Expression and Characterization of a β-Galactosidase from the Pacific Oyster, Crassostrea gigas, and Evaluation of Strategies for Testing Substrate Specificity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification and Expression of Putative β-Galactosidases from C. gigas
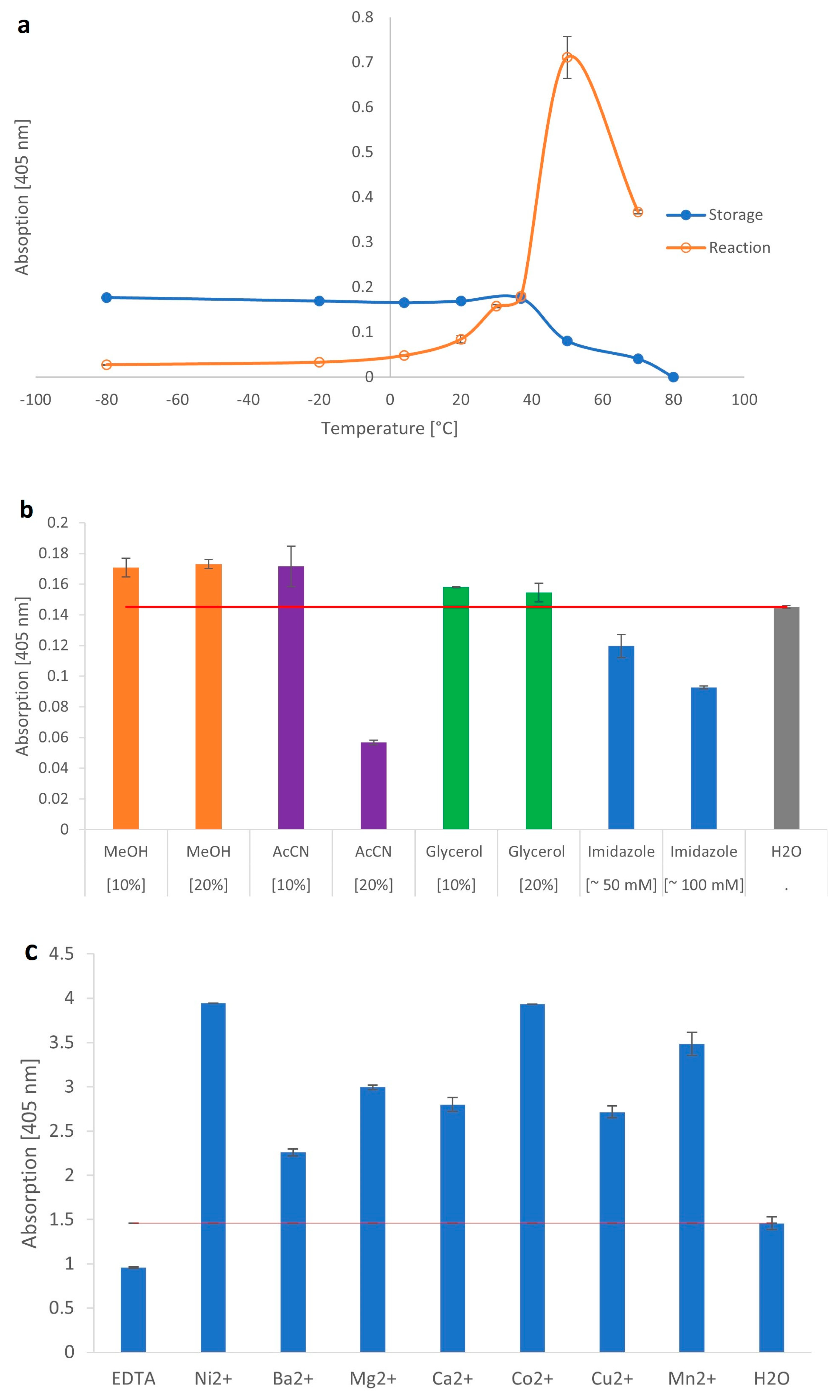
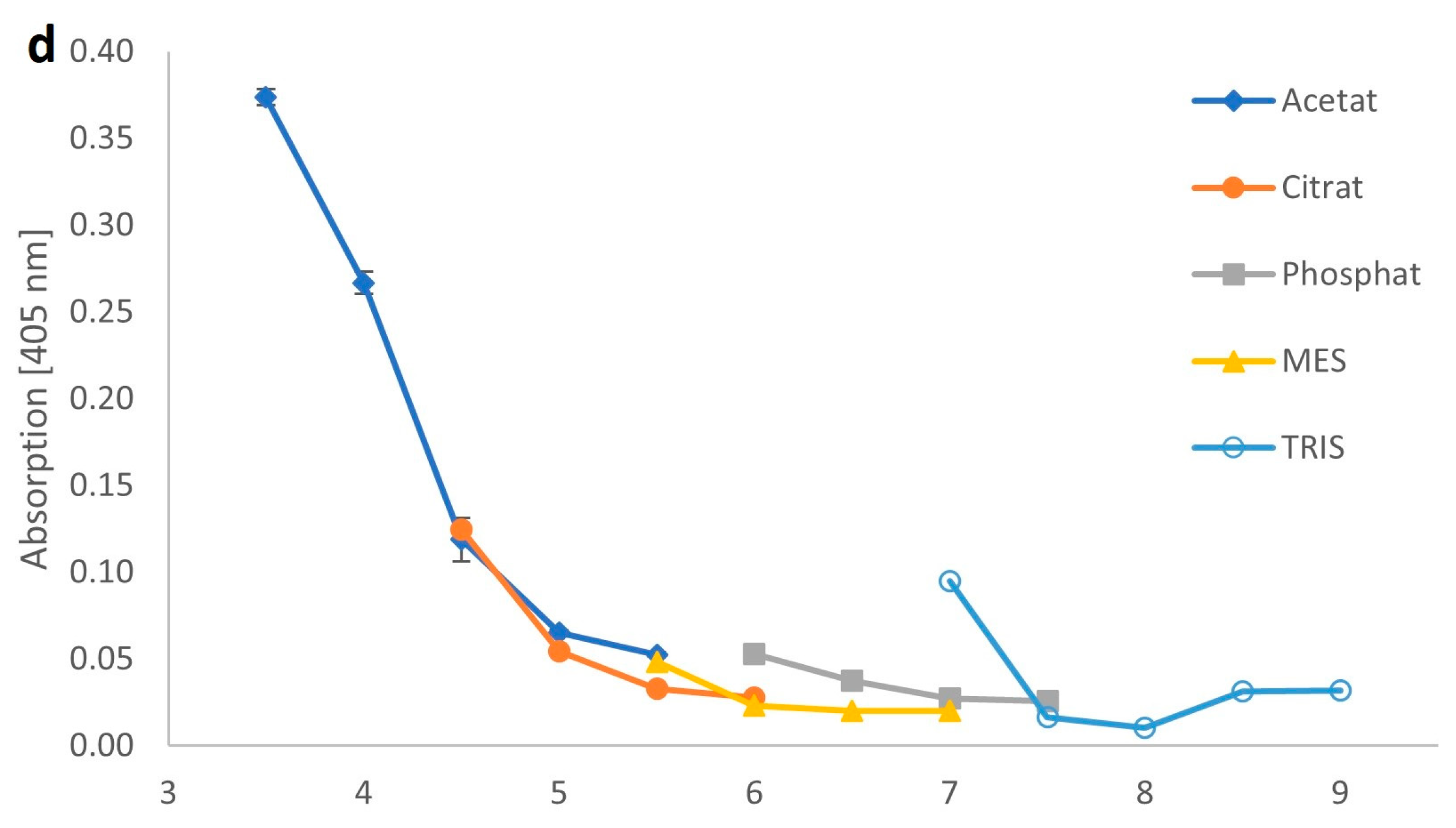
2.2. Biochemical Parameters of the Recombinant β-Galactosidase from C. gigas
2.3. Substrate Specificity of the Recombinant β-Galactosidase from C. gigas
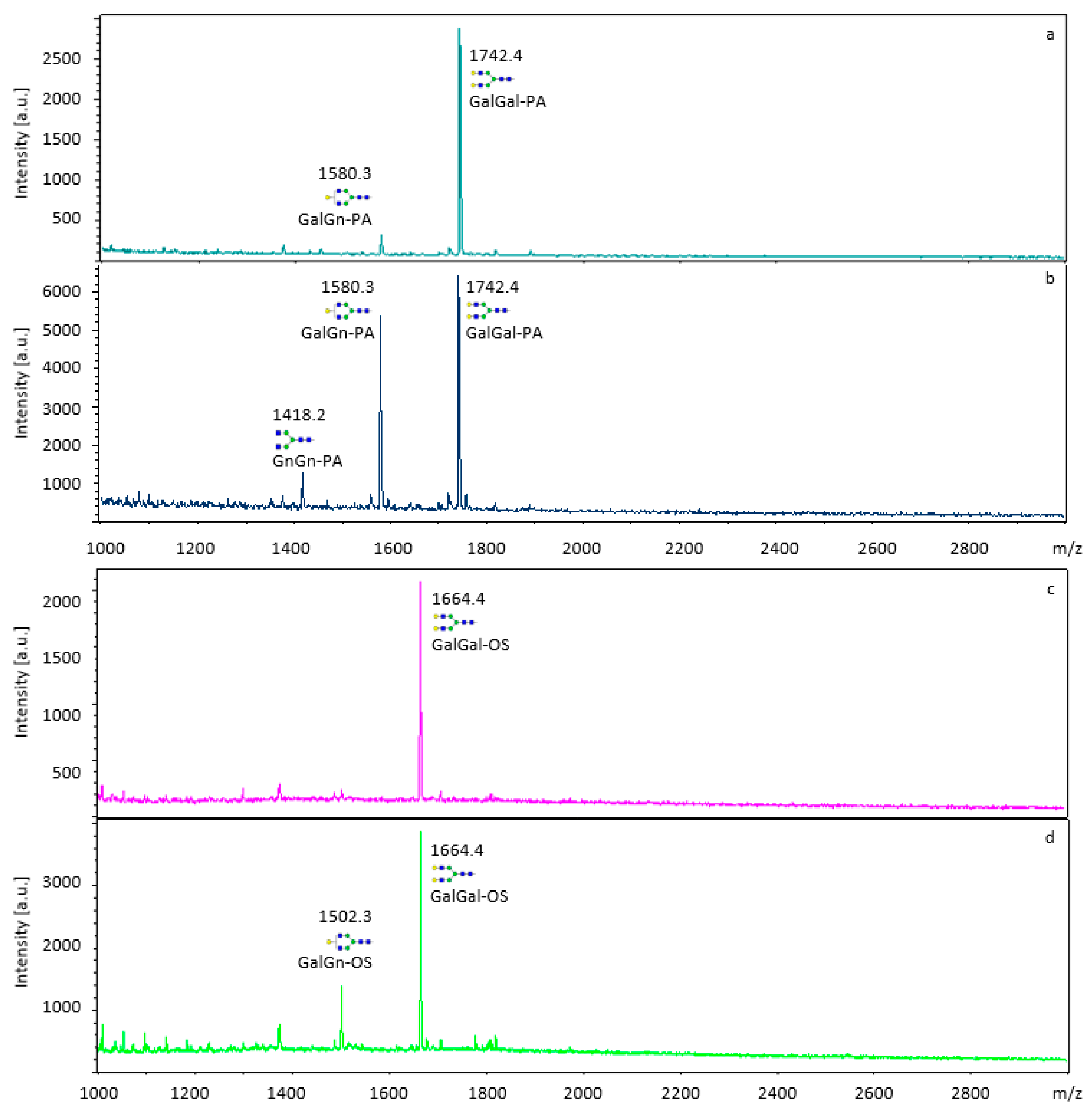
2.3.1. MALDI-TOF MS Analysis of Labeled and Unlabeled Substrates
2.3.2. Unlabeled Substrates Tested Using the Galactose Assay Kit (ab83382; ABCAM)
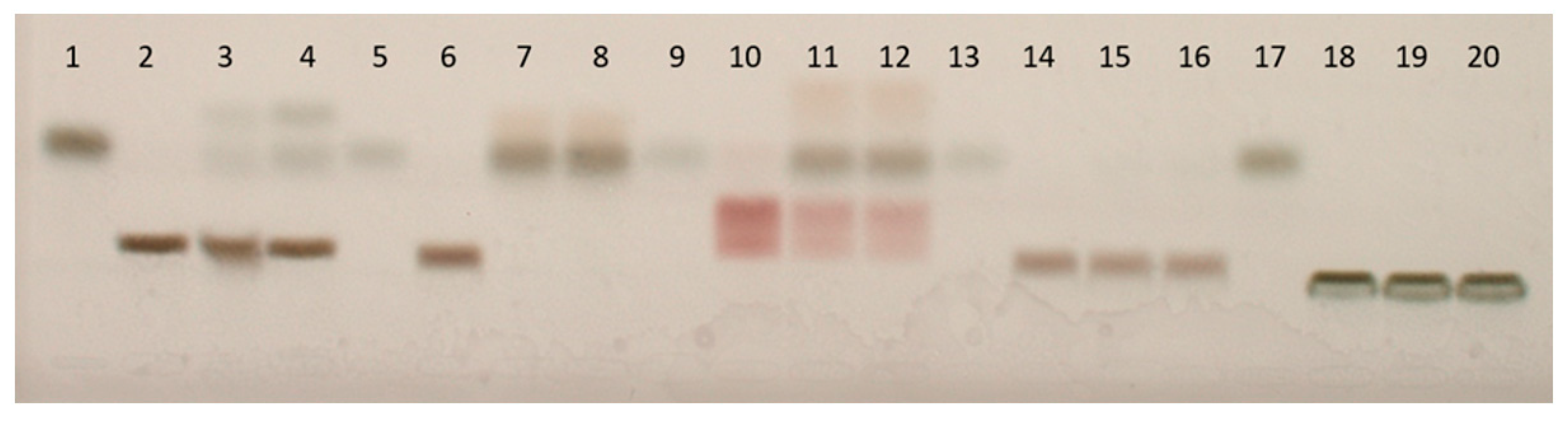
2.3.3. Unlabeled Substrates Tested Using High-Performance Thin-Layer Chromatography (HPTLC)
2.3.4. Labeled Substrates Tested Using High-Performance Liquid Chromatography (HPLC)
3. Materials and Methods
3.1. Materials
3.2. Expression of the Full-Length β-Galactosidase Gene from C. gigas
3.3. Determination of β-Galactosidase Activity from C. gigas
3.4. Galactan Hydrolysis
3.5. Substrate Specificity Assay
3.5.1. MALDI-TOF MS Analysis
3.5.2. Enzyme-Based Galactose Assay Kit (ab83382; ABCAM–Cambridge, UK)
3.5.3. High-Performance Thin-Layer Chromatography (HPTLC) Analysis
3.5.4. High-Performance Liquid Chromatography (HPLC) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varki, A. Biological roles of glycans. Glycobiology 2016, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Jin, D.K. GLB1-related disorders: GM1 gangliosidosis and Morquio B disease. J. Genet. Med. 2021, 18, 16–23. [Google Scholar] [CrossRef]
- Wanninger, A.; Wollesen, T. The evolution of molluscs. Biol. Rev. 2018, 94, 102–115. [Google Scholar] [CrossRef]
- Kumar, P.A. Review—On Molluscs as an Agricultural Pest and Their Control. Int. J. Food Sci. Agric. 2020, 4, 383–389. [Google Scholar] [CrossRef]
- Negara, B.F.S.P.; Mohibbullah, M.D.; Sohn, J.H.; Kim, J.S.; Choi, J.S. Nutritional Value and Potential Bioactivities of Pacific Oyster (Crassostrea gigas). Int. J. Food Sci. Technol. 2022, 57, 5732–5749. [Google Scholar] [CrossRef]
- Caurcel, C.; Laetsch, D.R.; Challis, R.; Kumar, S.; Gharbi, K.; Blaxter, M. MolluscDB: A genome and transcriptome database for molluscs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200157. [Google Scholar] [CrossRef]
- Tasumi, S.; Vasta, G.R. A Galectin of Unique Domain Organization from Hemocytes of the Eastern Oyster (Crassostrea virginica) Is a Receptor for the Protistan Parasite Perkinsus marinus. J. Immunol. 2007, 179, 3086–3098. [Google Scholar] [CrossRef]
- Staudacher, E. Mucin-Type O-Glycosylation in Invertebrates. Molecules 2015, 20, 10622–10640. [Google Scholar] [CrossRef]
- Staudacher, E. Mollusc N-glycosylation: Structures, Functions and Perspectives. Biomolecules 2021, 11, 1820. [Google Scholar] [CrossRef]
- Husain, Q. β-galactosidases and their potential applications: A review. Crit. Rev. Biotechnol. 2009, 30, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Saqib, S.; Akram, A.; Halim, S.A.; Tassaduq, R. Sources of β-galactosidase and its applications in food industry. 3 Biotech 2017, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.H.; Zhang, X.J.; DuBose, R.F.; Mattews, B.W. Three-dimensional structure of β-galactosidase from E. coli. Nature 1994, 369, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Juers, D.H.; Jacobson, R.H.; Wigley, D.; Zhang, X.J.; Huber, R.E.; Tronrud, D.E.; Mattews, B.W. High resolution refinement of -galactosidase in a new crystal from reveals multiple metal-binding sites and provides a structural basis for α-complementation. Protein Sci. 2000, 9, 1685–1699. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, S.; Banerjee, S.; Mandi, N. Screening of Bacterial Recombinants: Strategies and Preventing False Positives. In Molecular Cloning—Selected Applications in Medicine and Biology, 1st ed.; Brown, G., Ed.; InTech: Rijeka, Croatia, 2011; Chapter 1; pp. 3–20. [Google Scholar] [CrossRef]
- Sharma, S.K.; Sharma, S.P.; Leblanc, R.M. Methods of detection of β-galactosidase enzyme in living cells. Enzym. Microb. Technol. 2021, 150, 109885. [Google Scholar] [CrossRef]
- Xu, X.; Fan, X.; Fan, C.; Qin, X.; Liu, B.; Nie, C.; Sun, N.; Yao, Q.; Zhang, Y.; Zhang, W. Production Optimization of an Active β-Galactosidase of Bifidobacterium animalis in Heterologous Expression Systems. BioMed Res. Int. 2019, 2019, 8010635. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Yang, S.; Jiang, T.; Liang, T.; Li, Y.; Cai, S.; Wu, Q.; Zhang, J.; Chen, W.; Xie, X. Cloning, Expression, Purification, and Characterization of β-Galactosidase from Bifidobacterium longum and Bifidobacterium pseudocatenulatum. Molecules 2022, 27, 4497. [Google Scholar] [CrossRef]
- Thoma, J.; Stenitzer, D.; Grabherr, R.; Staudacher, E. Identification, Characterization, and Expression of a β-Galactosidase from Arion Species (Mollusca). Biomolecules 2022, 12, 1578. [Google Scholar] [CrossRef]
- Sampedro, J.; Gianzo, C.; Iglesias, N.; Guitian, E.; Revilla, G.; Zarra, I. AtBGAL10 Is the Main Xyloglucan b-Galactosidase in Arabidopsis, and Its Absence Results in Unusual Xyloglucan Subunits and Growth Defects. Plant Physiol. 2012, 158, 1146–1157. [Google Scholar] [CrossRef]
- Karasová, P.; Spiwok, V.; Malá, Š.; Králová, B.; Russell, N.J. Beta-galactosidase activity in psychrotrophic microorganisms and their potential use in food industry. Czech J. Food Sci. 2002, 20, 43–47. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, Q.; Zou, H.; Guo, B.; Ni, J. A Method for the Analysis of Low-Mass Molecules by MALDI-TOF Mass Spectrometry. Anal. Chem. 2002, 74, 1637–1641. [Google Scholar] [CrossRef] [PubMed]
- Wong-Madden, S.T.; Landry, D. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology 1995, 5, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Leparoux, S.; Padrines, M.; Placier, G.; Colas, B. Characterization of a strictly specific acid beta-galactosidase from Achatina achatina. Biochim. Biophys. Acta-Gen. Subj. 1997, 1336, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.R.; Ramana, K.V.; Sharma, R.K. β-galactosidase: Biotechnological applications in food processing. J. Food Biochem. 2018, 42, e12564. [Google Scholar] [CrossRef]
- Martinez-Bilbao, M.; Gaunt, M.T.; Huber, R.E. E461H-beta-galactosidase (Escherichia coli): Altered divalent metal specificity and slow but reversible metal inactivation. Biochemistry 1995, 34, 13437–13442. [Google Scholar] [CrossRef]
- Poltorak, O.M.; Chukhrai, E.S.; Pilipenko, O.S.; Atyaksheva, L.F.; Beregalov, A.E. A Comparative Study of the Structure and Properties of β-galactosidases. Biophys. Chem. 2007, 81, 808–812. [Google Scholar] [CrossRef]
- Got, R.; Marnay, A. Isolement, purification et quelques caractéristiques physicochimiques de deux beta-hexosidases du suc digestif d’Helix pomatia [Isolation, purification and some physicochemical characteristics of 2 beta-hexosidases of the digestive juice of Helix pomatia]. Eur. J. Biochem. 1968, 4, 240–246. [Google Scholar] [CrossRef]
- Colas, B. Kinetic studies on β-fucosidases of Achatina balteata. Biochim. Biophys. Acta—Enzymol. 1979, 613, 448–458. [Google Scholar] [CrossRef]
- Calvo, P.; Santamaria, M.G.; Melgar, M.J.; Cabezas, J.A. Kinetic evidence for two active sites in beta-D-fucosidase of Helicella ericetorum. Int. J. Biochem. 1983, 15, 685–693. [Google Scholar] [CrossRef]
- Summers, M.D.; Smith, G.E. A manual of methods for baculovirus vectors and insect cell culture procedures. Tex. Agric. Exp. Stn. Bull. 1987, 1555, 1–60. [Google Scholar]
- Kubelka, V.; Altmann, F.; Staudacher, E.; Tretter, V.; März, L.; Hård, K.; Kamerling, J.P.; Vliegenthart, J.F.G. Primary structures of the N-linked carbohydrate chains from honeybee venom phospholipase A2. Eur. J. Biochem. 1993, 213, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Stepan, H.; Staudacher, E. Optimization of monosaccharide determination using anthranilic acid and 1-phenyl-3-methyl-5-pyrazolone for gastropod analysis. Anal. Biochem. 2011, 418, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Oomens, A.G.; Blissard, G.W. Requirement for GP64 to Drive Efficient Budding of Autographa californica Multicapsid Nucleopolyhedrovirus. Virology 1999, 254, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Morlock, G.E.; Sabir, G. Comparison of two orthogonal liquid chromatographic methods for quantitation of sugars in food. J. Liq. Chromatogr. Relat. Technol. 2011, 34, 902–919. [Google Scholar] [CrossRef]

| HPTLC Unlabeled Substrate | HPLC–AA AA Labeled Substrate | HPLC–pNP pNP Labeled Substrate | |
|---|---|---|---|
| Galβ1,4Glc (Lactose) | Yes (partly) | Yes | Yes |
| Galβ1,3GalNAc (Galacto-N-biose) | Yes | No | Yes |
| Galβ1,4GlcNAc (N-Acetyllactosamine) | Yes (partly) | No | ND |
| Galβ1,6GlcNAc | No | No | ND |
| Fucα1,3[Galβ1,4]Glc (3-Fucosyllactose) | No | No | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thoma, J.; Grabherr, R.; Staudacher, E. Expression and Characterization of a β-Galactosidase from the Pacific Oyster, Crassostrea gigas, and Evaluation of Strategies for Testing Substrate Specificity. Int. J. Mol. Sci. 2023, 24, 15287. https://doi.org/10.3390/ijms242015287
Thoma J, Grabherr R, Staudacher E. Expression and Characterization of a β-Galactosidase from the Pacific Oyster, Crassostrea gigas, and Evaluation of Strategies for Testing Substrate Specificity. International Journal of Molecular Sciences. 2023; 24(20):15287. https://doi.org/10.3390/ijms242015287
Chicago/Turabian StyleThoma, Julia, Reingard Grabherr, and Erika Staudacher. 2023. "Expression and Characterization of a β-Galactosidase from the Pacific Oyster, Crassostrea gigas, and Evaluation of Strategies for Testing Substrate Specificity" International Journal of Molecular Sciences 24, no. 20: 15287. https://doi.org/10.3390/ijms242015287
APA StyleThoma, J., Grabherr, R., & Staudacher, E. (2023). Expression and Characterization of a β-Galactosidase from the Pacific Oyster, Crassostrea gigas, and Evaluation of Strategies for Testing Substrate Specificity. International Journal of Molecular Sciences, 24(20), 15287. https://doi.org/10.3390/ijms242015287






