Regulation Mechanism of Dopamine Receptor 1 in Low Temperature Response of Marsupenaeus japonicus
Abstract
1. Introduction
2. Results
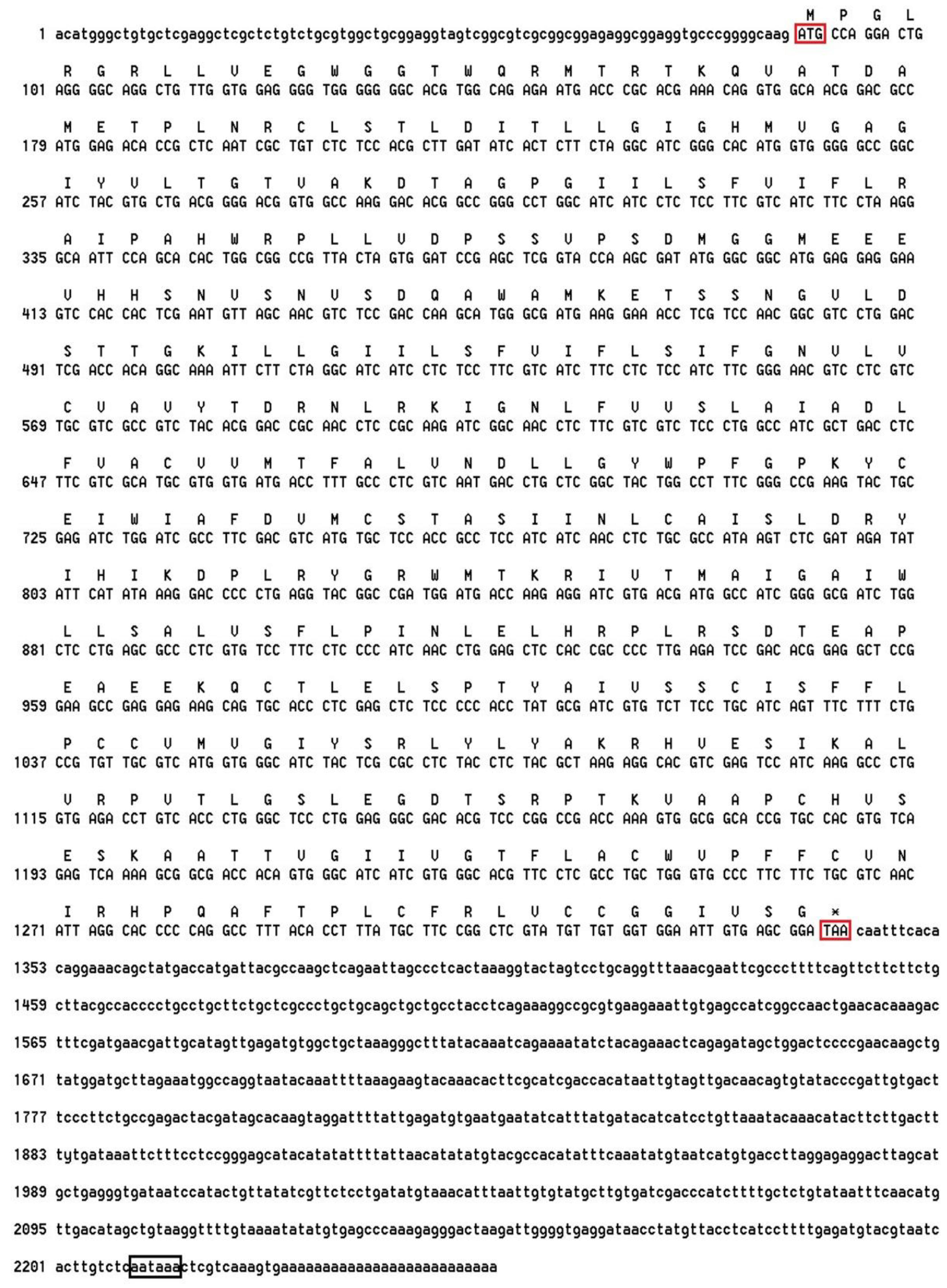
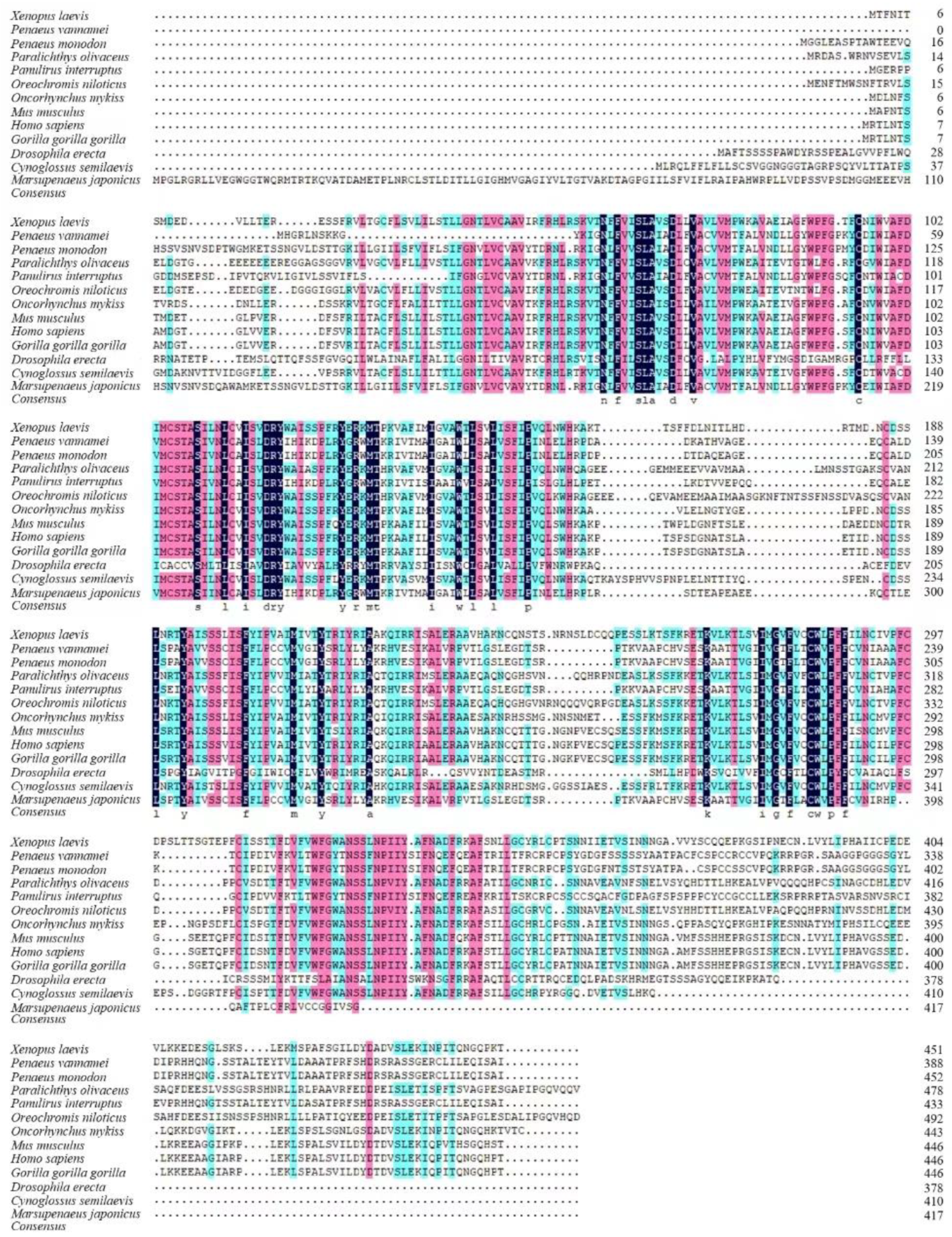
2.1. Characterization of the MjDAD1 Sequence
2.2. Tissue Expression of MjDAD1
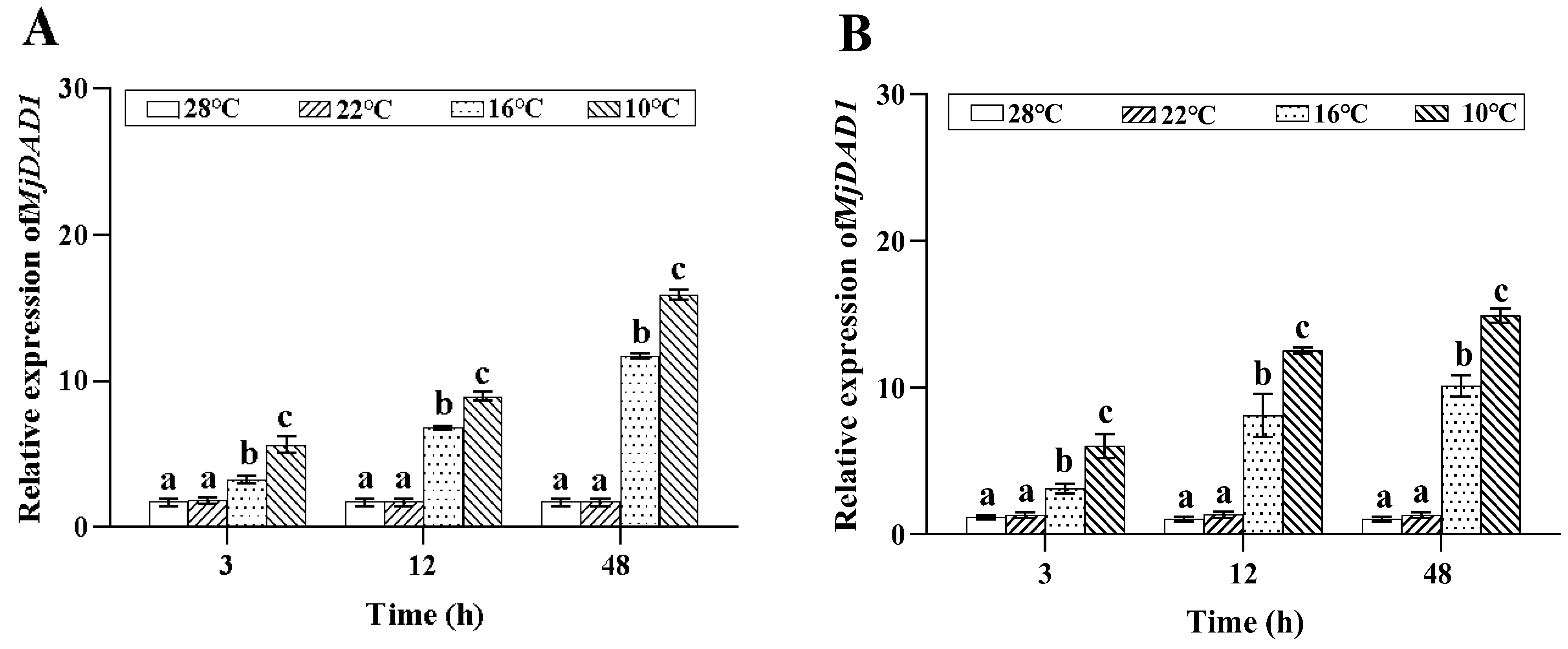
2.3. MjDAD1 mRNA Expression under Low Temperature Stress
2.4. Low Temperature Stress Effects on Intracellular Pathway Factors
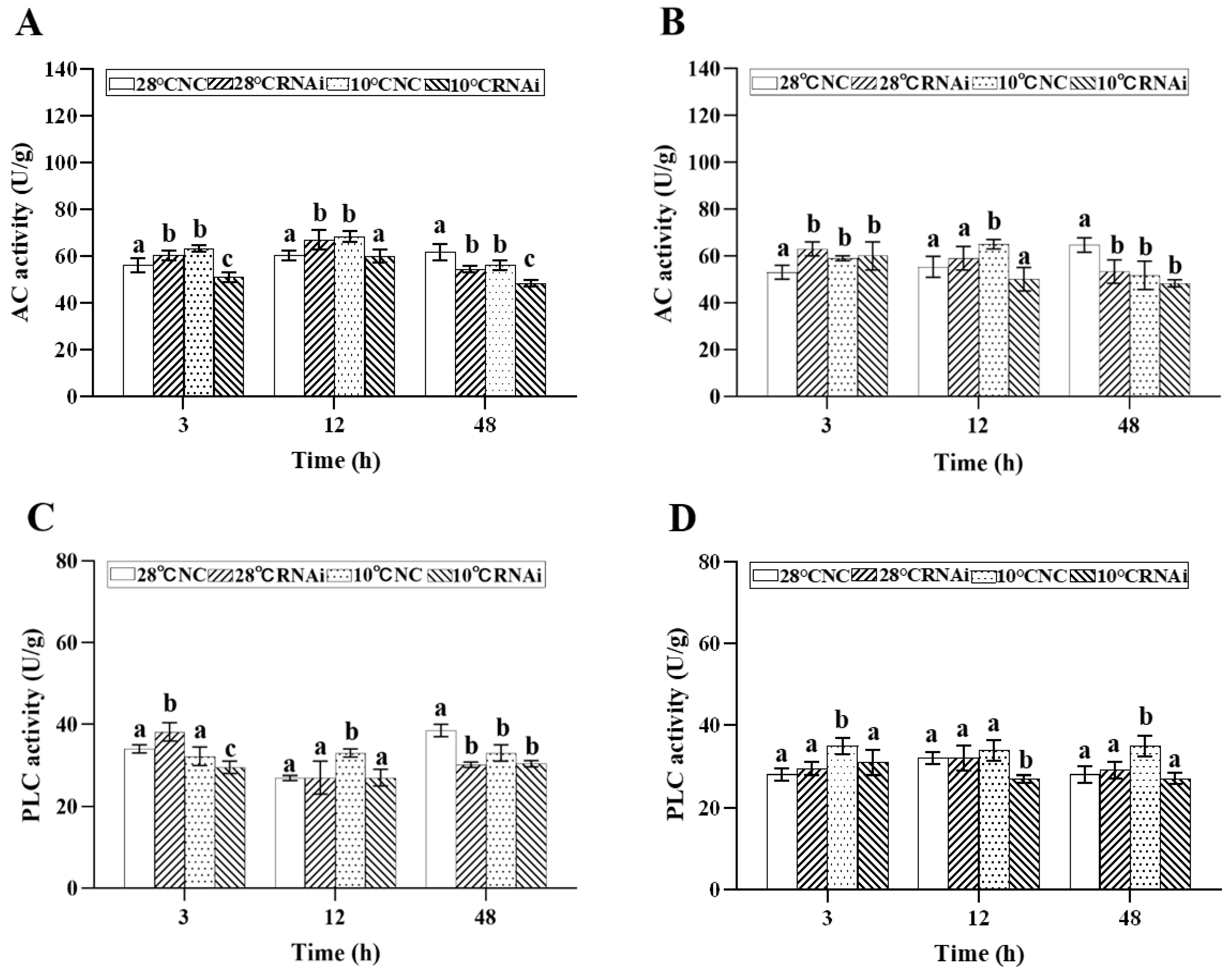
2.4.1. Effects of Low Temperature Stress on G Protein Effectors
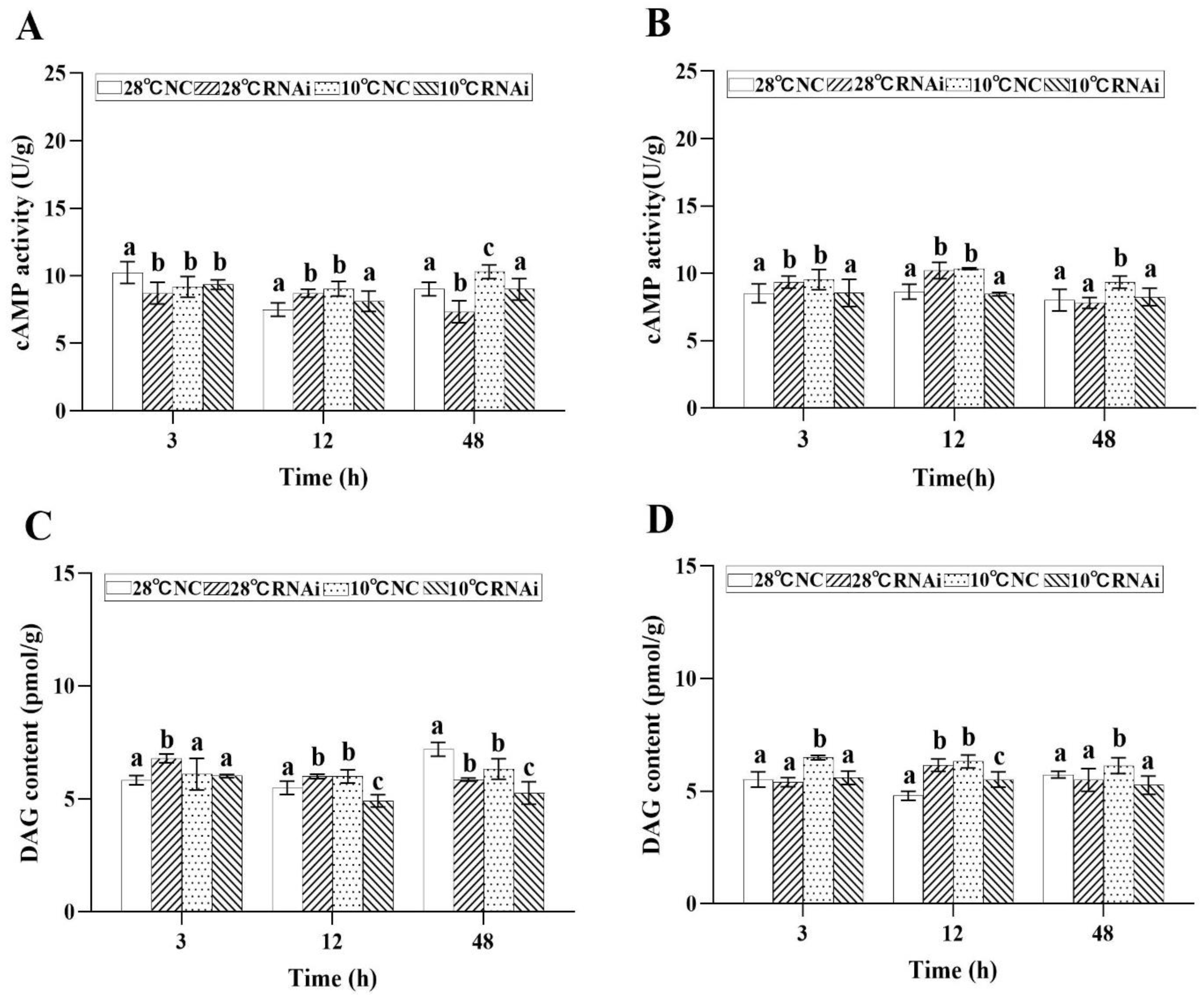
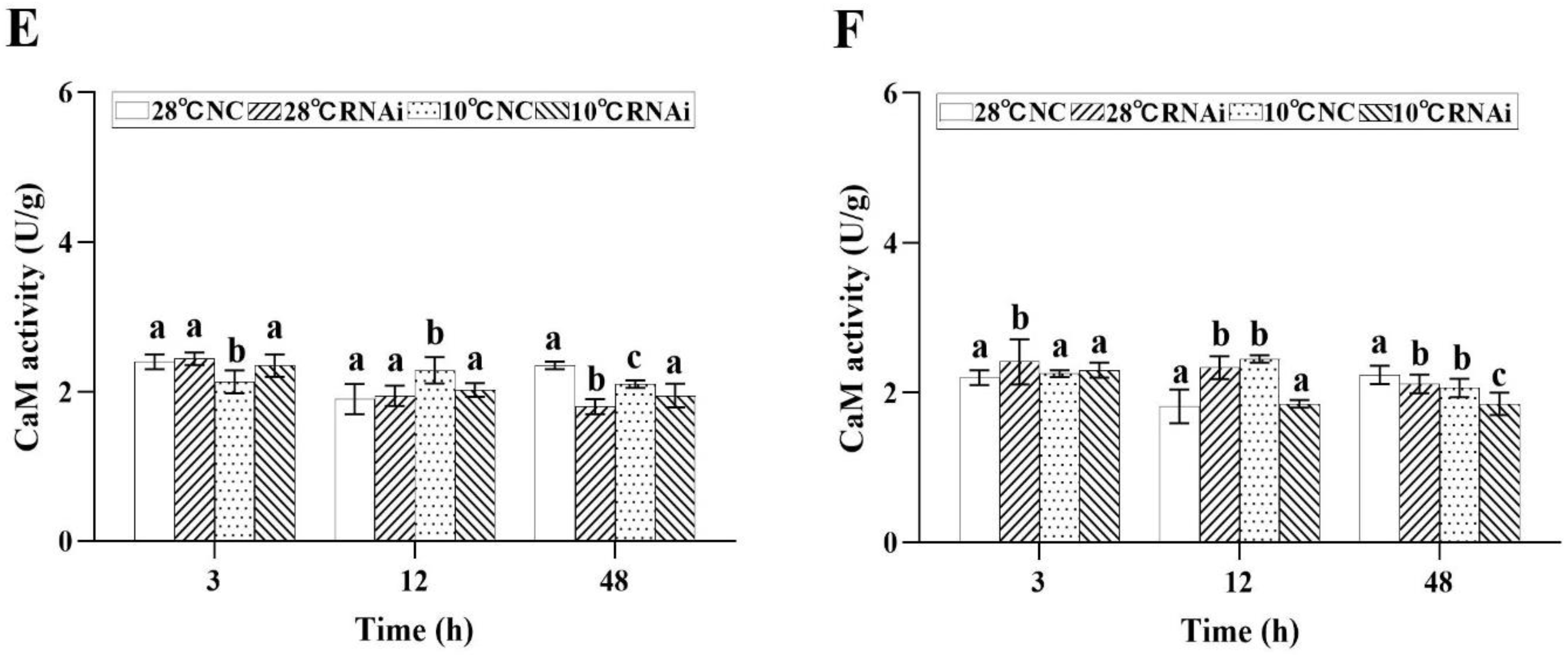
2.4.2. Effect of Low Temperature Stress on Second Messengers
2.4.3. Effect of Low Temperature Stress on Protein Kinases
2.5. The Relative Expression Ratio of MjDAD1 after MjDAD1 RNAi
2.6. Effect of MjDAD1 Interference on Tissue Damage of M. japonicus under Low Temperature Stress
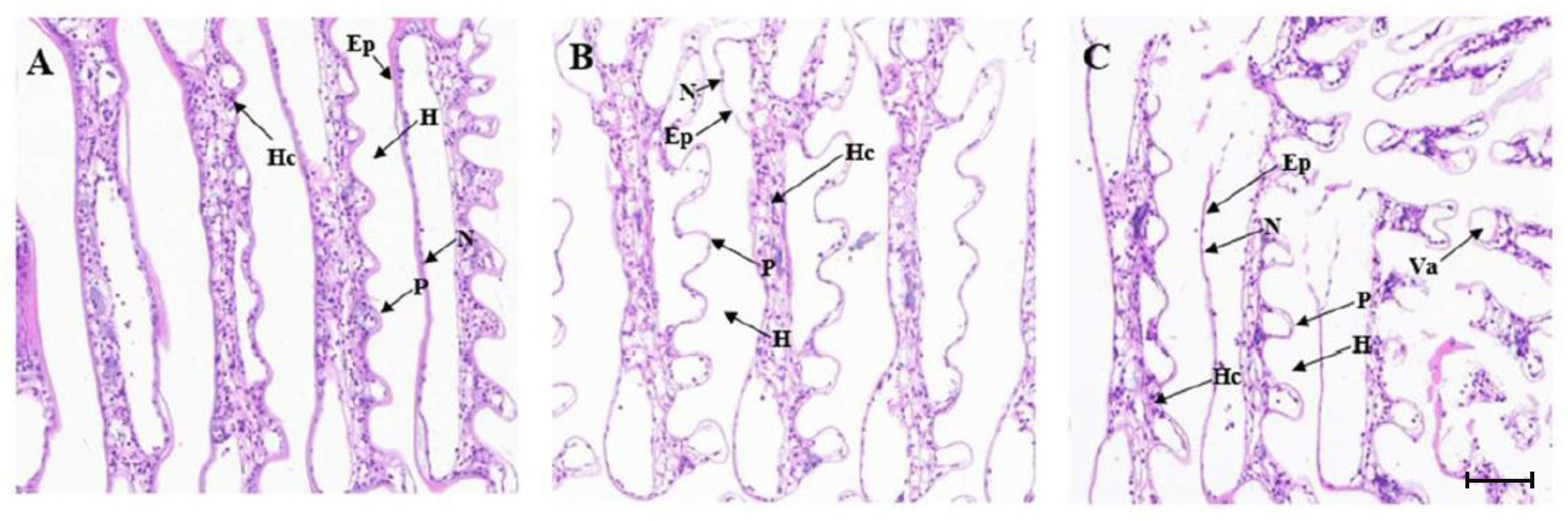
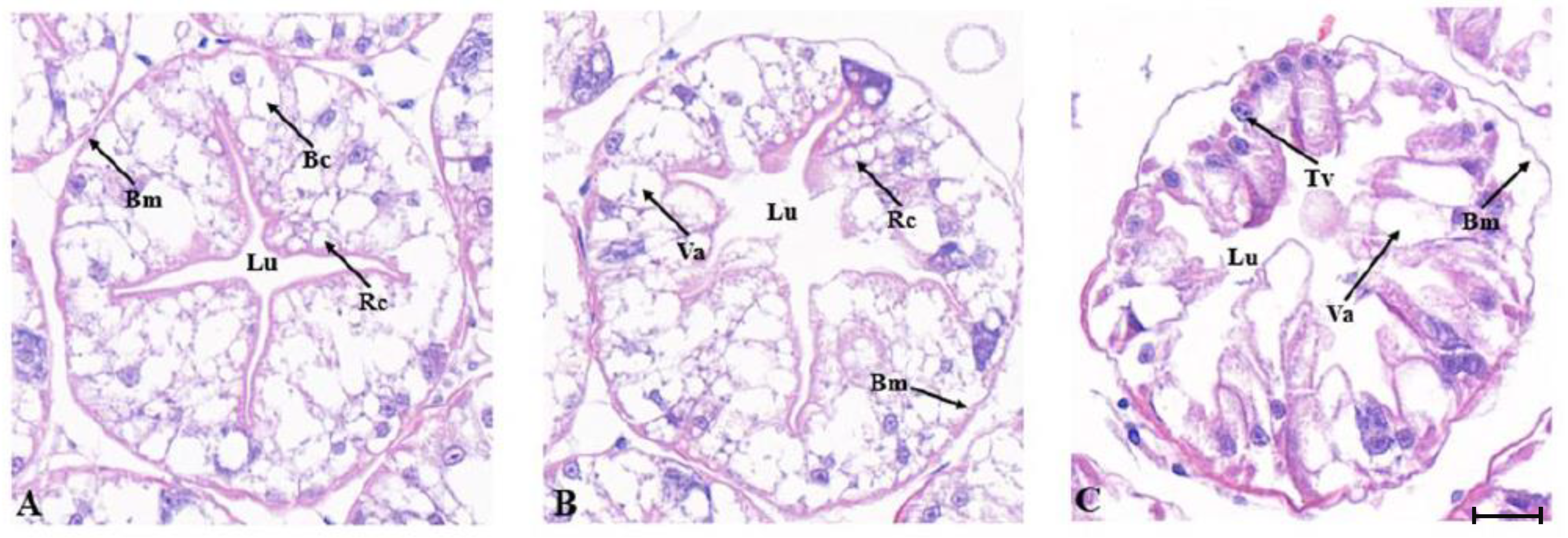
2.6.1. Histopathology of the Gills and Hepatopancreas after MjDAD1 Interference
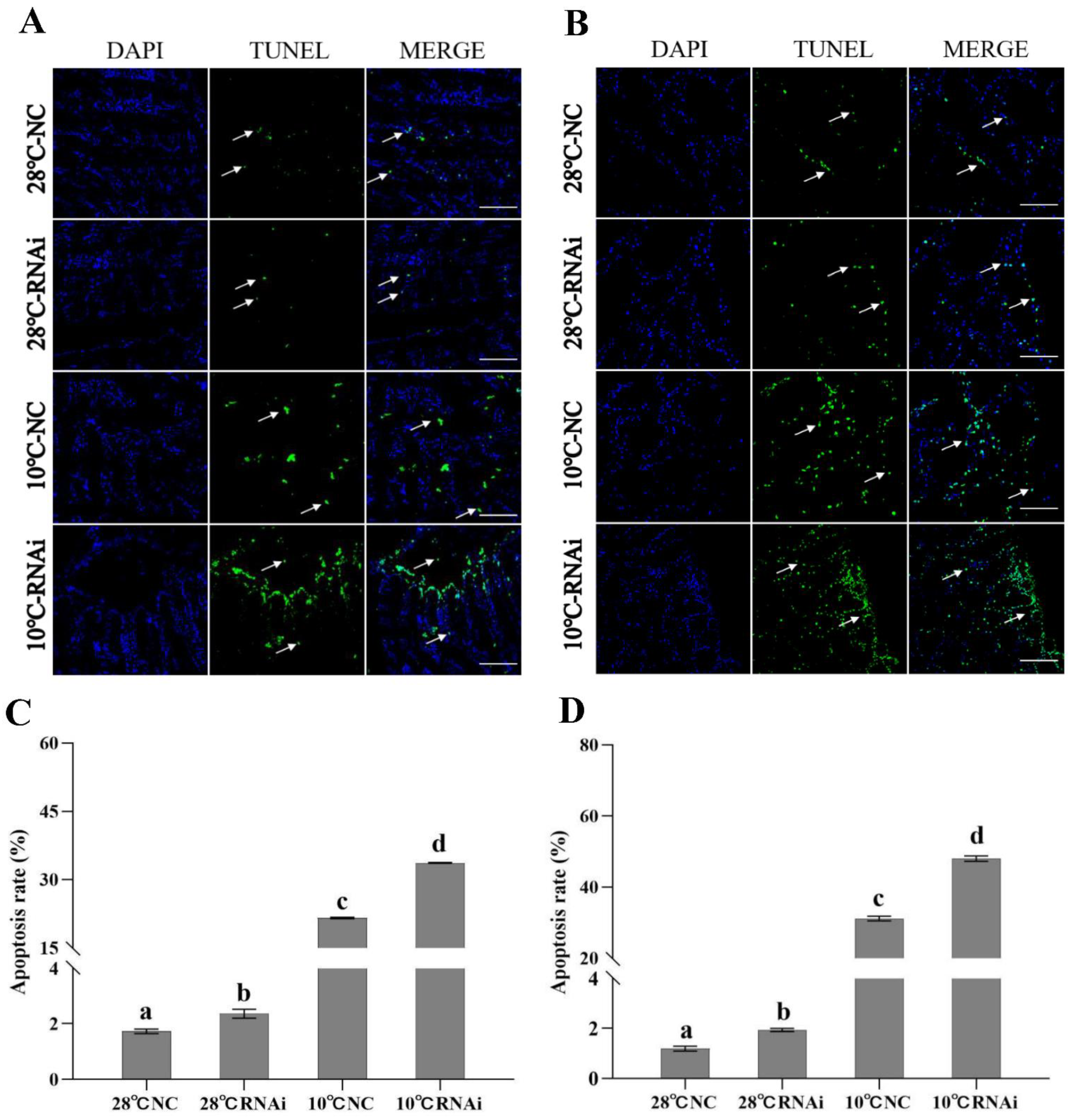
2.6.2. TUNEL Detection of Apoptosis after MjDAD1 Interference
2.7. MjDAD1 Silencing Affects Intracellular Signaling Pathway Factors in the Gills and Hepatopancreas under Low Temperature Stress
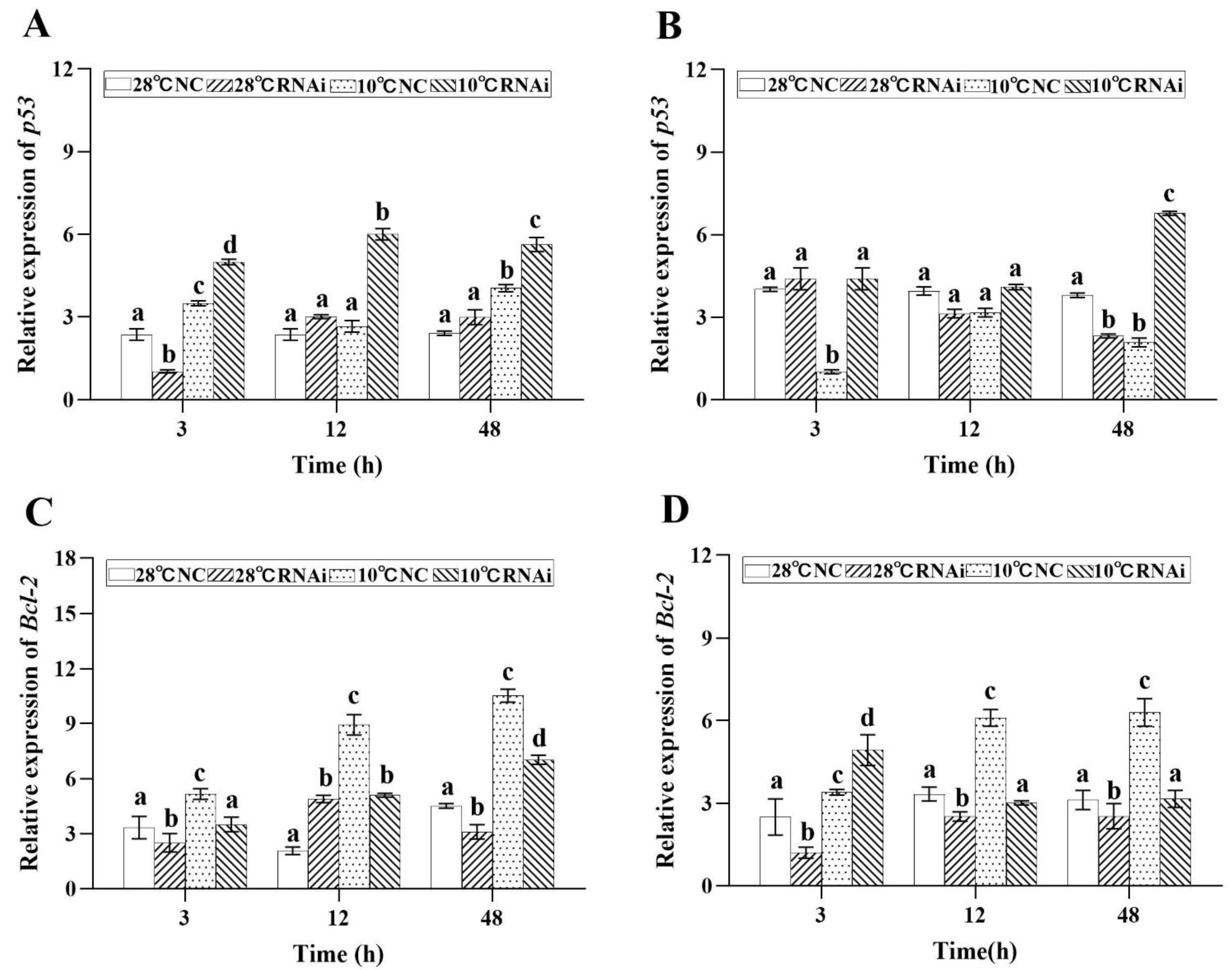
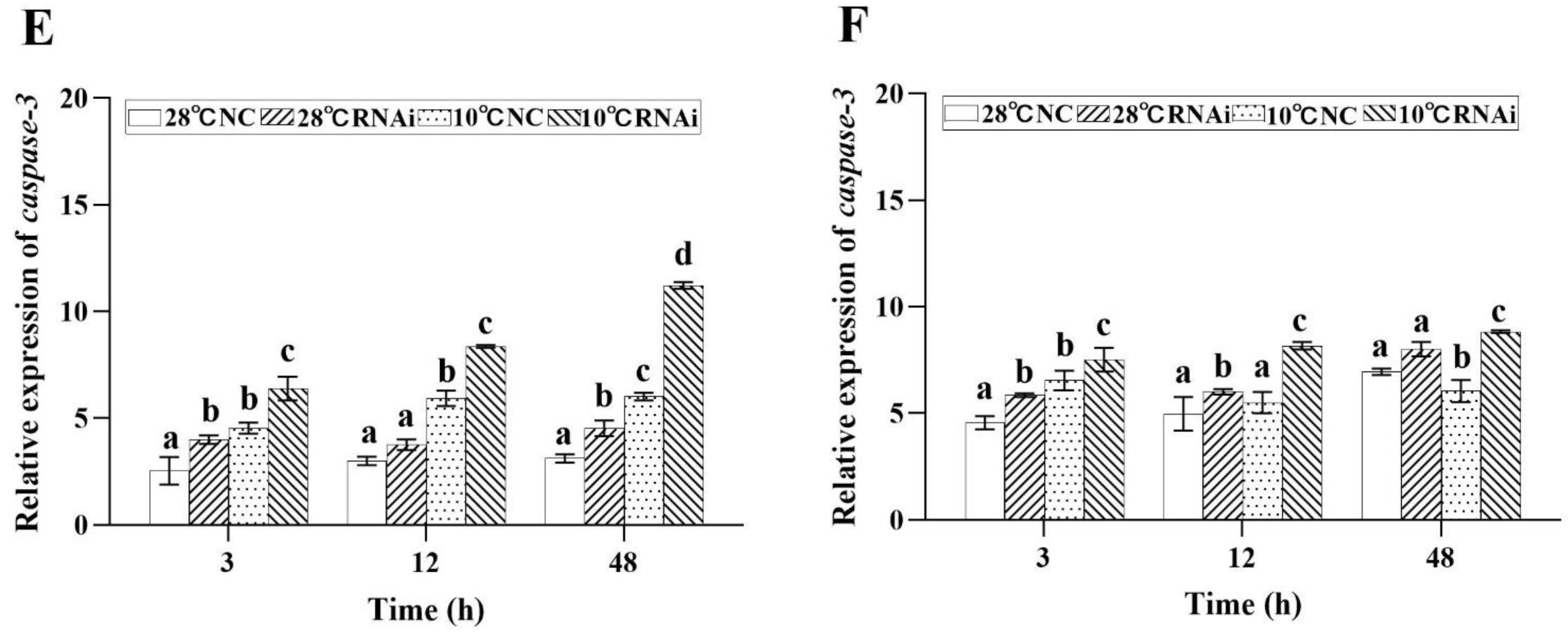
2.8. MjDAD1 Silencing Affects Apoptosis-related Gene Expression in M. japonicus under Low Temperature Stress
3. Discussion
4. Materials and Methods
4.1. Ethical Considerations
4.2. Experimental Animals
4.3. Application of Low Temperature Stress and Collection of Samples
4.4. Cloning of MjDAD1
4.4.1. Extraction of Total RNA and Full Length cDNA Cloning
4.4.2. Analysis of the Sequence and Phylogenetic Tree Construction
4.4.3. The Distribution of MjDAD1 in Tissues and Analysis of Its Expression Pattern
4.5. Short Interfering RNA (siRNA) Analysis
4.6. Histological Examination and TUNEL Assay
4.7. Protein Kinases and Intracellular Second Messengers Determination
4.7.1. Supernatant Preparations
4.7.2. Assay for Intracellular Signaling Transduction Factors
4.8. Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
4.9. Statistical Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GPCRs | G protein-coupled receptors |
| DARs | Dopamine receptors |
| DAD1 | D1-Dopamine receptor |
| DA | dopamine |
| AC | adenylate cyclase |
| PLC | phospholipase C |
| cAMP | cyclic adenosine monophosphate |
| cGMP | cyclic guanosine monophosphate |
| CaM | calmodulin |
| DAG | diacylglycerol |
| PKA | protein kinase A |
| PKC | protein kinases C |
| Bcl-2 | BCL2 apoptosis regulator |
| TM | transmembrane |
| TUNEL | Terminal deoxynulceotidyl transferase nick-end-labeling |
| qRT-PCR | quantitative real-time reverse transcription PCR |
| PBS | Phosphate-buffered saline |
References
- Liccardo, F.; Luini, A.; Di Martino, R. Endomembrane-Based Signaling by GPCRs and G-Proteins. Cells 2022, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Katritch, V.; Cherezov, V.; Stevens, R.C. Structure-Function of the G Protein–Coupled Receptor Superfamily. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 531–556. [Google Scholar] [CrossRef] [PubMed]
- Karam, C.S.; Jones, S.K.; Javitch, J.A. Come Fly with Me: An overview of dopamine receptors in Drosophila melanogaster. Basic Clin. Pharmacol. Toxicol. 2019, 126, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Gainetdinov, R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 2011, 63, 182–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liao, C.; Meng, W.; Huang, Q.; Li, D. Activation of D1-like dopamine receptors increases the NMDA-induced gain modulation through a PKA-dependent pathway in the premotor nucleus of adult zebra finches. Neurosci. Lett. 2015, 589, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, J.C.; Lee, J.S. G protein-coupled receptors (GPCRs) in rotifers and cladocerans: Potential applications in ecotoxicology, ecophysiology, comparative endocrinology, and pharmacology. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2022, 256, 109297. [Google Scholar] [CrossRef] [PubMed]
- Kawato, S.; Nishitsuji, K.; Arimoto, A.; Hisata, K.; Kawamitsu, M.; Nozaki, R.; Kondo, H.; Shinzato, C.; Ohira, T.; Satoh, N.; et al. Genome and transcriptome assemblies of the kuruma shrimp, Marsupenaeus japonicus. G3 2021, 11, jkab268. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture of the People’s Republic of China. China Agriculture Yearbook; China Agricultural Press: Beijing, China, 2022. [Google Scholar]
- Chen, R.; Xiao, M.; Buchberger, A.; Li, L. Quantitative Neuropeptidomics Study of the Effects of Temperature Change in the Crab Cancer borealis. J. Proteome Res. 2014, 13, 5767–5776. [Google Scholar] [CrossRef]
- Dong, C.L.; Zhu, F.; Du, Y.Z.; Lu, M.X. Depending on different apoptosis pathways, the effector Cscaspase-3 in Chilo suppressalis exposed to temperature and parasitic stress was induced. Int. J. Biol. Macromol. 2023, 238, 124270. [Google Scholar] [CrossRef]
- Yu, K.; Shi, C.; Ye, Y.; Li, R.; Mu, C.; Ren, Z.; Wang, C. The effects of overwintering temperature on the survival of female adult mud crab, Scylla paramamosain, under recirculating aquaculture systems as examined by histological analysis of the hepatopancreas and expression of apoptosis-related genes. Aquaculture 2023, 565, 739080. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, L.; Xu, L.; Si, L. Effects of ammonia-N exposure on the concentrations of neurotransmitters, hemocyte intracellular signaling pathways and immune responses in white shrimp Litopenaeus vannamei. Fish Shellfish. Immunol. 2018, 75, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, J.; Wei, B.; Cheng, Y.; Zhang, L.; Zhen, X. Comparative transcriptome analysis reveals osmotic-regulated genes in the gill of Chinese mitten crab (Eriocheir sinensis). PLoS ONE 2019, 14, e0210469. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.M.; Mykles, D.L.; Elizur, A.; Ventura, T. Characterization of G-protein coupled receptors from the blackback land crab Gecarcinus lateralis Y organ transcriptome over the molt cycle. BMC Genom. 2019, 20, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Huang, Y.; Liu, Z.; Cao, B.B.; Peng, Y.P.; Qiu, Y.H. Dopamine Receptors Modulate Cytotoxicity of Natural Killer Cells via cAMP-PKA-CREB Signaling Pathway. PLoS ONE 2013, 8, e65860. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Xu, P.; Mao, C.; Wang, L.; Krumm, B.; Zhou, X.E.; Huang, S.; Liu, H.; Cheng, X.; Huang, X.P.; et al. Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Cell 2021, 184, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Gaur, U.; Cao, Z.; Hou, S.T.; Zheng, W. Dopamine D1- and D2-like receptors oppositely regulate lifespan via a dietary restriction mechanism in Caenorhabditis elegans. BMC Biol. 2022, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rosikon, K.D.; Bone, M.C.; Lawal, H.O. Regulation and modulation of biogenic amine neurotransmission in Drosophila and Caenorhabditis elegans. Front. Physiol. 2023, 14, 970405. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, E.; Capuzzo, A. Cyclic AMP signaling in bivalve molluscs: An overview. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2010, 313, 179–200. [Google Scholar] [CrossRef]
- Tong, R.; Pan, L.; Zhang, X.; Li, Y. Neuroendocrine-immune regulation mechanism in crustaceans: A review. Rev. Aquac. 2022, 14, 378–398. [Google Scholar] [CrossRef]
- Rex, E.B.; Rankin, M.L.; Yang, Y.; Lu, Q.; Gerfen, C.R.; Jose, P.A.; Sibley, D.R. Identification of RanBP 9/10 as interacting partners for protein kinase C (PKC) γ/δ and the D1 dopamine receptor: Regulation of PKC-mediated receptor phosphorylation. Mol. Pharmacol. 2010, 78, 69–80. [Google Scholar] [CrossRef]
- Broome, S.T.; Louangaphay, K.; Keay, K.A.; Leggio, G.M.; Musumeci, G.; Castorina, A. Dopamine: An immune transmitter. Neural Regen. Res. 2020, 15, 2173. [Google Scholar] [CrossRef]
- Arnaldo, F.B.; Villar, V.A.M.; Konkalmatt, P.R.; Owens, S.A.; Asico, L.D.; Concepcion, G.P. D1-like dopamine receptors downregulate Na+-K+-ATPase activity and increase cAMP production in the posterior gills of the blue crab Callinectes sapidus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, 634–642. [Google Scholar] [CrossRef]
- Sánchez-Nuño, S.; Sanahuja, I.; Fernández-Alacid, L.; Ordóñez-Grande, B.; Fontanillas, R.; Fernández-Borràs, J.; Blasco, J.; Carbonell, T.; Ibarz, A. Redox Challenge in a Cultured Temperate Marine Species During Low Temperature and Temperature Recovery. Front. Physiol. 2018, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wu, J.; Sun, L.; Qin, X.; Fan, X.; Zheng, X. Combined stress of acute cold exposure and waterless duration at low temperature induces mortality of shrimp Litopenaeus vannamei through injuring antioxidative and immunological response in hepatopancreas tissue. J. Therm. Biol. 2021, 100, 103080. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.X.; Ren, X.Y.; Shao, H.X.; Liu, P.; Li, J. Effect of low temperature stress on antioxidant system and apoptosis of Marsupenaeus japonicus. Prog. Fish. Sci. 2022, 43, 157–166. [Google Scholar] [CrossRef]
- Yin, X.; Ren, Y.; Luo, W.; Liao, M.; Huang, L.; Zhuang, X.; Liu, Y.; Wang, W. Nemo-like kinase (NLK) gene regulates apoptosis via the p53 signaling pathway in Litopenaeus vannamei under low-temperature stress. Dev. Comp. Immunol. 2022, 131, 104378. [Google Scholar] [CrossRef] [PubMed]
- Pirger, Z.; Rácz, B.; Kiss, T. Dopamine-induced programmed cell death is associated with cytochrome c release and caspase-3 activation in snail salivary gland cells. Biol. Cell 2009, 101, 105–116. [Google Scholar] [CrossRef]
- Ren, X.; Lv, J.; Liu, M.; Wang, Q.; Shao, H.; Liu, P.; Li, J. A chromosome-level genome of the kuruma shrimp (Marsupenaeus japonicus) provides insights into its evolution and cold-resistance mechanism. Genomics 2022, 114, 110373. [Google Scholar] [CrossRef]
- Zemann, A.; De-Bekke, A.O.; Kiefmann, M.; Brosius, J.; Schmitz, J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006, 34, 2676–2685. [Google Scholar] [CrossRef][Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing evolutionary trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- An, Y.; Xing, H.; Zhang, Y.; Jia, P.; Gu, X.; Teng, X. The evaluation of potential immunotoxicity induced by environmental pollutant ammonia in broilers. Poult. Sci. 2019, 98, 3165–3175. [Google Scholar] [CrossRef]
- Shah, S.W.A.; Chen, J.; Han, Q.; Xu, Y.; Ishfaq, M.; Teng, X. Ammonia inhalation impaired immune function and mitochondrial integrity in the broilers bursa of fabricius: Implication of oxidative stress and apoptosis. Ecotoxicol. Environ. Saf. 2020, 190, 110078. [Google Scholar] [CrossRef]

| Primer | Sequence | Usage |
|---|---|---|
| MjDAD1 F1 | ATTCCCGACATCGTTTTCAAGGTGC | 3′ RACE |
| MjDAD1 F2 | CAGTTCTTCTTCTGCTTACGCCACCC | 3′ RACE |
| MjDAD1 R1 | GGCAAAGGTCATCACCACGCA | 5′ RACE |
| MjDAD1 R2 | AGGAAGATGACGAAGGAGAGGATGATGC | 5′ RACE |
| UPM(short) | CTAATACGACTCACTATAGGGC | RACE |
| UPM(long) | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | RACE |
| M13 F | GTAAAACGACGGCCAGT | colony PCR |
| M13 R | CAGGAAACAGCTATGAC | colony PCR |
| NUP | AAGCAGTGGTATCAACGCAGAGT | RACE |
| dsDAD1 F | GCUUGAUAUCAAUCUUCUATT | RNAi |
| dsDAD1 R | UAGAAGAGUGAUAUCAAGCTT | RNAi |
| NC F | UUCUCCGAACGUGUCACGUTT | RNAi |
| NC R | ACGUGACACGUUCGGAGAATT | RNAi |
| MjDAD1 F | CGCCTCCATCATCAACCTCT | qRT-PCR |
| MjDAD1 R | GCCATCGTCACGATCCTCTT | qRT-PCR |
| MjDAD2 R | AAGCAAGCACGTCGAAACTCC | qRT-PCR |
| MjBcl-2 F | TCTCAAAATGGCTCCCG | qRT-PCR |
| MjBcl-2 R | GTCACTGTCGCTCACACTAC | qRT-PCR |
| p53 F | CCAGTGGGTGGAGTATCA | qRT-PCR |
| p53 R | TTTGTGACGACCAGCCC | qRT-PCR |
| caspase-3 F | GCCTCTCACGACGCCTACAT | qRT-PCR |
| caspase-3 R | GTCGCTGTGGTCTCGTT | qRT-PCR |
| β-actin F | TCCACGAGACCACATACAAC | qRT-PCR |
| β-actin R | CACTTCCTGTGAACGATTGA | qRT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, X.; Bian, X.; Shao, H.; Jia, S.; Yu, Z.; Liu, P.; Li, J.; Li, J. Regulation Mechanism of Dopamine Receptor 1 in Low Temperature Response of Marsupenaeus japonicus. Int. J. Mol. Sci. 2023, 24, 15278. https://doi.org/10.3390/ijms242015278
Ren X, Bian X, Shao H, Jia S, Yu Z, Liu P, Li J, Li J. Regulation Mechanism of Dopamine Receptor 1 in Low Temperature Response of Marsupenaeus japonicus. International Journal of Molecular Sciences. 2023; 24(20):15278. https://doi.org/10.3390/ijms242015278
Chicago/Turabian StyleRen, Xianyun, Xueqiong Bian, Huixin Shao, Shaoting Jia, Zhenxing Yu, Ping Liu, Jian Li, and Jitao Li. 2023. "Regulation Mechanism of Dopamine Receptor 1 in Low Temperature Response of Marsupenaeus japonicus" International Journal of Molecular Sciences 24, no. 20: 15278. https://doi.org/10.3390/ijms242015278
APA StyleRen, X., Bian, X., Shao, H., Jia, S., Yu, Z., Liu, P., Li, J., & Li, J. (2023). Regulation Mechanism of Dopamine Receptor 1 in Low Temperature Response of Marsupenaeus japonicus. International Journal of Molecular Sciences, 24(20), 15278. https://doi.org/10.3390/ijms242015278





