking | 4 | Recessive | Broad-spectrum resistance | Encodes messenger RNA (mRNA) surveillance factor Pelota. Silencing of Pelota in a susceptible line rendered the transgenic plants highly resistant. | ||
| Ty-6 | S. chilense | 10 | Dominant | Complements the resistance conferred by Ty-3 and Ty-5 | Not reported. | ||
| Sw gene family (against orthotospoviruses) | Sw-1a | Lycopersicum pimpinellifolium | Not reported | Dominant | Some degree of resistance to specific TSWV | Not reported. | [10,50,52,53,54] |
| Sw-1b | L. pimpinellifolium | Not reported | Dominant | Some degree of resistance to specific TSWV | Not reported. | ||
| Sw-2 | L. pimpinellifolium | Not reported | Recessive | Some degree of resistance to specific TSWV | Not reported. | ||
| Sw-3 | L. pimpinellifolium | Not reported | Recessive | Some degree of resistance to specific TSWV | Not reported. | ||
| Sw-4 | L. pimpinellifolium | Not reported | Recessive | Some degree of resistance to specific TSWV | Not reported. | ||
| Sw-5 | S. peruvianum | 9 | Dominant | High level of resistance to a wide range of TSWV | Sw-5 belongs to nucleotide-binding leucine-rich repeat (NB-LRR) type R gene. Sw-5 confers resistance by recognizing a 21-amino-acid peptide region of the viral movement protein NSm, triggering immunity response. | ||
| Sw-6 | L. pimpinellifolium | Not reported | Incompletely Dominant | Some degree of resistance to specific TSWV | Not reported | ||
| Sw-7 | L. chilense | 12 | Dominant | Resistance to a wide range of TSWV | Involved in pathogenesis-related (PR) proteins PR1 and PR5-related resistance process. | ||
| Tm gene family (against tobamoviruses) | Tm-1 | S. habrochaites | 2 | Incompletely Dominant | TMV partial resistance | Tm-1 encodes a protein that binds ToMV replication proteins and inhibits the RNA-dependent RNA replication of ToMV. | [14,55,56,57,58,59,60] |
| Tm-2 | S. peruvianum | 9 | Dominant | TMV partial resistance | Tm-2 belongs to nucleotide-binding leucine-rich repeat (NB-LRR) type R gene, which can recognize movement proteins (MPs) of TMV and ToMV and activate a resistance response. | ||
| Tm-22 | S. peruvianum | 9 | Dominant | Confers a more effective TMV resistance | Tm-22 belongs to nucleotide-binding leucine-rich repeat (NB-LRR) type R gene, which can recognize movement proteins (MPs) of TMV and ToMV and activate a resistance response. | ||
| Pot-1 gene (against potyviruses) | Pot-1 | L. hirsutum | 3 | Recessive | PVY resistance | Tomato Pot-1 is the orthologue of the pepper pvr2-eIF4E gene, encoding the plant-susceptible eIF4E1 translation initiation factor protein. Duplicate recessive Pot-1 genes interrupt the interaction of the potyviruses VPg protein with the eIF4E1, suppressing virus replication. | [61,62,63] |
| Methods | Characteristics | Challenges | Suggestions and Future Prospects |
|---|---|---|---|
| MAS-based hybrid breeding |
|
|
|
| RNAi-based transgenic breeding |
|
|
|
| CRISPR/Cas-based gene editing |
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahriari, Z.; Su, X.; Zheng, K.; Zhang, Z. Advances and Prospects of Virus-Resistant Breeding in Tomatoes. Int. J. Mol. Sci. 2023, 24, 15448. https://doi.org/10.3390/ijms242015448
Shahriari Z, Su X, Zheng K, Zhang Z. Advances and Prospects of Virus-Resistant Breeding in Tomatoes. International Journal of Molecular Sciences. 2023; 24(20):15448. https://doi.org/10.3390/ijms242015448
Chicago/Turabian StyleShahriari, Zolfaghar, Xiaoxia Su, Kuanyu Zheng, and Zhongkai Zhang. 2023. "Advances and Prospects of Virus-Resistant Breeding in Tomatoes" International Journal of Molecular Sciences 24, no. 20: 15448. https://doi.org/10.3390/ijms242015448
APA StyleShahriari, Z., Su, X., Zheng, K., & Zhang, Z. (2023). Advances and Prospects of Virus-Resistant Breeding in Tomatoes. International Journal of Molecular Sciences, 24(20), 15448. https://doi.org/10.3390/ijms242015448






