Pigment Identification and Gene Expression Analysis during Erythrophore Development in Spotted Scat (Scatophagus argus) Larvae
Abstract
:1. Introduction
2. Results
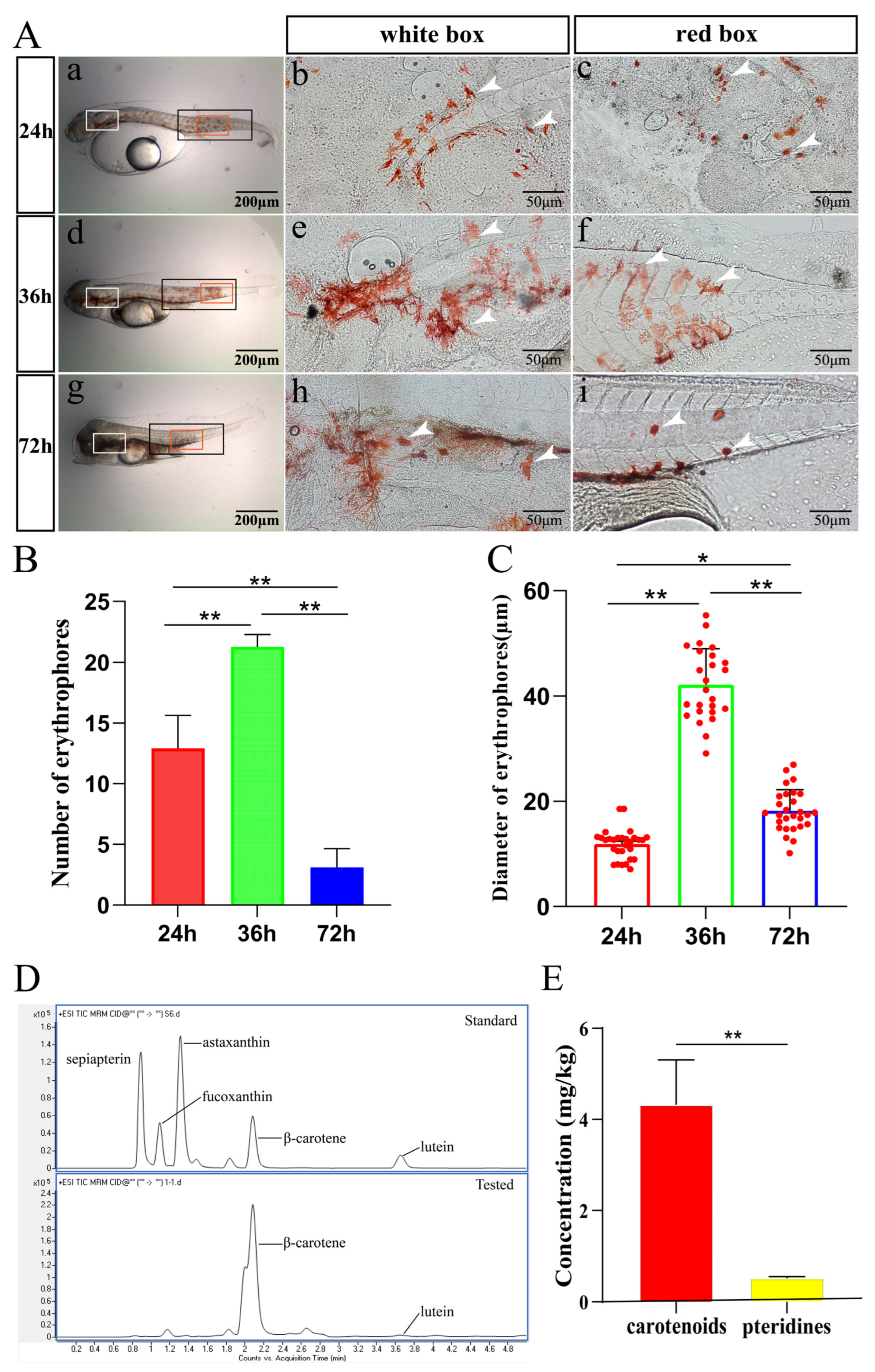
2.1. Microscopic Observation of Erythrophores in Spotted Scat during Hatching
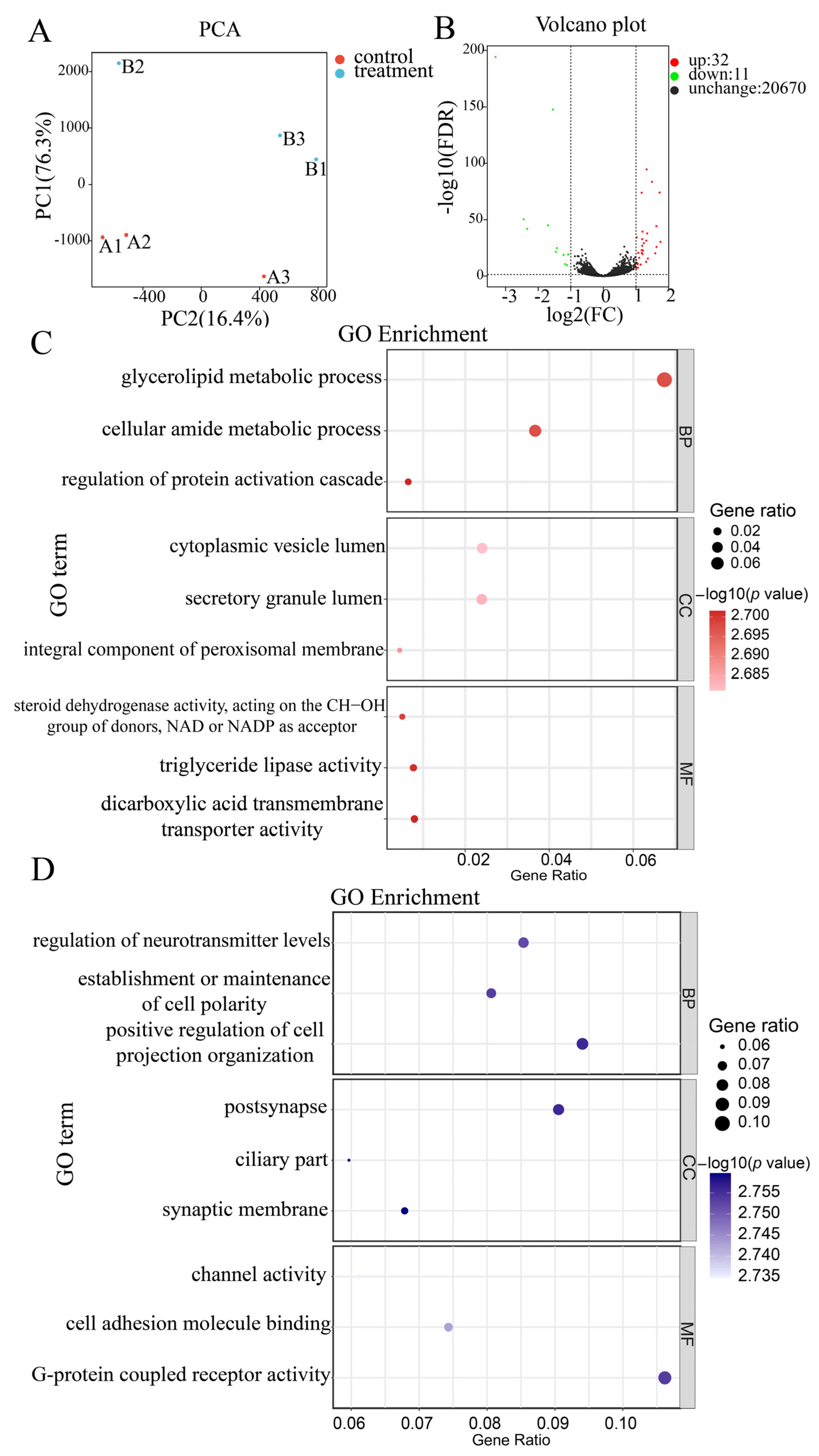
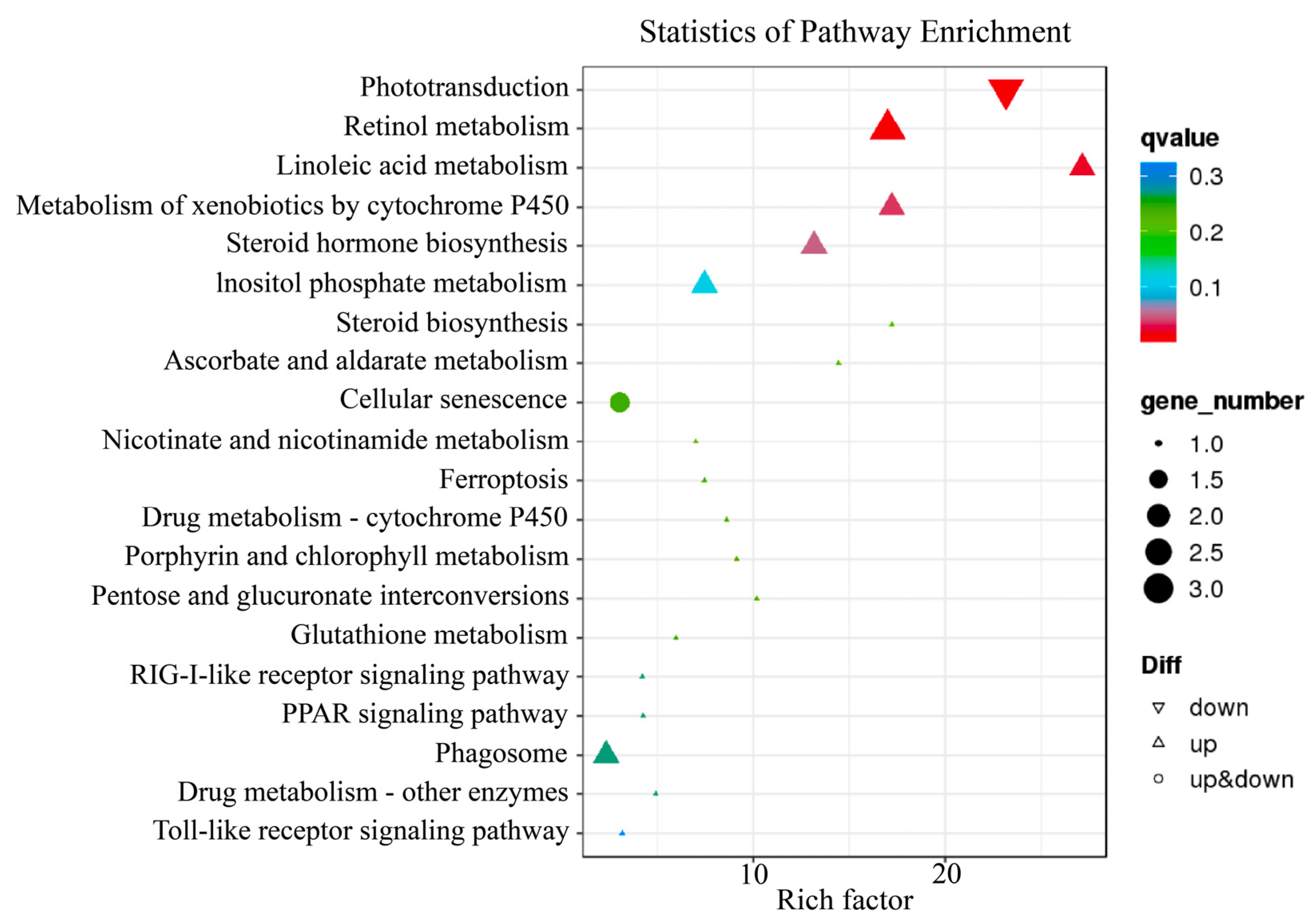
2.2. Illumina Sequencing and Annotation, Functional Enrichment Analyses of Differentially Expressed Genes (DEGs)
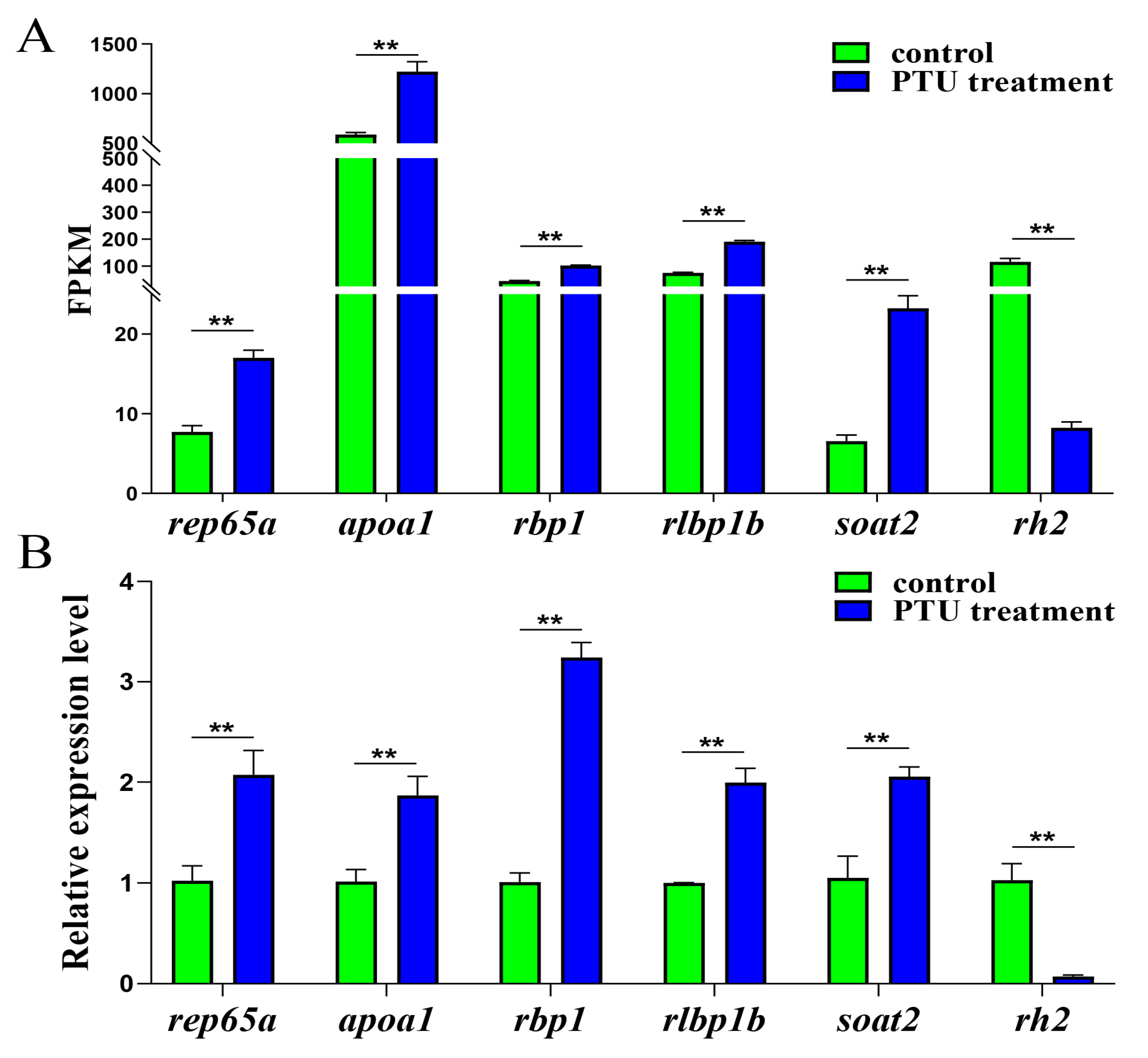
2.3. Validation of RNA-Seq Data with qRT-PCR
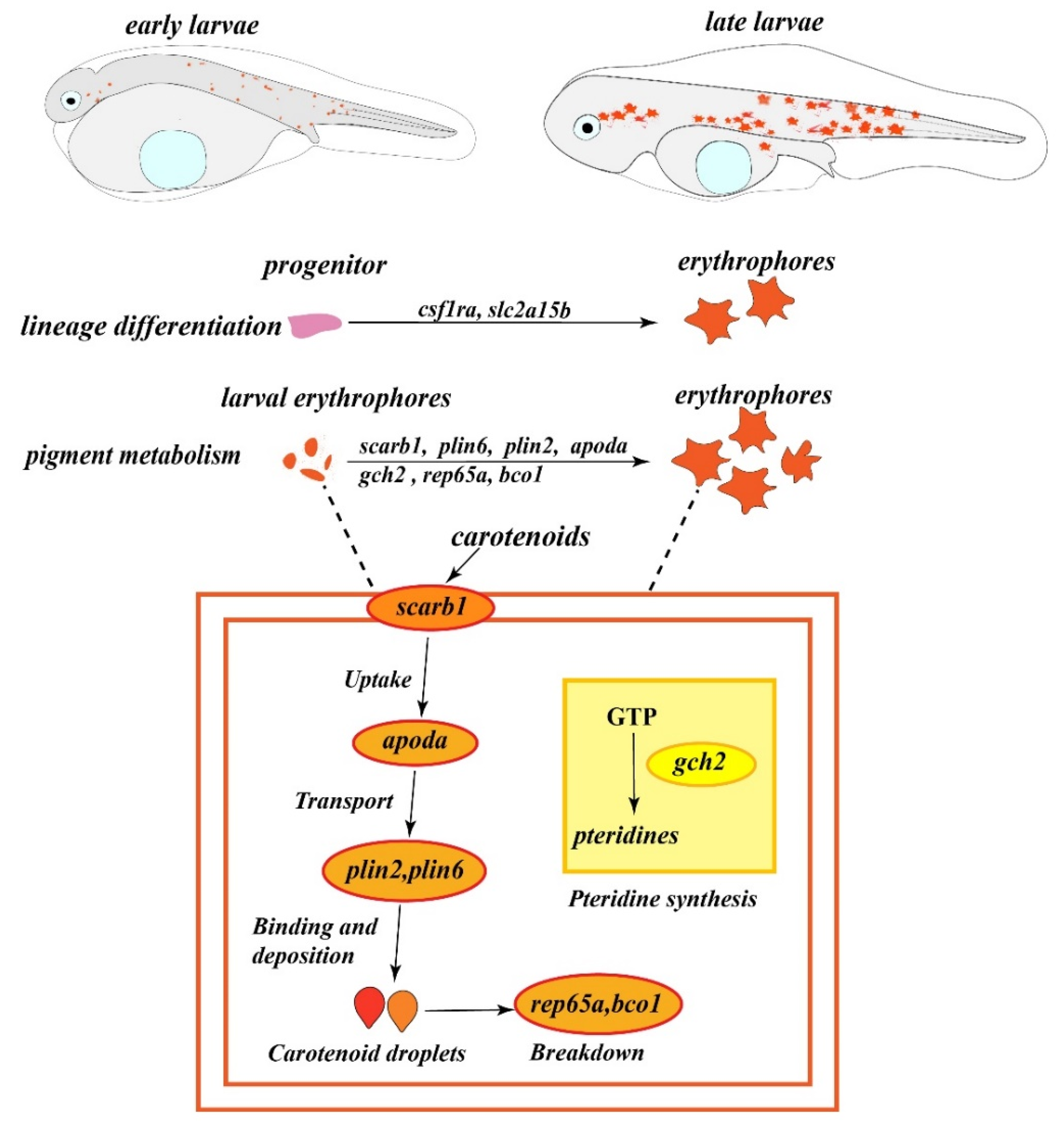
2.4. Characteristics and Pigments Responsible for Red Coloration in Spotted Scat Larvae
2.5. Expression of Genes Responsible for Red Coloration in Spotted Scat Larvae
3. Discussion
3.1. Effects of PTU on Melanin Pigment Formation and Gene Expression
3.2. Characteristics, Pigment Identification, and Expression of Genes Responsible for Red Coloration
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Design, Fish Treatment, and Sample Collection
4.3. Reagents and Preparation of Standard Stock Solutions
4.4. Pigment Extraction and Saponification Procedure
4.5. Identification of Pigments Using LC–MS
4.6. Spectrophotometry
4.7. Total RNA Extraction, Library Construction, and Sequencing
4.8. Data Filtering, Read Mapping, and Differential Expression Analysis
4.9. Validation of Transcriptome Data by qRT-PCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hubbard, J.K.; Uy, J.A.C.; Hauber, M.E.; Hoekstra, H.E.; Safran, R.J. Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. TIG 2010, 26, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Cal, L.; Suarez-Bregua, P.; Cerdá-Reverter, J.M.; Braasch, I.; Rotllant, J. Fish pigmentation and the melanocortin system. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2017, 211, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Lu, G.; Yin, H.; Wang, L.; Atuganile, M.; Dong, Z. Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev. Aquac. 2021, 13, 2395–2412. [Google Scholar] [CrossRef]
- Fujii, R. The Regulation of Motile Activity in Fish Chromatophores: Regulation of Fish Chromatophore Motility. Pigment Cell Res. 2000, 13, 300–319. [Google Scholar] [CrossRef]
- Silver, D.L.; Hou, L.; Pavan, W.J. The genetic regulation of pigment cell development. Adv. Exp. Med. Biol. 2006, 589, 155–169. [Google Scholar] [PubMed]
- Singh, A.P.; Nüsslein-Volhard, C. Zebrafish Stripes as a Model for Vertebrate Colour Pattern Formation. Curr. Biol. 2015, 25, R81–R92. [Google Scholar] [CrossRef]
- Rocha, M.; Singh, N.; Ahsan, K.; Beiriger, A.; Prince, V.E. Neural crest development: Insights from the zebrafish. Dev. Dyn. 2020, 249, 88–111. [Google Scholar] [CrossRef]
- Je, M.; Sm, H. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 2008, 317, 508–522. [Google Scholar]
- Curran, K.; Lister, J.A.; Kunkel, G.R.; Prendergast, A.; Parichy, D.M.; Raible, D.W. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 2010, 344, 107–118. [Google Scholar] [CrossRef]
- Lopes, S.S.; Yang, X.; Müller, J.; Carney, T.J.; McAdow, A.R.; Rauch, G.J.; Jacoby, A.S.; Hurst, L.D.; Delfino-Machín, M.; Haffter, P.; et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008, 4, e1000026. [Google Scholar] [CrossRef]
- Hoekstra, H.E. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 2006, 97, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, I.; McDonald, T.; Hesslinger, C.; Pelletier, I.; Boyle, P. Development of the pteridine pathway in the zebrafish, Danio rerio. J. Biol. Chem. 2000, 275, 18926–18932. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Hamamoto, A.; Takahashi, A.; Saito, Y. Dimerization of melanocortin receptor 1 (MC1R) and MC5R creates a ligand-dependent signal modulation: Potential participation in physiological color change in the flounder. Gen. Comp. Endocrinol. 2016, 230–231, 103–109. [Google Scholar] [CrossRef]
- Matsuda, N.; Kasagi, S.; Nakamaru, T.; Masuda, R.; Takahashi, A.; Tagawa, M. Left-right pigmentation pattern of Japanese flounder corresponds to expression levels of melanocortin receptors (MC1R and MC5R), but not to agouti signaling protein 1 (ASIP1) expression. Gen. Comp. Endocrinol. 2018, 262, 90–98. [Google Scholar] [CrossRef]
- Liang, Y.; Grauvogl, M.; Meyer, A.; Kratochwil, C.F. Functional conservation and divergence of color-pattern-related agouti family genes in teleost fishes. J. Exp. Zoolog. B Mol. Dev. Evol. 2021, 336, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, R.D.; Ozakinci, G.; Perrett, D.I. Attractive skin coloration: Harnessing sexual selection to improve diet and health. Evol. Psychol. Int. J. Evol. Approaches Psychol. Behav. 2012, 10, 842–854. [Google Scholar] [CrossRef]
- McGraw, K.J.; Ardia, D.R. Carotenoids, immunocompetence, and the information content of sexual colors: An experimental test. Am. Nat. 2003, 162, 704–712. [Google Scholar] [CrossRef]
- Blount, J.D.; McGraw, K.J. Carotenoids: Natural Functions. In Signal Functions of Carotenoid Colouration; Britton, G., Liaaen-Jensen, S., Pfander, H., Eds.; Birkhäuser: Basel, Switzerland, 2008; Volume 4, pp. 213–236. [Google Scholar]
- Dale, J.; Dey, C.J.; Delhey, K.; Kempenaers, B.; Valcu, M. The effects of life history and sexual selection on male and female plumage colouration. Nature 2015, 527, 367–370. [Google Scholar] [CrossRef]
- Walsh, N.; Dale, J.; McGraw, K.J.; Pointer, M.A.; Mundy, N.I. Candidate genes for carotenoid coloration in vertebrates and their expression profiles in the carotenoid-containing plumage and bill of a wild bird. Proceedings. Proc. Biol. Sci. 2012, 279, 58–66. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R.; Caramujo, M.J. Carotenoids in Aquatic Ecosystems and Aquaculture: A Colorful Business with Implications for Human Health. Front. Mar. Sci. 2017, 4, 93. [Google Scholar] [CrossRef]
- Toews, D.P.L.; Hofmeister, N.R.; Taylor, S.A. The Evolution and Genetics of Carotenoid Processing in Animals. Trends Genet. 2017, 33, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Sakudoh, T.; Kuwazaki, S.; Iizuka, T.; Narukawa, J.; Yamamoto, K.; Uchino, K.; Sezutsu, H.; Banno, Y.; Tsuchida, K. CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori. J. Lipid Res. 2013, 54, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Borel, P.; Lietz, G.; Goncalves, A.; Szabo de Edelenyi, F.; Lecompte, S.; Curtis, P.; Goumidi, L.; Caslake, M.J.; Miles, E.A.; Packard, C.; et al. CD36 and SR-BI are involved in cellular uptake of provitamin A carotenoids by Caco-2 and HEK cells, and some of their genetic variants are associated with plasma concentrations of these micronutrients in humans. J. Nutr. 2013, 143, 448–456. [Google Scholar] [CrossRef]
- Toomey, M.B.; Lopes, R.J.; Araújo, P.M.; Johnson, J.D.; Gazda, M.A.; Afonso, S.; Mota, P.G.; Koch, R.E.; Hill, G.E.; Corbo, J.C.; et al. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. USA 2017, 114, 5219–5224. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, P.; Larson, A.J.; Frederick, J.M.; Southwick, K.; Thulin, C.D.; Bernstein, P.S. Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J. Biol. Chem. 2004, 279, 49447–49454. [Google Scholar] [CrossRef]
- Li, B.; Vachali, P.; Frederick, J.M.; Bernstein, P.S. Identification of StARD3 as a lutein-binding protein in the macula of the primate retina. Biochemistry 2011, 50, 2541–2549. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, H.; Tan, K.; Ma, H.; Li, S.; Zheng, H. Identification of a key gene StAR-like-3 responsible for carotenoids accumulation in the noble scallop Chlamys nobilis. Food Chem. Mol. Sci. 2022, 4, 100072. [Google Scholar] [CrossRef]
- Granneman, J.G.; Kimler, V.A.; Zhang, H.; Ye, X.; Luo, X.; Postlethwait, J.H.; Thummel, R. Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. eLife 2017, 6, e21771. [Google Scholar] [CrossRef]
- Eriksson, J.; Larson, G.; Gunnarsson, U.; Bed’hom, B.; Tixier-Boichard, M.; Strömstedt, L.; Wright, D.; Jungerius, A.; Vereijken, A.; Randi, E.; et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008, 4, e1000010. [Google Scholar] [CrossRef]
- Borel, P. Genetic variations involved in interindividual variability in carotenoid status. Mol. Nutr. Food Res. 2012, 56, 228–240. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, M.; Yan, C.; Yang, H.; Wu, Z.; Liu, Y.; Su, N.; Hou, J.; Zhang, J.; Yang, F.; et al. CRISPR/Cas9-mediated deletion of β, β-carotene 9’, 10’-oxygenase gene (EcBCO2) from Exopalaemon carinicauda. Int. J. Biol. Macromol. 2020, 151, 168–177. [Google Scholar] [CrossRef]
- Lin, X.; Tian, C.; Huang, Y.; Shi, H.; Li, G. Comparative Transcriptome Analysis Identifies Candidate Genes Related to Black-Spotted Pattern Formation in Spotted Scat (Scatophagus argus). Animals 2021, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, J.R. An analysis of melanogenesis in differentiating pigment cells of ascidian embryos. Dev. Biol. 1966, 14, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J. Melanogenesis in amphibians. I. A study of the fine structure of the normal and phenylthiourea-treated pigmented epithelium in Rana pipiens tadpole eyes. Z. Zellforsch. Mikrosk. Anat. 1970, 103, 238–246. [Google Scholar] [CrossRef]
- Karlsson, J.; von Hofsten, J.; Olsson, P.E. Generating Transparent Zebrafish: A Refined Method to Improve Detection of Gene Expression During Embryonic Development. Mar. Biotechnol. 2001, 3, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Mourabit, S.; Edenbrow, M.; Croft, D.P.; Kudoh, T. Embryonic development of the self-fertilizing mangrove killifish Kryptolebias marmoratus. Dev. Dyn. 2011, 240, 1694–1704. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ptak, D.; Zhang, L.; Walls, E.; Zhong, W.; Leung, Y. Phenylthiourea Specifically Reduces Zebrafish Eye Size. PLoS ONE 2012, 7, e40132. [Google Scholar] [CrossRef]
- Antinucci, P.; Hindges, R. crystal-clear zebrafish for in vivo imaging. Sci. Rep. 2016, 6, 29490. [Google Scholar] [CrossRef]
- Chang, N.Y.; Chan, Y.J.; Ding, S.T.; Lee, Y.H.; HuangFu, W.C.; Liu, I.H. Sterol O-Acyltransferase 2 Contributes to the Yolk Cholesterol Trafficking during Zebrafish Embryogenesis. PLoS ONE 2016, 11, e0167644. [Google Scholar] [CrossRef]
- Dias, I.H.K.; Polidori, M.C.; Li, L.; Weber, D.; Stahl, W.; Nelles, G.; Grune, T.; Griffiths, H.R. Plasma levels of HDL and carotenoids are lower in dementia patients with vascular comorbidities. J. Alzheimers Dis. JAD 2014, 40, 399–408. [Google Scholar] [CrossRef]
- Qu, J.; Ko, C.W.; Tso, P.; Bhargava, A. Apolipoprotein A-IV: A Multifunctional Protein Involved in Protection against Atherosclerosis and Diabetes. Cells 2019, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- van der Vorst, E.P.C. High-Density Lipoproteins and Apolipoprotein A1. Subcell. Biochem. 2020, 94, 399–420. [Google Scholar] [PubMed]
- Chen, X.K.; Kwan, J.S.K.; Chang, R.C.C.; Ma, A.C.H. 1-phenyl 2-thiourea (PTU) activates autophagy in zebrafish embryos. Autophagy 2021, 17, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Pietzsch-Rohrschneider, I. The influence of phenylthiourea (PTU) on the pigment epithelium and the photoreceptor cells in the developing retina of Haplochromis burtoni (Cichlidae, Teleostei). Cell Tissue Res. 1977, 178, 279–283. [Google Scholar] [CrossRef]
- Musilova, Z.; Cortesi, F. The evolution of the green-light-sensitive visual opsin genes (RH2) in teleost fishes. Vis. Res. 2023, 206, 108204. [Google Scholar] [CrossRef]
- Hart, N.S.; Lamb, T.D.; Patel, H.R.; Chuah, A.; Natoli, R.C.; Hudson, N.J.; Cutmore, S.C.; Davies, W.I.L.; Collin, S.P.; Hunt, D.M. Visual Opsin Diversity in Sharks and Rays. Mol. Biol. Evol. 2020, 37, 811–827. [Google Scholar] [CrossRef]
- Goodman, D.S. Plasma retinol-binding protein. Ann. N. Y. Acad. Sci. 1980, 348, 378–390. [Google Scholar] [CrossRef]
- Goodman, D.S. Retinoid-binding proteins in plasma and in cells. Ann. N. Y. Acad. Sci. 1981, 359, 69–78. [Google Scholar] [CrossRef]
- Schlegel, D.K.; Ramkumar, S.; von Lintig, J.; Neuhauss, S.C. Disturbed retinoid metabolism upon loss of rlbp1a impairs cone function and leads to subretinal lipid deposits and photoreceptor degeneration in the zebrafish retina. eLife 2021, 10, e71473. [Google Scholar] [CrossRef]
- Saari, J.C.; Nawrot, M.; Kennedy, B.N.; Garwin, G.G.; Hurley, J.B.; Huang, J.; Possin, D.E.; Crabb, J.W. Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron 2001, 29, 739–748. [Google Scholar] [CrossRef]
- Flach, H.; Basten, T.; Schreiner, C.; Dietmann, P.; Greco, S.; Nies, L.; Roßmanith, N.; Walter, S.; Kühl, M.; Kühl, S.J. Retinol binding protein 1 affects Xenopus anterior neural development via all-trans retinoic acid signaling. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2021, 250, 1096–1112. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Bao, B.; Xie, Z.; Chen, X.; Li, B.; Jia, X.; Yao, Q.; Ortí, G.; Li, W.; Li, X.; et al. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat. Genet. 2017, 49, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, B.L.; Gallina, D.; Kahana, A. Phenothiourea sensitizes zebrafish cranial neural crest and extraocular muscle development to changes in retinoic acid and IGF signaling. PLoS ONE 2011, 6, e22991. [Google Scholar] [CrossRef]
- Ghosh, S.; Kumar, T.T.A.; Balasubramanian, T. Embryology of Maldives clownfish, Amphiprion nigripes (Amphiprioninae). J. Ocean Univ. China 2012, 11, 174–180. [Google Scholar] [CrossRef]
- Du, J.; Chen, H.; Mandal, B.K.; Wang, J.; Shi, Z.; Lu, G.; Wang, C. HDL receptor/Scavenger receptor B1-Scarb1 and Scarb1-like mediate the carotenoid-based red coloration in fish. Aquaculture 2021, 545, 737208. [Google Scholar] [CrossRef]
- Mookkan, M.; Muniyandi, K.; Palaniyandi, S. Carotenoid composition in wild-caught spotted scat (Scatophagus argus) broodstocks: Effects on gonad development. Fish Physiol. Biochem. 2022, 48, 991–1009. [Google Scholar] [CrossRef] [PubMed]
- Grether, G.F.; Cummings, M.E.; Hudon, J. Countergradient variation in the sexual coloration of guppies (Poecilia reticulata): Drosopterin synthesis balances carotenoid availability. Evolution 2005, 59, 175–188. [Google Scholar]
- Twyman, H.; Prager, M.; Mundy, N.I.; Andersson, S. Expression of a carotenoid-modifying gene and evolution of red coloration in weaverbirds (Ploceidae). Mol. Ecol. 2018, 27, 449–458. [Google Scholar] [CrossRef]
- Gazda, M.A.; Toomey, M.B.; Araújo, P.M.; Lopes, R.J.; Afonso, S.; Myers, C.A.; Serres, K.; Kiser, P.D.; Hill, G.E.; Corbo, J.C.; et al. Genetic Basis of De Novo Appearance of Carotenoid Ornamentation in Bare Parts of Canaries. Mol. Biol. Evol. 2020, 37, 1317–1328. [Google Scholar] [CrossRef]
- McGraw, K.J.; Toomey, M.B. Carotenoid accumulation in the tissues of zebra finches: Predictors of integumentary pigmentation and implications for carotenoid allocation strategies. Physiol. Biochem. Zool. PBZ 2010, 83, 97–109. [Google Scholar] [CrossRef]
- McLean, C.A.; Lutz, A.; Rankin, K.J.; Elliott, A.; Moussalli, A.; Stuart-Fox, D. Red carotenoids and associated gene expression explain colour variation in frillneck lizards. Proc. Biol. Sci. 2019, 286, 20191172. [Google Scholar] [CrossRef] [PubMed]
- Andrade, P.; Pinho, C.; Pérez I. de Lanuza, G.; Afonso, S.; Brejcha, J.; Rubin, C.J.; Wallerman, O.; Pereira, P.; Sabatino, S.J.; Bellati, A.; et al. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. USA 2019, 116, 5633–5642. [Google Scholar] [CrossRef] [PubMed]
- Sundvold, H.; Helgeland, H.; Baranski, M.; Omholt, S.W.; Våge, D.I. Characterisation of a novel paralog of scavenger receptor class B member I (SCARB1) in Atlantic salmon (Salmo salar). BMC Genet. 2011, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, S.J.; Garver, K.A.; Richard, J.; Devlin, R.H.; Lajoie, C.; Pitcher, T.E.; Heath, D.D. Significant differences in maternal carotenoid provisioning and effects on offspring fitness in Chinook salmon colour morphs. J. Evol. Biol. 2018, 31, 1876–1893. [Google Scholar] [CrossRef]
- Tyndale, S.; Letcher, R.; Heath, J.; Heath, D. Why are salmon eggs red? Egg carotenoids and early life survival of Chinook salmon (Oncorhynchus tshawytscha). Evol. Ecol. Res. 2008, 10, 1187–1199. [Google Scholar]
- Wilkins, L.G.E.; Marques da Cunha, L.; Menin, L.; Ortiz, D.; Vocat-Mottier, V.; Hobil, M.; Nusbaumer, D.; Wedekind, C. Maternal allocation of carotenoids increases tolerance to bacterial infection in brown trout. Oecologia 2017, 185, 351–363. [Google Scholar] [CrossRef]
- Roulin, A.; Ducrest, A.L. Genetics of colouration in birds. Semin. Cell Dev. Biol. 2013, 24, 594–608. [Google Scholar] [CrossRef]
- During, A.; Dawson, H.D.; Harrison, E.H. Carotenoid transport is decreased and expression of the lipid transporters SR-BI, NPC1L1, and ABCA1 is downregulated in Caco-2 cells treated with ezetimibe. J. Nutr. 2005, 135, 2305–2312. [Google Scholar] [CrossRef]
- Mazeikiene, A.; Karciauskaite, D.; Burokiene, N.; Daunoravicius, Z.; Kucinskiene, Z.A.; Kucinskas, V. Association of serum carotenoid concentration with single nucleotide polymorphisms in APOE, BCMO1, NPC1L1, CD36, GSTP1, ABCG5, FABP1, FABP2 and LIPC genes. Atherosclerosis 2016, 252, e79. [Google Scholar] [CrossRef]
- Huang, D.; Lewis, V.M.; Foster, T.N.; Toomey, M.B.; Corbo, J.C.; Parichy, D.M. Development and genetics of red coloration in the zebrafish relative Danio albolineatus. eLife 2021, 10, e70253. [Google Scholar] [CrossRef]
- Tripathy, P.S.; Devi, N.C.; Parhi, J.; Priyadarshi, H.; Patel, A.B.; Pandey, P.K.; Mandal, S.C. Molecular Mechanisms of Natural Carotenoid-based Pigmentation of Queen Loach, Botia dario (Hamilton, 1822) Under Captive Condition. Sci. Rep. 2019, 9, 12585. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.C.; Tripathy, P.S.; Khatei, A.; Behera, D.U.; Ghosh, A.; Pandey, P.K.; Parhi, J. Genetics of colour variation in wild versus cultured queen loach, Botia dario (Hamilton, 1822). Genomics 2020, 112, 3256–3267. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, E.; Uppal, S.; Rogozin, I.B.; Gentleman, S.; Redmond, T.M. Evolutionary aspects and enzymology of metazoan carotenoid cleavage oxygenases. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2020, 1865, 158665. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Peng, L.; Ren, L.; Zhang, H.; Zou, L.; Liu, W.; Xiao, Y. Comparative transcriptome analysis of molecular mechanism underlying gray-to-red body color formation in red crucian carp (Carassius auratus, red var.). Fish Physiol. Biochem. 2017, 43, 1387–1398. [Google Scholar] [CrossRef]
- Lister, J.A. Larval but not adult xanthophore pigmentation in zebrafish requires GTP cyclohydrolase 2 ( gch2 ) function. Pigment Cell Melanoma Res. 2019, 32, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Lewis, V.M.; Saunders, L.M.; Larson, T.A.; Bain, E.J.; Sturiale, S.L.; Gur, D.; Chowdhury, S.; Flynn, J.D.; Allen, M.C.; Deheyn, D.D.; et al. Fate plasticity and reprogramming in genetically distinct populations of Danio leucophores. Proc. Natl. Acad. Sci. USA 2019, 116, 11806–11811. [Google Scholar] [CrossRef]
- Lencer, E.; Prekeris, R.; Artinger, K.B. Single-cell RNA analysis identifies pre-migratory neural crest cells expressing markers of differentiated derivatives. eLife 2021, 10, e66078. [Google Scholar] [CrossRef]
- Dm, P.; Jm, T. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Dev. Camb. Engl. 2003, 130, 817–833. [Google Scholar]
- Quigley, I.K.; Manuel, J.L.; Roberts, R.A.; Nuckels, R.J.; Herrington, E.R.; MacDonald, E.L.; Parichy, D.M. Evolutionary diversification of pigment pattern in Danio fishes: Differential fms dependence and stripe loss in D. albolineatus. Dev. Camb. Engl. 2005, 132, 89–104. [Google Scholar]
- Arrizabalaga-Larrañaga, A.; Rodríguez, P.; Medina, M.; Santos, F.J.; Moyano, E. Simultaneous analysis of natural pigments and E-141i in olive oils by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 5577–5591. [Google Scholar] [CrossRef]
- Clotfelter, E.D.; Ardia, D.R.; McGraw, K. Red fish, blue fish: Trade-offs between pigmentation and immunity in Betta splendens. Behav. Ecol. 2007, 18, 1139–1145. [Google Scholar] [CrossRef]
- Grether, G.F.; Hudon, J.; Endler, J.A. Carotenoid Scarcity, Synthetic Pteridine Pigments and the Evolution of Sexual Coloration in Guppies (Poecilia reticulata). Proc. Biol. Sci. 2001, 268, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]


| Samples | Clean Reads | Clean Bases | GC Content (%) | ≥Q30 |
|---|---|---|---|---|
| A1 | 21,068,282 | 6,286,267,334 | 48.78% | 95.11% |
| A2 | 20,859,439 | 6,220,965,958 | 48.84% | 95.21% |
| A3 | 19,835,579 | 5,915,579,956 | 48.76% | 95.22% |
| B1 | 20,520,674 | 6,120,618,944 | 48.76% | 95.41% |
| B2 | 21,126,146 | 6,301,031,704 | 48.96% | 95.24% |
| B3 | 20,894,446 | 6,232,585,618 | 48.71% | 95.28% |
| Gene ID | Gene Symbol | log2FC | Description |
|---|---|---|---|
| XM_046383247.1 | soat2 | 1.625596728 | sterol O-acyltransferase 2 |
| XM_046394003.1 | rlbp1b | 1.326384862 | retinaldehyde-binding protein 1b |
| XM_046408526.1 | rbp1 | 1.175138631 | retinol-binding protein 1 |
| XM_046389465.1 | apoa1 | 1.023536727 | apolipoprotein A-I-like |
| XM_046408744.1 | rpe65a | 1.006035995 | retinoid isomerohydrolase RPE65 a |
| XM_046383565.1 | rh2 | −3.307257368 | green-sensitive opsin-like |
| Gene ID | Gene Symbol | FPKM | Description |
|---|---|---|---|
| XM_046380961.1 | plin2 | 18.11774 | perilipin 2 |
| XM_046374291.1 | scarb1 | 14.03952 | scavenger receptor class B, member 1 |
| XM_046371628.1 | bco1 | 4.338945 | beta-carotene oxygenase 1 |
| XM_046398563.1 | plin6 | 7.46246 | perilipin 6 |
| XM_046405845.1 | csf1ra | 3.745435 | colony-stimulating factor 1 receptor, a |
| XM_046376452.1 | gch2 | 3.497639 | GTP cyclohydrolase 2 |
| XM_046405324.1 | slc2a15b | 3.810482 | solute carrier family 2 member 15b |
| XM_046408801.1 | apoda | 15.3108 | apolipoprotein Da, duplicate 1 |
| Tested Object | Monitoring Ion Pairs (m/z) | Fragmentation Voltage (V) | Collision Energy (eV) |
|---|---|---|---|
| astaxanthin | 598/173.4 | 100 | 30 |
| 598/147.3 a | 100 | 25 | |
| lutein | 570/133.4 | 120 | 20 |
| 570/89.2 a | 120 | 30 | |
| fucoxanthin | 660/642.8 | 100 | 5 |
| 660/109.3 a | 100 | 30 | |
| β-carotene | 538/300.7 | 120 | 20 |
| 538/286.2 a | 120 | 30 | |
| l-sepiapterin | 238/192.1 a | 100 | 10 |
| 238/165.1 | 100 | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, Y.; Shi, H.; Han, T.; Jiang, D.; Lu, B.; Shi, G.; Zhu, C.; Li, G. Pigment Identification and Gene Expression Analysis during Erythrophore Development in Spotted Scat (Scatophagus argus) Larvae. Int. J. Mol. Sci. 2023, 24, 15356. https://doi.org/10.3390/ijms242015356
Liao Y, Shi H, Han T, Jiang D, Lu B, Shi G, Zhu C, Li G. Pigment Identification and Gene Expression Analysis during Erythrophore Development in Spotted Scat (Scatophagus argus) Larvae. International Journal of Molecular Sciences. 2023; 24(20):15356. https://doi.org/10.3390/ijms242015356
Chicago/Turabian StyleLiao, Yongguan, Hongjuan Shi, Tong Han, Dongneng Jiang, Baoyue Lu, Gang Shi, Chunhua Zhu, and Guangli Li. 2023. "Pigment Identification and Gene Expression Analysis during Erythrophore Development in Spotted Scat (Scatophagus argus) Larvae" International Journal of Molecular Sciences 24, no. 20: 15356. https://doi.org/10.3390/ijms242015356
APA StyleLiao, Y., Shi, H., Han, T., Jiang, D., Lu, B., Shi, G., Zhu, C., & Li, G. (2023). Pigment Identification and Gene Expression Analysis during Erythrophore Development in Spotted Scat (Scatophagus argus) Larvae. International Journal of Molecular Sciences, 24(20), 15356. https://doi.org/10.3390/ijms242015356







