Integrated Analysis of Transcriptome and Proteome of the Human Cornea and Aqueous Humor Reveal Novel Biomarkers for Corneal Endothelial Cell Dysfunction
Abstract
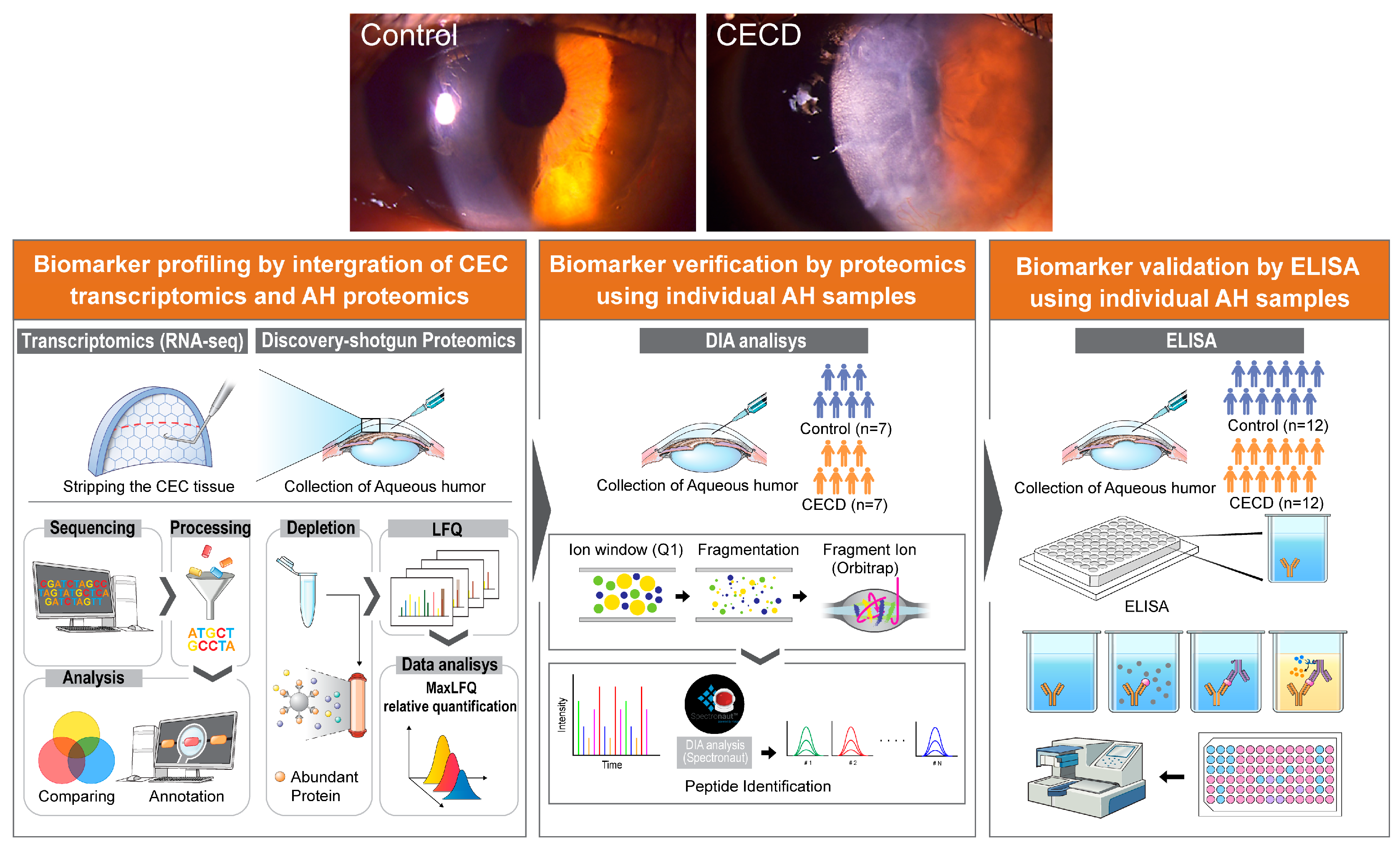
1. Introduction
2. Results
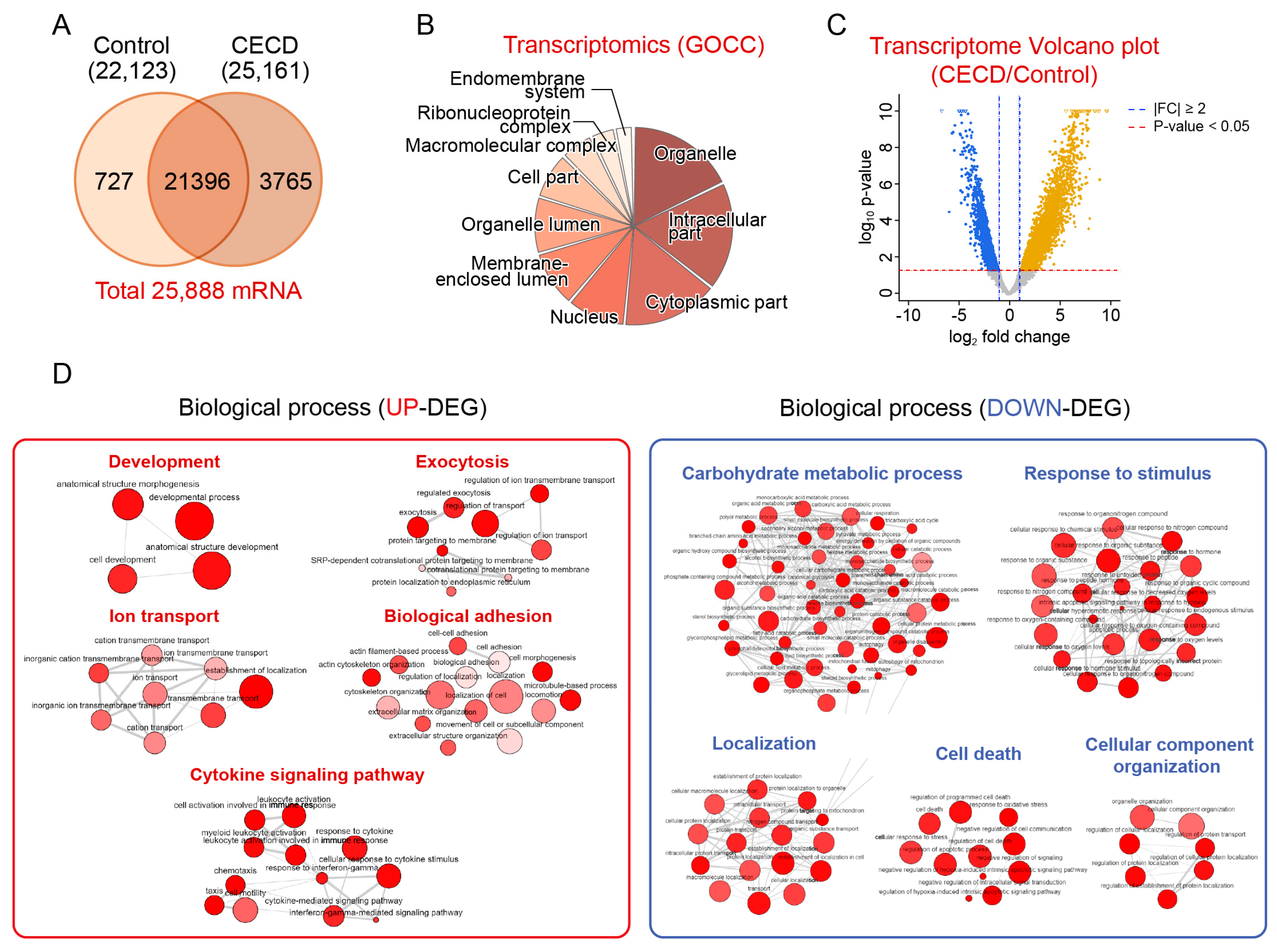
2.1. Transcriptomic Analysis of Dysfunctional CECs
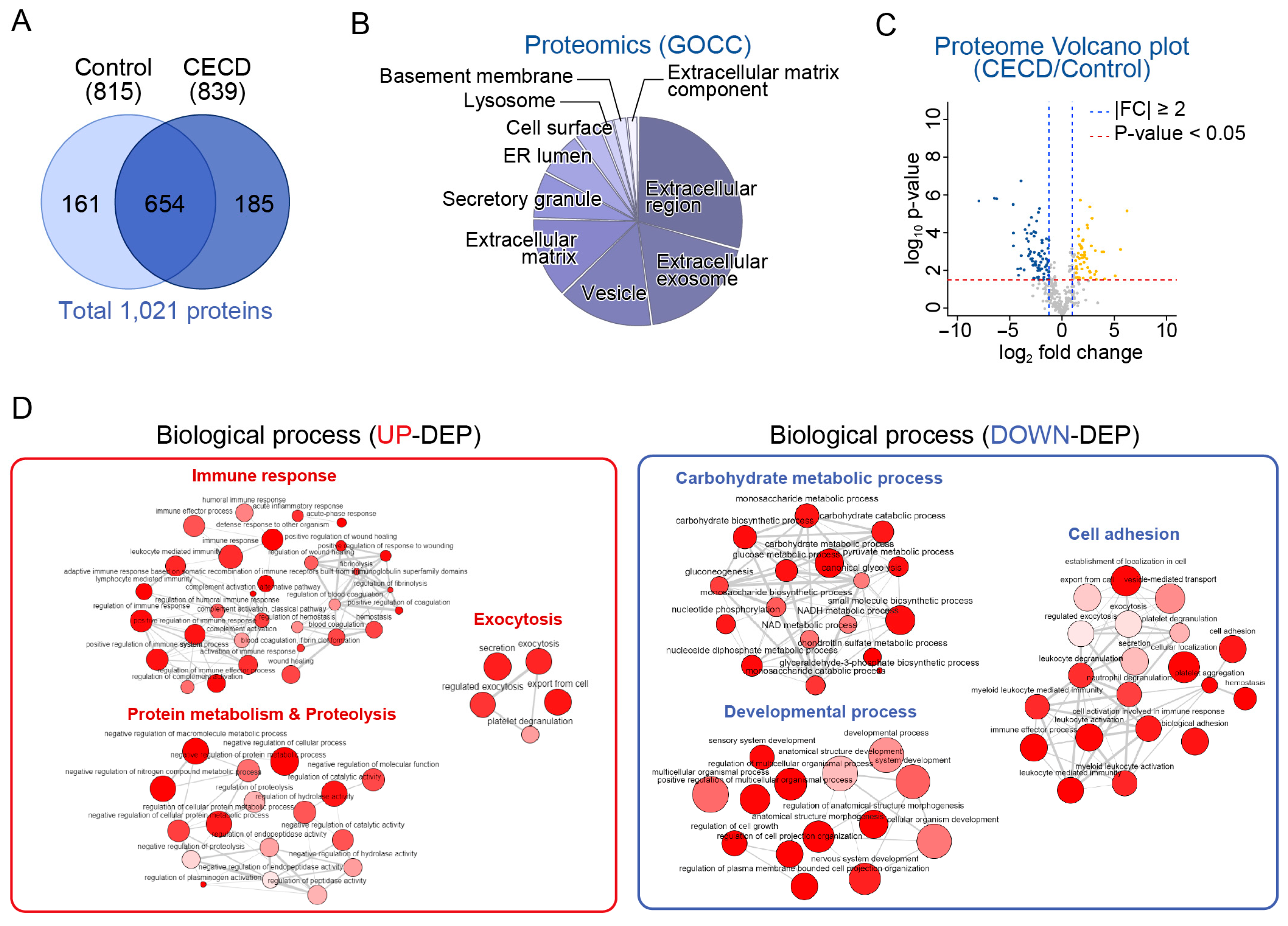
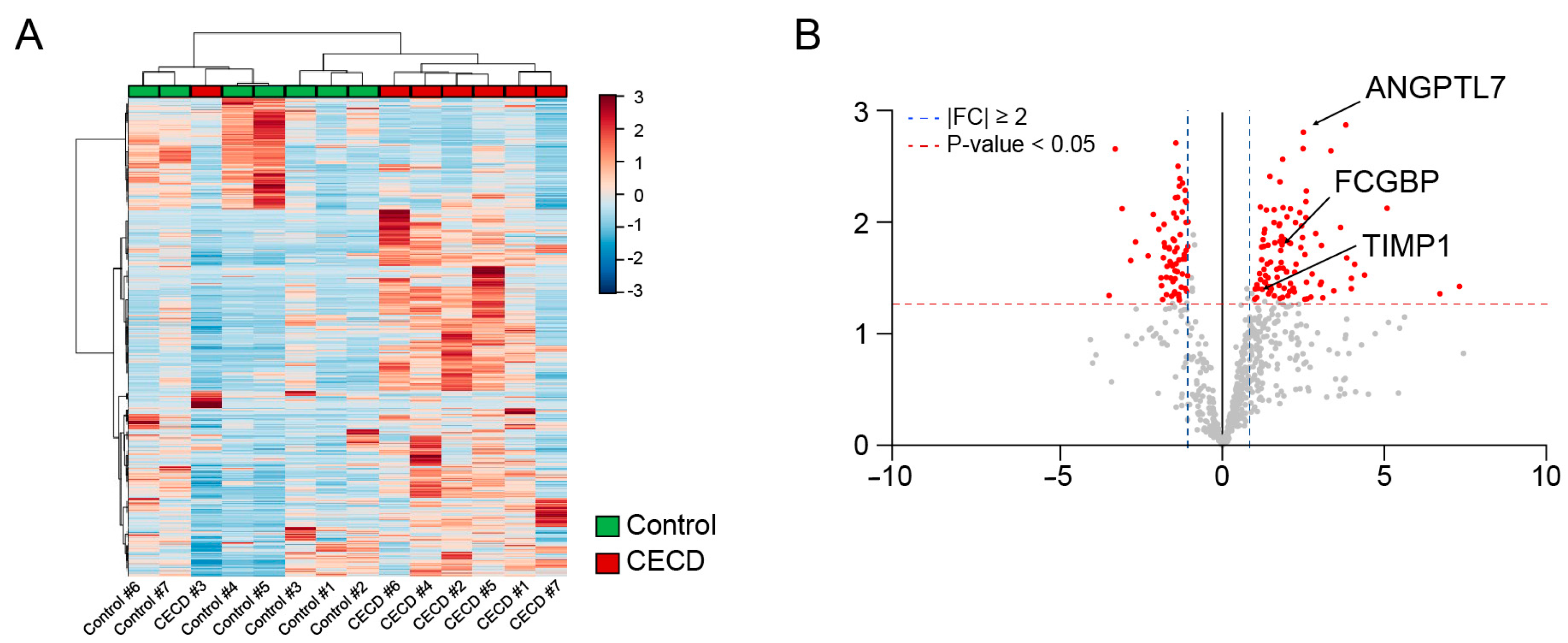
2.2. Global Proteome Profiling of AH from Patients with CECD
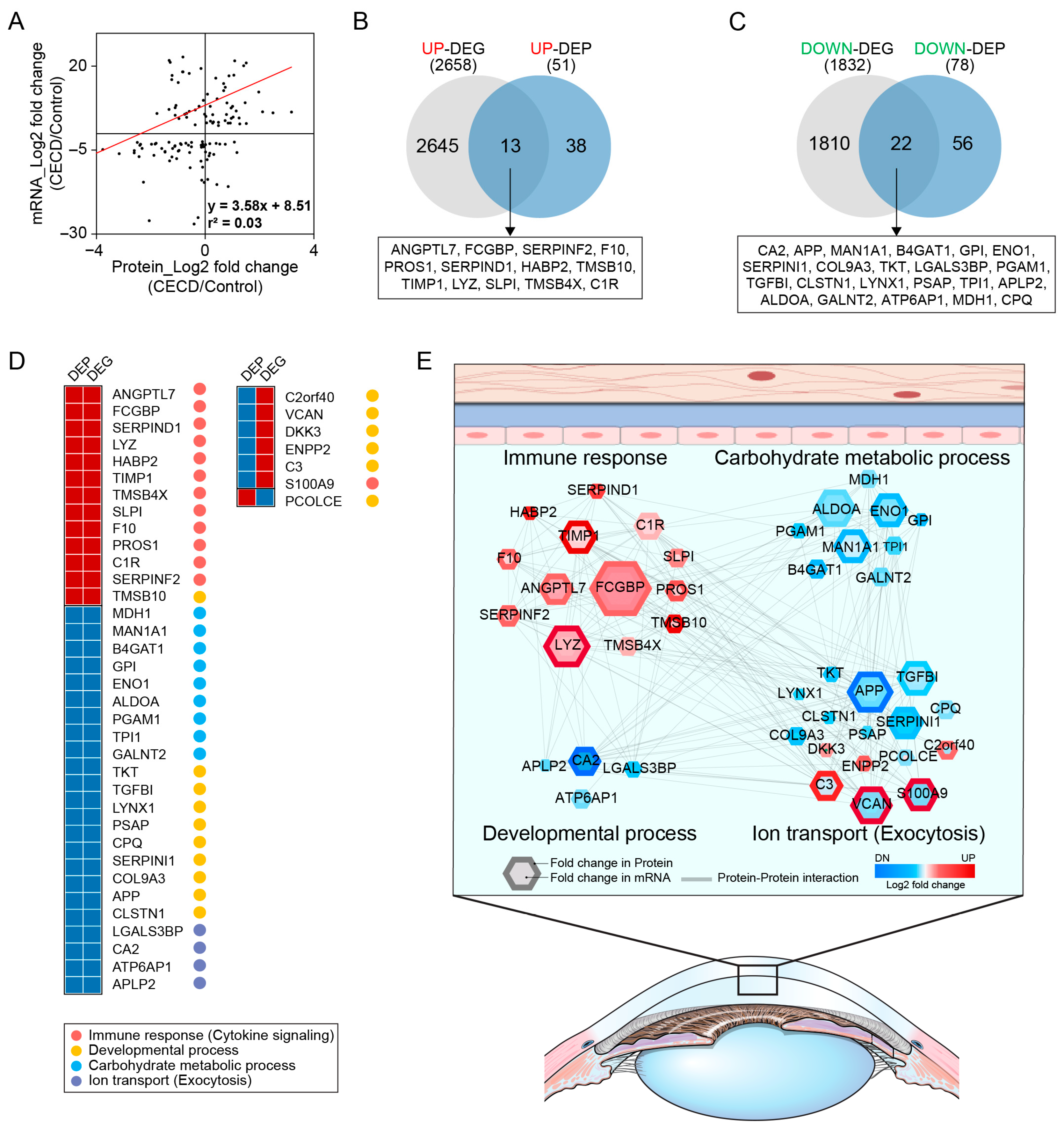
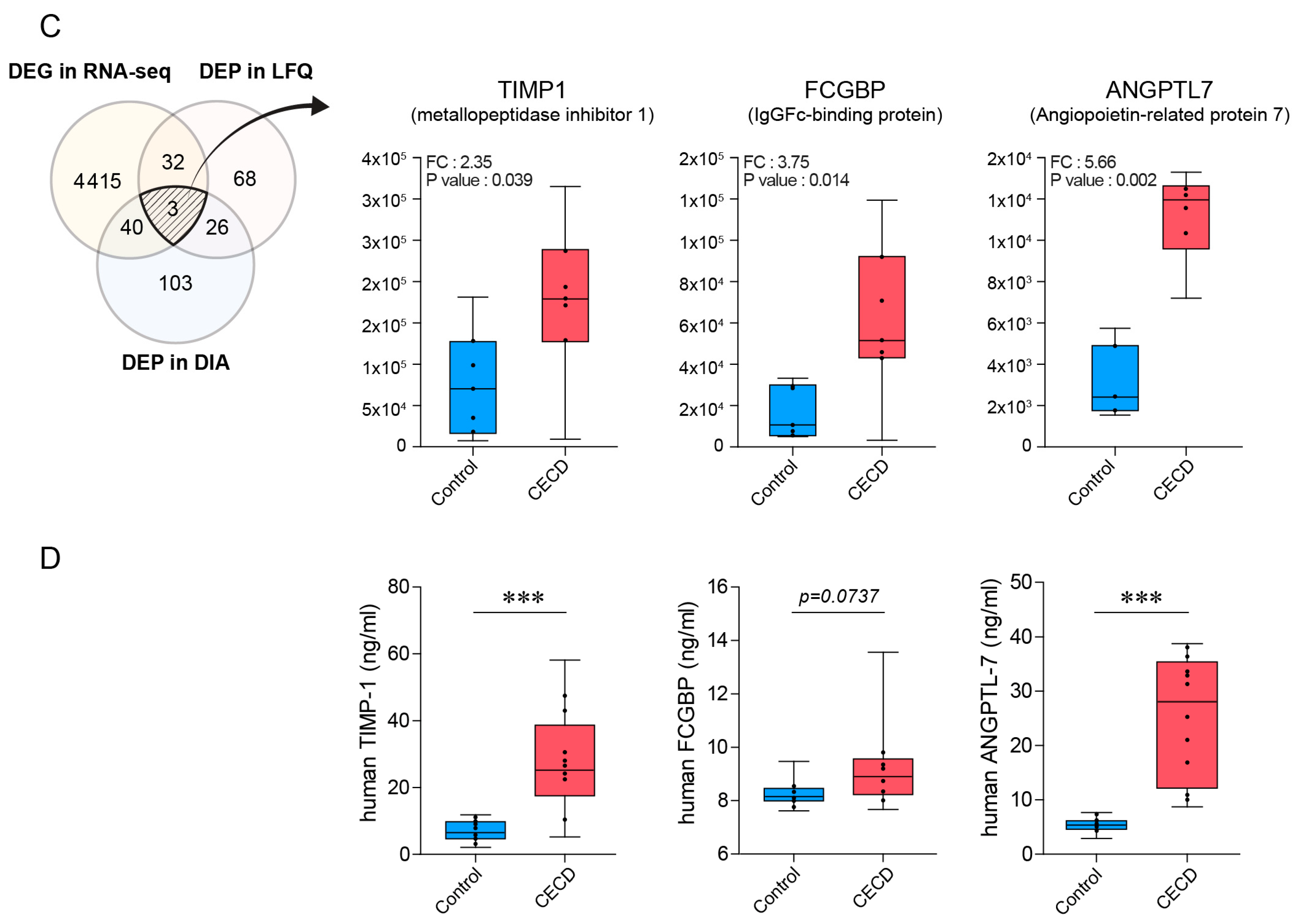
2.3. AH Proteome Alterations Are Associated with CEC Transcriptome Changes in CECD
2.4. Individual AH Analysis for Marker Verification via DIA Proteomics in CECD
2.5. Validation of AH Marker Candidates for CECD
3. Discussion
4. Materials and Methods
4.1. Participants and Sample Collection
4.2. RNA Preparation and Sequencing of CECs
4.3. Sample Preparation for Proteomic Analysis
4.4. Quantitative Global Profiling
4.5. Individual Aqueous Humor Analysis and Data Processing
4.6. Database Search and Quantitative Analysis
4.7. Enrichment Analysis Using Gene Ontology
4.8. Enzyme-Linked Immunosorbent Assay (ELISA)
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joyce, N.C. Proliferative capacity of the corneal endothelium. Prog. Retin. Eye Res. 2003, 22, 359–389. [Google Scholar] [CrossRef]
- Bourne, W.M. Clinical estimation of corneal endothelial pump function. Trans. Am. Ophthalmol. Soc. 1998, 96, 229–239; discussion 239–242. [Google Scholar] [PubMed]
- Bonanno, J.A. Molecular mechanisms underlying the corneal endothelial pump. Exp. Eye Res. 2012, 95, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.; Alvarado, J.; Juster, R.; Maglio, M. Prenatal and postnatal cellularity of the human corneal endothelium. A quantitative histologic study. Investig. Ophthalmol. Vis. Sci. 1984, 25, 312–322. [Google Scholar]
- Hollingsworth, J.; Perez-Gomez, I.; Mutalib, H.A.; Efron, N. A population study of the normal cornea using an in vivo, slit-scanning confocal microscope. Optom. Vis. Sci. 2001, 78, 706–711. [Google Scholar] [CrossRef]
- Joyce, N.C. Proliferative capacity of corneal endothelial cells. Exp. Eye Res. 2012, 95, 16–23. [Google Scholar] [CrossRef]
- Keenan, T.D.; Jones, M.N.; Rushton, S.; Carley, F.M.; National Health Service Blood; Transplant Ocular Tissue Advisory Group; Contributing Ophthalmologists. Trends in the indications for corneal graft surgery in the United Kingdom: 1999 through 2009. Arch. Ophthalmol. 2012, 130, 621–628. [Google Scholar] [CrossRef]
- Shimazaki, J.; Shinozaki, N.; Shimmura, S.; Holland, E.J.; Tsubota, K. Efficacy and safety of international donor sharing: A single-center, case-controlled study on corneal transplantation. Transplantation 2004, 78, 216–220. [Google Scholar] [CrossRef]
- Alfawaz, A.M.; Holland, G.N.; Yu, F.; Margolis, M.S.; Giaconi, J.A.; Aldave, A.J. Corneal Endothelium in Patients with Anterior Uveitis. Ophthalmology 2016, 123, 1637–1645. [Google Scholar] [CrossRef]
- Lass, J.H.; Benetz, B.A.; Gal, R.L.; Kollman, C.; Raghinaru, D.; Dontchev, M.; Mannis, M.J.; Holland, E.J.; Chow, C.; McCoy, K.; et al. Donor age and factors related to endothelial cell loss 10 years after penetrating keratoplasty: Specular Microscopy Ancillary Study. Ophthalmology 2013, 120, 2428–2435. [Google Scholar] [CrossRef]
- Bertelmann, E.; Pleyer, U.; Rieck, P. Risk factors for endothelial cell loss post-keratoplasty. Acta Ophthalmol. Scand. 2006, 84, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Aldrich, B.T.; Schlotzer-Schrehardt, U.; Skeie, J.M.; Burckart, K.A.; Schmidt, G.A.; Reed, C.R.; Zimmerman, M.B.; Kruse, F.E.; Greiner, M.A. Mitochondrial and Morphologic Alterations in Native Human Corneal Endothelial Cells Associated with Diabetes Mellitus. Investig. Ophthalmol. Vis. Sci. 2017, 58, 2130–2138. [Google Scholar] [CrossRef] [PubMed]
- Tamhane, M.; Cabrera-Ghayouri, S.; Abelian, G.; Viswanath, V. Review of Biomarkers in Ocular Matrices: Challenges and Opportunities. Pharm. Res. 2019, 36, 40. [Google Scholar] [CrossRef] [PubMed]
- Freddo, T.F. A contemporary concept of the blood-aqueous barrier. Prog. Retin. Eye Res. 2013, 32, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Tomida, D.; Yagi-Yaguchi, Y.; Higa, K.; Satake, Y.; Shimazaki, J.; Yamaguchi, T. Correlations between tear fluid and aqueous humor cytokine levels in bullous keratopathy. Ocul. Surf. 2020, 18, 801–807. [Google Scholar] [CrossRef]
- Kwok, L.S. Calculation and application of the anterior surface area of a model human cornea. J. Theor. Biol. 1984, 108, 295–313. [Google Scholar] [CrossRef]
- Murthy, K.R.; Rajagopalan, P.; Pinto, S.M.; Advani, J.; Murthy, P.R.; Goel, R.; Subbannayya, Y.; Balakrishnan, L.; Dash, M.; Anil, A.K.; et al. Proteomics of human aqueous humor. OMICS 2015, 19, 283–293. [Google Scholar] [CrossRef]
- Kornblueth, W.; Tenenbaum, E. The inhibitory effect of aqueous humor on the growth of cells in tissue cultures. Am. J. Ophthalmol. 1956, 42, 70–74. [Google Scholar] [CrossRef]
- Taylor, A.W.; Alard, P.; Yee, D.G.; Streilein, J.W. Aqueous humor induces transforming growth factor-beta (TGF-beta)-producing regulatory T-cells. Curr. Eye Res. 1997, 16, 900–908. [Google Scholar] [CrossRef]
- Weinsieder, A.; Reddan, J.; Wilson, D. Aqueous humor in lens repair and cell proliferation. Exp. Eye Res. 1976, 23, 355–363. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, L.; Ye, E.A.; Wang, A.; Li, G.; Chen, L. Aqueous humor induces lymphatic regression on the ocular surface. Ocul. Surf. 2020, 18, 505–510. [Google Scholar] [CrossRef]
- Wierenga, A.P.A.; Cao, J.; Mouthaan, H.; van Weeghel, C.; Verdijk, R.M.; van Duinen, S.G.; Kroes, W.G.M.; Dogrusöz, M.; Marinkovic, M.; van der Burg, S.S.H.; et al. Aqueous Humor Biomarkers Identify Three Prognostic Groups in Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4740–4747. [Google Scholar] [CrossRef]
- Sharma, S.; Bollinger, K.E.; Kodeboyina, S.K.; Zhi, W.; Patton, J.; Bai, S.; Edwards, B.; Ulrich, L.; Bogorad, D.; Sharma, A. Proteomic Alterations in Aqueous Humor from Patients with Primary Open Angle Glaucoma. Investig. Ophthalmol. Vis. Sci. 2018, 59, 2635–2643. [Google Scholar] [CrossRef]
- Zhang, B.N.; Wu, X.; Dai, Y.; Qi, B.; Fan, C.; Huang, Y. Proteomic analysis of aqueous humor from cataract patients with retinitis pigmentosa. J. Cell Physiol. 2020, 236, 2659–2668. [Google Scholar] [CrossRef]
- Qu, S.C.; Xu, D.; Li, T.T.; Zhang, J.F.; Liu, F. iTRAQ-based proteomics analysis of aqueous humor in patients with dry age-related macular degeneration. Int. J. Ophthalmol. 2019, 12, 1758–1766. [Google Scholar] [CrossRef]
- Kliuchnikova, A.A.; Samokhina, N.I.; Ilina, I.Y.; Karpov, D.S.; Pyatnitskiy, M.A.; Kuznetsova, K.G.; Toropygin, I.Y.; Kochergin, S.A.; Alekseev, I.B.; Zgoda, V.G.; et al. Human aqueous humor proteome in cataract, glaucoma, and pseudoexfoliation syndrome. Proteomics 2016, 16, 1938–1946. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Higa, K.; Yagi-Yaguchi, Y.; Ueda, K.; Noma, H.; Shibata, S.; Nagai, T.; Tomida, D.; Yasu-Mimura, R.; Ibrahim, O.; et al. Pathological processes in aqueous humor due to iris atrophy predispose to early corneal graft failure in humans and mice. Sci. Adv. 2020, 6, eaaz5195. [Google Scholar] [CrossRef] [PubMed]
- Yagi-Yaguchi, Y.; Yamaguchi, T.; Higa, K.; Suzuki, T.; Aketa, N.; Dogru, M.; Satake, Y.; Shimazaki, J. Association between corneal endothelial cell densities and elevated cytokine levels in the aqueous humor. Sci. Rep. 2017, 7, 13603. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Yamaguchi, T.; Shibata, S.; Nagai, T.; Noma, H.; Tsubota, K.; Shimazaki, J. Cytokine Levels in the Aqueous Humor Are Associated With Corneal Thickness in Eyes With Bullous Keratopathy. Am. J. Ophthalmol. 2019, 198, 174–180. [Google Scholar] [CrossRef]
- Myer, C.; Perez, J.; Abdelrahman, L.; Mendez, R.; Khattri, R.B.; Junk, A.K.; Bhattacharya, S.K. Differentiation of soluble aqueous humor metabolites in primary open angle glaucoma and controls. Exp. Eye Res. 2020, 194, 108024. [Google Scholar] [CrossRef] [PubMed]
- Botling Taube, A.; Konzer, A.; Alm, A.; Bergquist, J. Proteomic analysis of the aqueous humour in eyes with pseudoexfoliation syndrome. Br. J. Ophthalmol. 2019, 103, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef]
- Forshed, J. Experimental Design in Clinical ‘Omics Biomarker Discovery. J. Proteome Res. 2017, 16, 3954–3960. [Google Scholar] [CrossRef]
- Rao, G.; Santhoshkumar, P.; Sharma, K.K. Anti-chaperone betaA3/A1(102-117) peptide interacting sites in human alphaB-crystallin. Mol. Vis. 2008, 14, 666–674. [Google Scholar] [PubMed]
- Janson, B.J.; Alward, W.L.; Kwon, Y.H.; Bettis, D.I.; Fingert, J.H.; Provencher, L.M.; Goins, K.M.; Wagoner, M.D.; Greiner, M.A. Glaucoma-associated corneal endothelial cell damage: A review. Surv. Ophthalmol. 2018, 63, 500–506. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, J.M.; Choi, C.Y.; Park, K.H. Changes in corneal endothelial cell density in patients with normal-tension glaucoma. Jpn. J. Ophthalmol. 2009, 53, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Galgauskas, S.; Krasauskaite, D.; Pajaujis, M.; Juodkaite, G.; Asoklis, R.S. Central corneal thickness and corneal endothelial characteristics in healthy, cataract, and glaucoma patients. Clin. Ophthalmol. 2012, 6, 1195–1199. [Google Scholar] [CrossRef][Green Version]
- Predović, J.; Balog, T.; Marotti, T.; Gabrić, N.; Bohac, M.; Romac, I.; Dekaris, I. The expression of human corneal MMP-2, MMP-9, proMMP-13 and TIMP-1 in bullous keratopathy and keratoconus. Coll. Antropol. 2008, 32 (Suppl. 2), 15–19. [Google Scholar]
- Saika, S.; Kawashima, Y.; Okada, Y.; Tanaka, S.I.; Yamanaka, O.; Ohnishi, Y.; Ooshima, A. Recombinant TIMP-1 and -2 enhance the proliferation of rabbit corneal epithelial cells in vitro and the spreading of rabbit corneal epithelium in situ. Curr. Eye Res. 1998, 17, 47–52. [Google Scholar] [CrossRef]
- Kuchtey, J.; Källberg, M.E.; Gelatt, K.N.; Rinkoski, T.; Komàromy, A.M.; Kuchtey, R.W. Angiopoietin-like 7 secretion is induced by glaucoma stimuli and its concentration is elevated in glaucomatous aqueous humor. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3438–3448. [Google Scholar] [CrossRef]
- Tanigawa, Y.; Wainberg, M.; Karjalainen, J.; Kiiskinen, T.; Venkataraman, G.; Lemmelä, S.; Turunen, J.A.; Graham, R.R.; Havulinna, A.S.; Perola, M.; et al. Rare protein-altering variants in ANGPTL7 lower intraocular pressure and protect against glaucoma. PLoS Genet. 2020, 16, e1008682. [Google Scholar] [CrossRef] [PubMed]
- Toyono, T.; Usui, T.; Yokoo, S.; Taketani, Y.; Nakagawa, S.; Kuroda, M.; Yamagami, S.; Amano, S. Angiopoietin-like 7 is an anti-angiogenic protein required to prevent vascularization of the cornea. PLoS ONE 2015, 10, e0116838. [Google Scholar] [CrossRef] [PubMed]
- Tausif, H.N.; Johnson, L.; Titus, M.; Mavin, K.; Chandrasekaran, N.; Woodward, M.A.; Shtein, R.M.; Mian, S.I. Corneal donor tissue preparation for Descemet’s membrane endothelial keratoplasty. J. Vis. Exp. 2014, 17, 51919. [Google Scholar] [CrossRef]
- Woodward, M.A.; Titus, M.; Mavin, K.; Shtein, R.M. Corneal donor tissue preparation for endothelial keratoplasty. J. Vis. Exp. 2012, 64, e3847. [Google Scholar] [CrossRef]
- Birbal, R.S.; Sikder, S.; Lie, J.T.; Groeneveld-van Beek, E.A.; Oellerich, S.; Melles, G.R.J. Donor Tissue Preparation for Descemet Membrane Endothelial Keratoplasty: An Updated Review. Cornea 2018, 37, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Terry, M.A. Step-by-step Descemet’s membrane endothelial keratoplasty surgery. Taiwan J. Ophthalmol. 2019, 9, 18–26. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Nasiri-Ansari, N.; Spilioti, E.; Kyrou, I.; Kalotychou, V.; Chatzigeorgiou, A.; Sanoudou, D.; Dahlman-Wright, K.; Randeva, H.S.; Papavassiliou, A.G.; Moutsatsou, P.; et al. Estrogen Receptor Subtypes Elicit a Distinct Gene Expression Profile of Endothelial-Derived Factors Implicated in Atherosclerotic Plaque Vulnerability. Int. J. Mol. Sci. 2022, 23, 10960. [Google Scholar] [CrossRef]
- Abu-Farha, M.; Cherian, P.; Al-Khairi, I.; Madhu, D.; Tiss, A.; Warsam, S.; Alhubail, A.; Sriraman, D.; Al-Refaei, F.; Abubaker, J. Plasma and adipose tissue level of angiopoietin-like 7 (ANGPTL7) are increased in obesity and reduced after physical exercise. PLoS ONE 2017, 12, e0173024. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
| (A) | ||
| Control (Donor) (n = 5) | CECD (n = 5) | |
| Age, y | 43.75 ± 6.55 | 69.25 ± 3.86 * |
| Sex, n (Female:Male) | 2:3 | 2:3 |
| CEC density, /mm2 | 2998.25 ± 112.87 | Not available |
| Corneal thickness, μm | 555.25 ± 5.44 | Bullous |
| (B) | ||
| Control (n = 5) | CECD (n = 5) | |
| Age, y | 61.54 ± 9.22 | 69.25 ± 3.86 |
| Sex, n (Female:Male) | 2:3 | 2:3 |
| CEC density, /mm2 | 2523.32 ± 114.42 | Not available |
| Corneal thickness, μm | 546.50 ± 6.32 | Bullous |
| (C) | ||
| Control (n = 7) | CECD (n = 7) | |
| Age, y | 68.14 ± 6.36 | 69.42 ± 7.04 |
| Sex, n (Female:Male) | 3:4 | 3:4 |
| CEC density, /mm2 | 2611.32 ± 122.32 | Not available |
| Corneal thickness, μm | 548.25 ± 5.23 | Bullous |
| (D) | ||
| Control (Donor) (n = 12) | CECD (n = 12) | |
| Age, y | 60.5 ± 4.19 | 55.17 ± 12.91 |
| Sex, n (Female:Male) | 7:5 | 7:5 |
| CEC density, /mm2 | 2549.43 ± 103.67 | Not available |
| Corneal thickness, μm | 551.14 ± 4.27 | Bullous |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, C.-E.; Kim, C.H.; Jung, J.H.; Cho, Y.J.; Choi, K.Y.; Han, K.; Seo, K.Y.; Lee, H.K.; Ji, Y.W. Integrated Analysis of Transcriptome and Proteome of the Human Cornea and Aqueous Humor Reveal Novel Biomarkers for Corneal Endothelial Cell Dysfunction. Int. J. Mol. Sci. 2023, 24, 15354. https://doi.org/10.3390/ijms242015354
Moon C-E, Kim CH, Jung JH, Cho YJ, Choi KY, Han K, Seo KY, Lee HK, Ji YW. Integrated Analysis of Transcriptome and Proteome of the Human Cornea and Aqueous Humor Reveal Novel Biomarkers for Corneal Endothelial Cell Dysfunction. International Journal of Molecular Sciences. 2023; 24(20):15354. https://doi.org/10.3390/ijms242015354
Chicago/Turabian StyleMoon, Chae-Eun, Chang Hwan Kim, Jae Hun Jung, Young Joo Cho, Kee Yong Choi, Kyusun Han, Kyoung Yul Seo, Hyung Keun Lee, and Yong Woo Ji. 2023. "Integrated Analysis of Transcriptome and Proteome of the Human Cornea and Aqueous Humor Reveal Novel Biomarkers for Corneal Endothelial Cell Dysfunction" International Journal of Molecular Sciences 24, no. 20: 15354. https://doi.org/10.3390/ijms242015354
APA StyleMoon, C.-E., Kim, C. H., Jung, J. H., Cho, Y. J., Choi, K. Y., Han, K., Seo, K. Y., Lee, H. K., & Ji, Y. W. (2023). Integrated Analysis of Transcriptome and Proteome of the Human Cornea and Aqueous Humor Reveal Novel Biomarkers for Corneal Endothelial Cell Dysfunction. International Journal of Molecular Sciences, 24(20), 15354. https://doi.org/10.3390/ijms242015354







