One Earth: The Equilibrium between the Human and the Bacterial Worlds †
Abstract
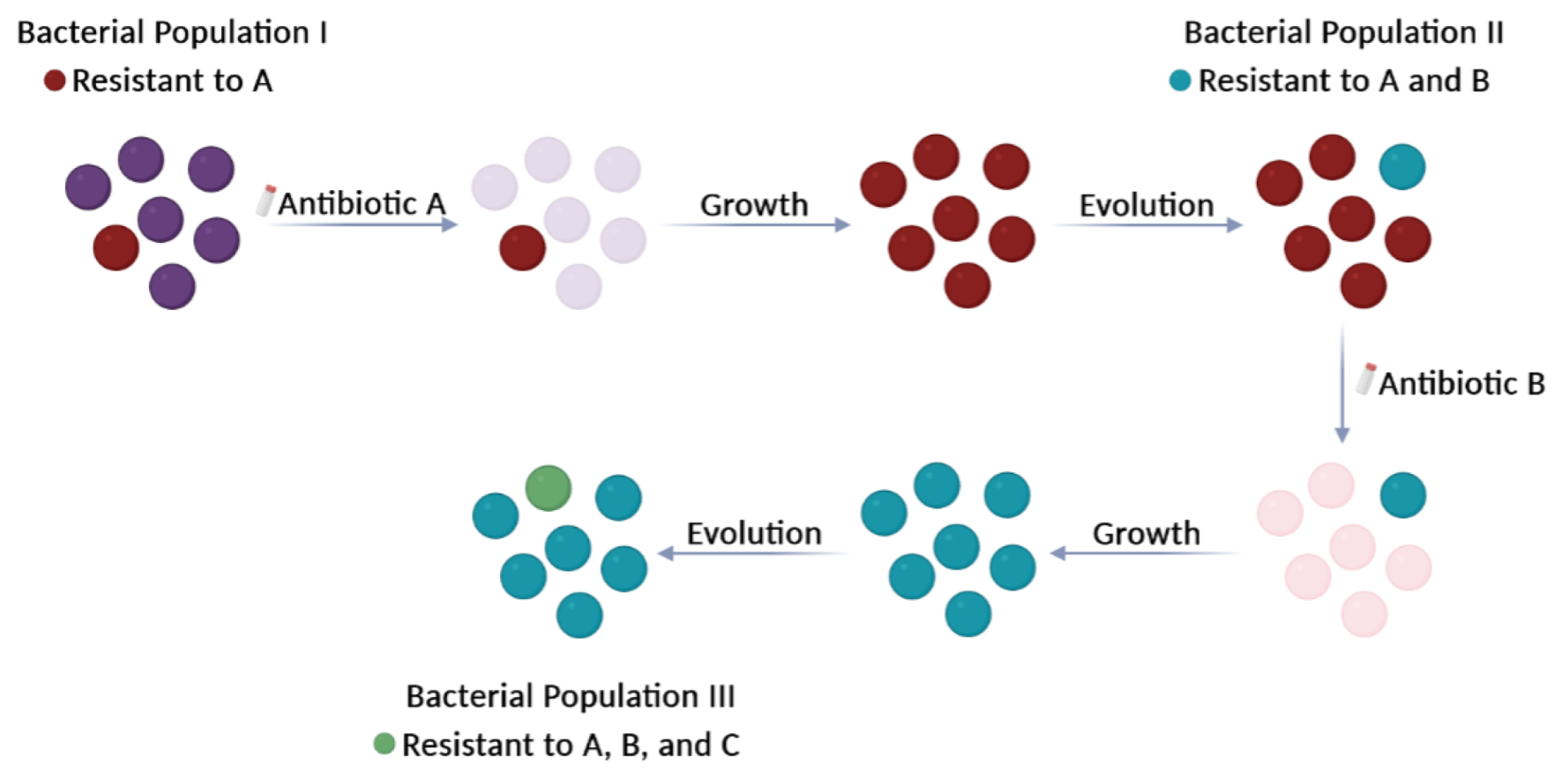
1. Introduction
2. The Antibiotic Resistome
3. The Mobilome
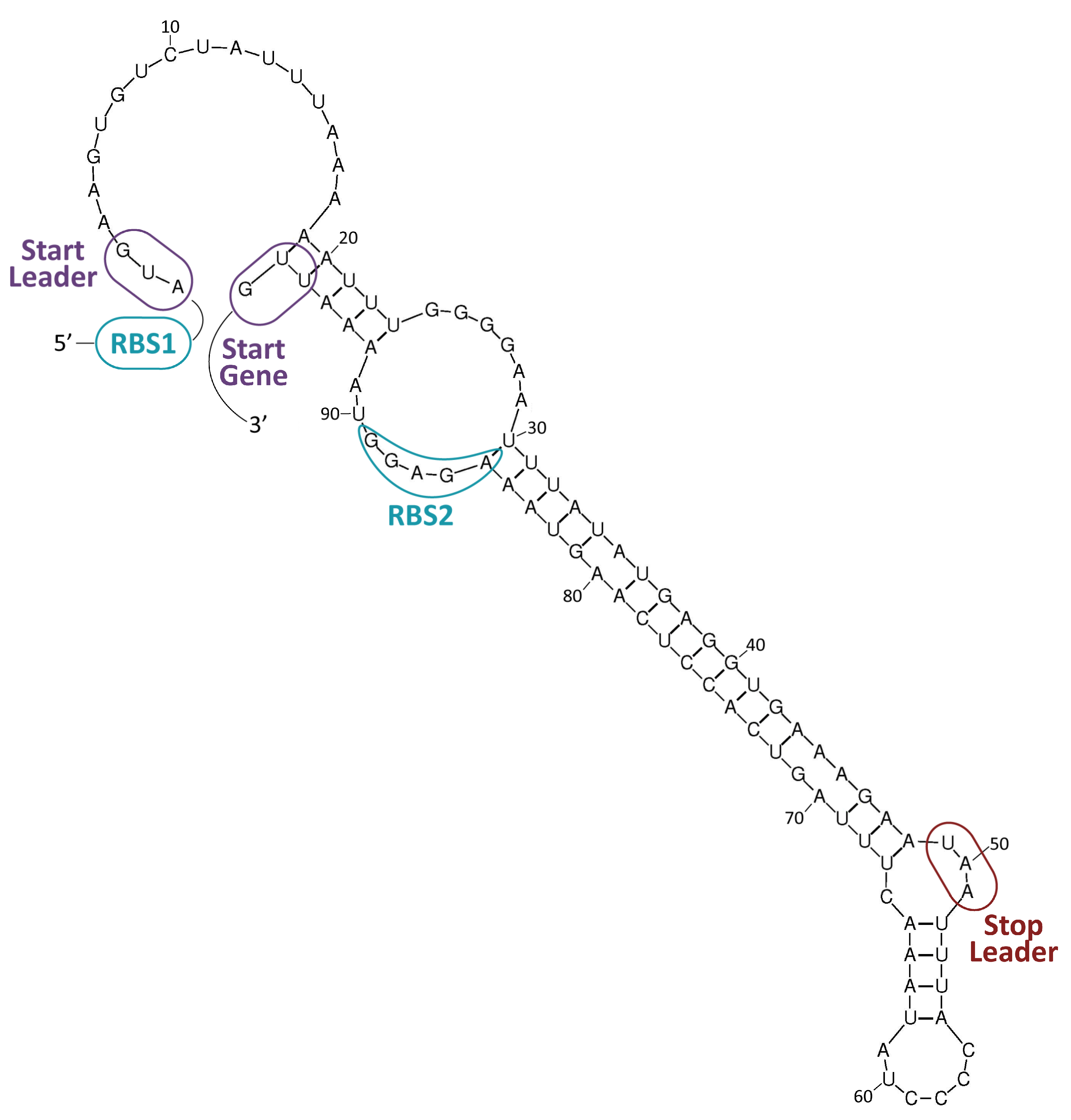
3.1. Mobilization Mechanisms
3.2. Transfer of MGEs
3.3. Inhibition of HGT Processes
4. The Nichome
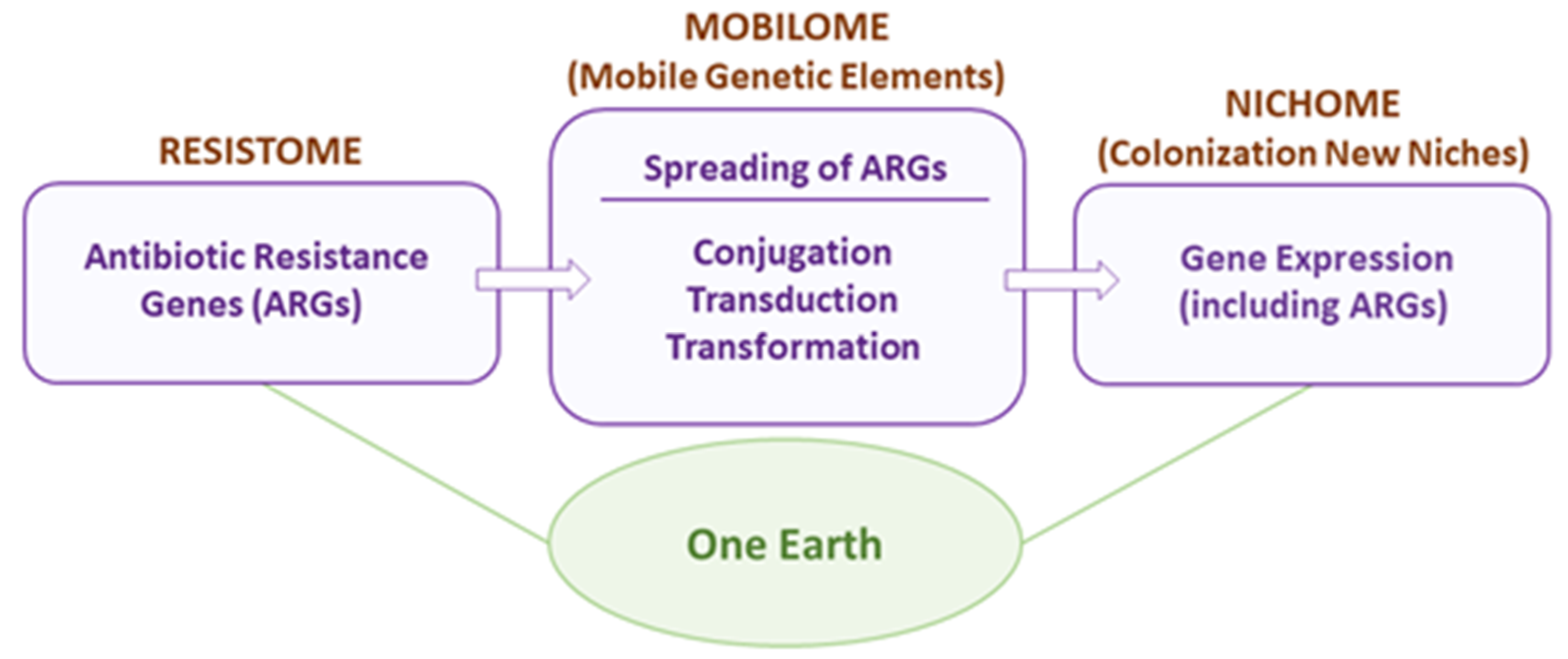
5. One Earth: Final Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Waglechner, N.; Wright, G.D. Antibiotic resistance: It’s bad, but why isn’t it worse? BMC Biol. 2017, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Hobson, C.; Chan, A.N.; Wright, G.D. The antibiotic resistome: A guide for the discovery of natural products as antimicrobial agents. Chem. Rev. 2021, 121, 3464–3494. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.; Zhu, K.; Surujon, D.; Rosconi, F.; Ortiz-Marquez, J.C.; van Opijnen, T. A Pangenomic perspective on the emergence, maintenance, and predictability of antibiotic resistance. In The Pangenome: Diversity, Dynamics and Evolution of Genomes; Tettelin, H., Medini, D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 169–202. [Google Scholar]
- Nadeem, S.F.; Gohar, U.F.; Tahir, S.F.; Mukhtar, H.; Pornpukdeewattana, S.; Nukthamna, P.; Moula Ali, A.M.; Bavisetty, S.C.B.; Massa, S. Antimicrobial resistance: More than 70 years of war between humans and bacteria. Crit. Rev. Microbiol. 2020, 46, 578–599. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Ruiz-Cruz, S.; Alkorta, I.; Espinosa, M. When humans met superbugs: Strategies to tackle bacterial resistances to antibiotics. BMC 2018, 9, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Vaillancourt, M.; Jorth, P. The unrecognized threat of secondary bacterial infections with COVID-19. mBio 2020, 11, e01806-20. [Google Scholar] [CrossRef]
- Taylor, L. COVID-19: Antimicrobial misuse in Americas sees drug resistant infections surge, says WHO. BMJ 2021, 375, n2845. [Google Scholar] [CrossRef]
- Daoud, Z. Editorial: Global dissemination and evolution of epidemic multidrug-resistant gram-negative bacterial pathogens: Surveillance, diagnosis, and treatment. Front. Microbiol. 2022, 13, 1028288. [Google Scholar] [CrossRef]
- Smith, D.F.Q.; Casadevall, A. Disaster Microbiology—A New Field of Study. mBio 2022, 13, e01680-22. [Google Scholar] [CrossRef]
- Stanton, I.C.; Murray, A.K.; Zhang, L.; Snape, J.; Gaze, W.H. Evolution of antibiotic resistance at low antibiotic concentrations including selection below the minimal selective concentration. Commun. Biol. 2020, 3, 467. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Pluta, R.; Lorenzo-Díaz, F.; Bravo, A.; Espinosa, M. The facts and family secrets of plasmids that replicate via the rolling-circle mechanism. Microbiol. Mol. Biol. Rev. 2022, 86, e00222-20. [Google Scholar] [CrossRef] [PubMed]
- Maccaro, J. Be mindful of your metaphors about microbes. mSphere 2021, 6, e00431-21. [Google Scholar] [CrossRef]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiologyopen 2021, 10, e1197. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef]
- Gschwind, R.; Ugarcina Perovic, S.; Weiss, M.; Petitjean, M.; Lao, J.; Coelho, L.P.; Ruppé, E. ResFinderFG v2.0: A database of antibiotic resistance genes obtained by functional metagenomics. Nucleic Acids Res. 2023, 51, W493–W500. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Barbosa da Costa, N.; Oliva, A.; Huot, Y.; Walsh, D.A. A resistome survey across hundreds of freshwater bacterial communities reveals the impacts of veterinary and human antibiotics use. Front. Microbiol. 2022, 13, 995418. [Google Scholar] [CrossRef]
- Liu, G.; Catacutan, D.B.; Rathod, K.; Swanson, K.; Jin, W.; Mohammed, J.C.; Chiappino-Pepe, A.; Syed, S.A.; Fragis, M.; Rachwalski, K.; et al. Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023. [Google Scholar] [CrossRef]
- Peplow, M. Skeleton crew. Nature 2023, 618, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Maasch, J.R.M.A.; Torres, M.D.T.; Melo, M.C.R.; de la Fuente-Nunez, C. Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning. Cell Host Microbe 2023, 31, 1260–1274.e6. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Virulence Mech. Bact. Pathog. 2016, 4, 481–511. [Google Scholar]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, resistome and resistance mechanisms: A bacterial perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef] [PubMed]
- Ojkic, N.; Serbanescu, D.; Banerjee, S.; Harwood, C.S. Antibiotic resistance via bacterial cell shape-shifting. mBio 2022, 13, e00659-22. [Google Scholar] [CrossRef] [PubMed]
- Pasqua, M.; Bonaccorsi di Patti, M.C.; Fanelli, G.; Utsumi, R.; Eguchi, Y.; Trirocco, R.; Prosseda, G.; Grossi, M.; Colonna, B. Host-Bacterial pathogen communication: The wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 2021, 8, 723274. [Google Scholar] [CrossRef] [PubMed]
- Guay, G.G.; Khan, S.A.; Rothstein, D.M. The tet(K) gene of plasmid pT181 of Staphylococcus aureus encodes an efflux protein that contains 14 transmembrane helices. Plasmid 1993, 30, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Rowe, S.J.; Mecaskey, R.J.; Nasef, M.; Talton, R.C.; Sharkey, R.E.; Halliday, J.C.; Dunkle, J.A. Shared requirements for key residues in the antibiotic resistance enzymes ErmC and ErmE suggest a common mode of RNA recognition. J. Biol. Chem. 2020, 295, 17476–17485. [Google Scholar] [CrossRef]
- Osterman, I.A.; Chervontseva, Z.S.; Evfratov, S.A.; Sorokina, A.V.; Rodin, V.A.; Rubtsova, M.P.; Komarova, E.S.; Zatsepin, T.S.; Kabilov, M.R.; Bogdanov, A.A.; et al. Translation at first sight: The influence of leading codons. Nucleic Acids Res. 2020, 48, 6931–6942. [Google Scholar] [CrossRef]
- Lovett, P.S.; Rogers, E.J. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996, 60, 366. [Google Scholar] [CrossRef]
- Ballester, S.; Alonso, J.C.; López, P.; Espinosa, M. Comparative expression of the pC194-cat gene in Streptococcus pneumoniae, Bacillus subtilis and Escherichia coli. Gene 1990, 86, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Crofts, T.S.; Gasparrini, A.J.; Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 2017, 15, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-W.; Cha, C.-J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Carballa, L.; Navarro-López, V.; Martinez-Garcia, M. A Resistome roadmap: From the human body to pristine environments. Front. Microbiol. 2022, 13, 858831. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M. The Horizontal Gene Pool; Harwood Academic Publishers: Amsterdam, The Netherlands, 2000; p. 419. [Google Scholar]
- Dimitriu, T. Evolution of horizontal transmission in antimicrobial resistance plasmids. Microbiology 2022, 168, 1214. [Google Scholar] [CrossRef] [PubMed]
- Siefert, J.L. Defining the mobilome. Methods Mol. Biol. 2009, 532, 13–27. [Google Scholar] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- De Sousa, J.A.M.; Fillol-Salom, A.; Penadés, J.R.; Rocha, E.P.C. Identification and characterization of thousands of bacteriophage satellites across bacteria. Nucleic Acids Res. 2023, 51, 2759–2777. [Google Scholar] [CrossRef]
- Carr, V.R.; Shkoporov, A.; Hill, C.; Mullany, P.; Moyes, D.L. Probing the Mobilome: Discoveries in the dynamic microbiome. Trends Microbiol. 2021, 29, 158–170. [Google Scholar] [CrossRef]
- Domenech, M.; Ruiz, S.; Moscoso, M.; García, E. In vitro biofilm development of Streptococcus pneumoniae and formation of choline-binding protein–DNA complexes. Environ. Microbiol. Rep. 2015, 7, 715–727. [Google Scholar] [CrossRef]
- Carvalho, G.; Fouchet, D.; Danesh, G.; Godeux, A.-S.; Laaberki, M.-H.; Pontier, D.; Charpentier, X.; Venner, S. Bacterial transformation buffers environmental fluctuations through the reversible integration of mobile genetic elements. mBio 2020, 11, e02443-19. [Google Scholar] [CrossRef]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of antibiotic resistance genes in biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Garcia-Quintanilla, M.; Ramos-Morales, F.; Casadesus, J. Conjugal Transfer of the Salmonella enterica Virulence Plasmid in the Mouse Intestine. J. Bacteriol. 2008, 190, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penadés, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207. [Google Scholar] [CrossRef] [PubMed]
- Igler, C.; Schwyter, L.; Gehrig, D.; Wendling, C.C. Conjugative plasmid transfer is limited by prophages but can be overcome by high conjugation rates. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20200470. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Yang, Y.; Xu, X.; Břinda, K.; Polz, M.F.; Hanage, W.P.; Zhang, T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2008731118. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Grande, J.; Garcillan-Barcia, M.P.; de la Cruz, F.; Fernandez-Lopez, R. Fundamental parameters governing the transmission of conjugative plasmids. bioRxiv 2023. [Google Scholar] [CrossRef]
- Igler, C.; Huisman, J.S.; Siedentop, B.; Bonhoeffer, S.; Lehtinen, S. Plasmid co-infection: Linking biological mechanisms to ecological and evolutionary dynamics. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20200478. [Google Scholar] [CrossRef]
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef]
- Thoma, L.; Dobrowinski, H.; Finger, C.; Guezguez, J.; Linke, D.; Sepulveda, E.; Muth, G. A multiprotein DNA translocation complex directs intramycelial plasmid spreading during Streptomyces conjugation. mBio 2015, 6, e02559-14. [Google Scholar] [CrossRef]
- Thoma, L.; Muth, G. Conjugative DNA transfer in Streptomyces by TraB: Is one protein enough? FEMS Microbiol. Lett. 2012, 337, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Thoma, L.; Muth, G. Conjugative DNA-transfer in Streptomyces, a mycelial organism. Plasmid 2016, 87–88, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gray, T.A.; Derbyshire, K.M. Blending genomes: Distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol. Microbiol. 2018, 108, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Tang, C.M.; Liu, G.-Y. Towards a better understanding of antimicrobial resistance dissemination: What can be learnt from studying model conjugative plasmids? Mil. Med. Res. 2022, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Tyler, B.M.; Goldberg, R.B. Transduction of chromosomal genes between enteric bacteria by bacteriophage P1. J. Bacteriol. 1976, 125, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Deichelbohrer, I.; Alonso, J.C.; Lüder, G.; Trautner, T.A. Plasmid transduction by Bacillus subtilis bacteriophage SPP1: Effects of DNA homology between plasmid and bacteriophage. J. Bacteriol. 1985, 162, 1238–1243. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Edelman, I.; Lofdahl, S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J. Mol. Biol. 1986, 192, 209–220. [Google Scholar] [CrossRef]
- Brady, A.; Felipe-Ruiz, A.; Gallego del Sol, F.; Marina, A.; Quiles-Puchalt, N.; Penadés, J.R. Molecular Basis of Lysis–Lysogeny Decisions in Gram-Positive Phages. Annu. Rev. Microbiol. 2021, 75, 563–581. [Google Scholar] [CrossRef]
- Humphrey, S.; Fillol-Salom, A.; Quiles-Puchalt, N.; Ibarra-Chávez, R.; Haag, A.F.; Chen, J.; Penadés, J.R. Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements. Nat. Commun. 2021, 12, 6509. [Google Scholar] [CrossRef]
- Hall, J.P.J. Is the bacterial chromosome a mobile genetic element? Nat. Commun. 2021, 12, 6400. [Google Scholar] [CrossRef]
- Godeux, A.-S.; Svedholm, E.; Barreto, S.; Potron, A.; Venner, S.; Charpentier, X.; Laaberki, M.-H.; Bonomo Robert, A. Interbacterial transfer of carbapenem resistance and large antibiotic resistance islands by natural transformation in pathogenic Acinetobacter. mBio 2022, 13, e02631-21. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Stiefel, U. The impact of natural transformation on the acquisition of antibiotic resistance determinants. mBio 2022, 13, e00336-22. [Google Scholar] [CrossRef] [PubMed]
- Claverys, J.P.; Havarstein, L.S. Cannibalism and fratricide: Mechanisms and raisons d’être. Nat. Rev. Microbiol. 2007, 5, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Dubnau, D.; Blokesch, M. Mechanisms of DNA uptake by naturally competent bacteria. Annu. Rev. Genet. 2019, 53, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Havarstein, L.S.; Martin, B.; Johnsborg, O.; Granadel, C.; Claverys, J.P. New insights into pneumococcal fratricide: Relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 2006, 59, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Dubnau, D.; Losick, R. Bistability in bacteria. Mol. Microbiol. 2006, 61, 564–572. [Google Scholar] [CrossRef]
- González-Pastor, J.E.; Hobbs, E.C.; Losick, R. Cannibalism by sporulating bacteria. Science 2003, 301, 510–513. [Google Scholar] [CrossRef]
- Claverys, J.P.; Prudhomme, M.; Martin, B. Induction of competence regulons as general stress responses in Gram-positive bacteria. Annu. Rev. Microbiol. 2006, 60, 451–475. [Google Scholar] [CrossRef]
- Ambur, O.H.; Engelstädter, J.; Johnsen, P.J.; Miller, E.L.; Rozen, D.E. Steady at the wheel: Conservative sex and the benefits of bacterial transformation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150528. [Google Scholar] [CrossRef]
- Stefanic, P.; Belcijan, K.; Kraigher, B.; Kostanjšek, R.; Nesme, J.; Madsen, J.; Stenløkke; Kovac, J.; Sørensen, S.J.; Vos, M.; et al. Kin discrimination promotes horizontal gene transfer between unrelated strains in Bacillus subtilis. Nat. Commun. 2021, 12, 3457. [Google Scholar] [CrossRef]
- Croucher, N.J.; Mostowy, R.; Wymant, C.; Turner, P.; Bentley, S.D.; Fraser, C. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. 2016, 14, e1002394. [Google Scholar] [CrossRef] [PubMed]
- Khedkar, S.; Smyshlyaev, G.; Letunic, I.; Maistrenko, O.M.; Coelho, L.P.; Orakov, A.; Forslund, S.K.; Hildebrand, F.; Luetge, M.; Schmidt, T.S.B.; et al. Landscape of mobile genetic elements and their antibiotic resistance cargo in prokaryotic genomes. Nucleic Acids Res. 2022, 50, 3155–3168. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Lanza, V.F.; de la Cruz, F.; Baquero, F. Genomic and metagenomic technologies to explore the antibiotic resistance mobilome. Ann. N. Y. Acad. Sci. 2017, 1388, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Orejas, R.; Espinosa, M.; Yeo, C.C. The importance of the expendable: Toxin–Antitoxin genes in plasmids and chromosomes. Front. Microbiol. 2017, 8, 1479. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Martínez, J.L.; Baquero, F.; Andersson, D.I. Beyond serial passages: New methods for predicting the emergence of resistance to novel antibiotics. Curr. Opin. Pharmacol. 2011, 11, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Arriaga, A.M.; Espinosa, M.; del Solar, G. Fitness of the pMV158 replicon in Streptococcus Pneumoniae. Plasmid 2012, 67, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Bobay, L.-M.; Rocha, E.P.C. The chromosomal accommodation and domestication of mobile genetic elements. Curr. Opin. Microbiol. 2014, 22, 22–29. [Google Scholar] [CrossRef]
- Lorenzo-Díaz, F.; Fernández-Lopez, C.; Douarre, P.-E.; Baez-Ortega, A.; Flores, C.; Glaser, P.; Espinosa, M. Streptococcal group B integrative and mobilizable element IMESag-rpsI encodes a functional relaxase involved in its transfer. Open Biol. 2016, 6, 160084. [Google Scholar] [CrossRef]
- Fernandez-Lopez, R.; Machon, C.; Longshaw, C.M.; Martin, S.; Molin, S.; Zechner, E.L.; Espinosa, M.; Lanka, E.; de la Cruz, F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 2005, 151, 3517–3526. [Google Scholar] [CrossRef]
- Lin, A.; Jimenez, J.; Derr, J.; Vera, P.; Manapat, M.L.; Esvelt, K.M.; Villanueva, L.; Liu, D.R.; Chen, I.A. Inhibition of bacterial conjugation by phage M13 and its protein g3p: Quantitative analysis and model. PLoS ONE 2011, 6, e19991. [Google Scholar] [CrossRef] [PubMed]
- Cabezón, E.; de la Cruz, F.; Arechaga, I. Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front. Microbiol. 2017, 8, 2329. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Jurado, P.; González-Pérez, B.; Moncalián, G.; Fernández, L.A.; de la Cruz, F. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 2007, 63, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Getino, M.; de la Cruz, F. Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol. Spectr. 2018, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Palencia-Gándara, C.; Getino, M.; Moyano, G.; Redondo, S.; Fernández-López, R.; González-Zorn, B.; de la Cruz, F. Conjugation inhibitors effectively prevent plasmid transmission in natural environments. mBio 2021, 12, e01277-21. [Google Scholar] [CrossRef] [PubMed]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Rodríguez, I.; Arana, L.; Ugarte-Uribe, B.; Gómez-Rubio, E.; Martín-Santamaría, S.; Garbisu, C.; Alkorta, I. Type IV Coupling Proteins as potential targets to control the dissemination of antibiotic resistance. Front. Mol. Biosci. 2020, 7, 201. [Google Scholar] [CrossRef] [PubMed]
- Domenech, A.; Brochado, A.R.; Sender, V.; Hentrich, K.; Henriques-Normark, B.; Typas, A.; Veening, J.-W. Proton motive force disruptors block bacterial competence and horizontal gene transfer. Cell Host Microbe 2020, 27, 544–555.e3. [Google Scholar] [CrossRef]
- Chan, W.T.; Balsa, D.; Espinosa, M. One cannot rule them all: Are bacterial toxins-antitoxins druggable? FEMS Microbiol. Rev. 2015, 39, 522–540. [Google Scholar] [CrossRef]
- García-Contreras, R.; Martínez-Vázquez, M.; González-Pedrajo, B.; Castillo-Juárez, I. Editorial: Alternatives to combat bacterial infections. Front. Microbiol. 2022, 13, 909866. [Google Scholar] [CrossRef]
- Walker-Sünderhauf, D.; Klümper, U.; Pursey, E.; Westra, E.R.; Gaze, W.H.; van Houte, S. Removal of AMR plasmids using a mobile, broad host-range CRISPR-Cas9 delivery tool. Microbiology 2023, 169, 1334. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, Y.; Xie, F.; Olsen, R.H.; Shi, L.; Li, L. Inhibition of plasmid conjugation in Escherichia coli by targeting rbsB gene using CRISPRi system. Int. J. Mol. Sci. 2023, 24, 10585. [Google Scholar] [CrossRef] [PubMed]
- Kharga, K.; Kumar, L.; Patel, S.K.S. Recent advances in monoclonal antibody-based approaches in the management of bacterial sepsis. Biomedicines 2023, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Rouli, L.; Merhej, V.; Fournier, P.E.; Raoult, D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015, 7, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Vernikos, G.; Medini, D.; Riley, D.R.; Tettelin, H. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 2015, 23, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Sempere, J.; Llamosí, M.; Román, F.; Lago, D.; González-Camacho, F.; Pérez-García, C.; Yuste, J.; Domenech, M. Clearance of mixed biofilms of Streptococcus pneumoniae and methicillin-susceptible/resistant Staphylococcus aureus by antioxidants N-acetyl-l-cysteine and cysteamine. Sci. Rep. 2022, 12, 6668. [Google Scholar] [CrossRef]
- Douglas, G.M.; Shapiro, B.J. Genic selection within prokaryotic pangenomes. Genome Biol. Evol. 2021, 13, evab234. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Kumar, L.; Patel, S.K.S.; Kharga, K.; Kumar, R.; Kumar, P.; Pandohee, J.; Kulshresha, S.; Harjai, K.; Chhibber, S. Molecular mechanisms and applications of N-Acyl Homoserine Lactone-mediated Quorum Sensing in bacteria. Molecules 2022, 27, 7584. [Google Scholar] [CrossRef] [PubMed]
- Wellington, S.; Greenberg, E.P. Quorum Sensing signal selectivity and the potential for interspecies cross talk. mBio 2019, 10, e00146-19. [Google Scholar] [CrossRef]
- Azimi, S.; Lewin, G.R.; Whiteley, M. The biogeography of infection revisited. Nat. Rev. Microbiol. 2022, 20, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Vial, L.; Hommais, F. Plasmid-chromosome cross-talks. Environ. Microbiol. 2020, 22, 540–556. [Google Scholar] [CrossRef] [PubMed]
- Billane, K.; Harrison, E.; Cameron, D.; Brockhurst, M.A. Why do plasmids manipulate the expression of bacterial phenotypes? Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20200461. [Google Scholar] [CrossRef]
- Tierney, A.R.P.; Rather, P.N. Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 2019, 14, 533–552. [Google Scholar] [CrossRef]
- Bhagirath, A.Y.; Li, Y.; Patidar, R.; Yerex, K.; Ma, X.; Kumar, A.; Duan, K. Two component regulatory systems and antibiotic resistance in Gram-negative pathogens. Int. J. Mol. Sci. 2019, 20, 1781. [Google Scholar] [CrossRef]
- Stupar, M.; Furness, J.; De Voss, C.J.; Tan, L.; West, N.P. Two-component sensor histidine kinases of Mycobacterium tuberculosis: Beacons for niche navigation. Mol. Microbiol. 2022, 117, 973–985. [Google Scholar] [CrossRef]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef]
- Kato, A.; Groisman, E.A. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004, 18, 2302–2313. [Google Scholar] [CrossRef]
- Firon, A.; Tazi, A.; Da Cunha, V.; Brinster, S.; Sauvage, E.; Dramsi, S.; Golenbock, D.T.; Glaser, P.; Poyart, C.; Trieu-Cuot, P. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog. 2013, 9, e1003179. [Google Scholar] [CrossRef]
- Mitchell, S.L.; Ismail, A.M.; Kenrick, S.A.; Camilli, A. The VieB auxiliary protein negatively regulates the VieSA signal transduction system in Vibrio cholerae. BMC Microbiol. 2015, 15, 59. [Google Scholar] [CrossRef][Green Version]
- Mitrophanov, A.Y.; Groisman, E.A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008, 22, 2601–2611. [Google Scholar] [CrossRef]
- Pi, H.; Weiss, A.; Laut, C.L.; Grunenwald, C.M.; Lin, H.K.; Yi, X.I.; Stauff, D.L.; Skaar, E.P. An RNA-binding protein acts as a major post-transcriptional modulator in Bacillus anthracis. Nat. Commun. 2022, 13, 1491. [Google Scholar] [CrossRef]
- Wang, X.; Yu, D.; Chen, L. Antimicrobial resistance and mechanisms of epigenetic regulation. Front. Cell. Infect. Microbiol. 2023, 13, 1199646. [Google Scholar] [CrossRef]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Atlung, T.; Ingmer, H. H-NS: A modulator of environmentally regulated gene expression. Mol. Microbiol. 1997, 24, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Amit, R.; Oppenheim, A.B.; Stavans, J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003, 84, 2467–2473. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, H.; Kenney, L.J.; Yan, J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010, 24, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Winardhi, R.S.; Yan, J.; Kenney, L.J. H-NS regulates gene expression and compacts the nucleoid: Insights from single-molecule experiments. Biophys. J. 2015, 109, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, T.N.; Schmidt, H.; Madrid, C.; Juárez, A.; Bernadó, P.; Griesinger, C.; García, J.; Pons, M. Indirect DNA readout by an H-NS related protein: Structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog. 2011, 7, e1002380. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Wyman, C.; Wurm, R.; Wagner, R.; Goosen, N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1 J. Biol. Chem. 2002, 277, 2146–2150. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Lee, S.Y.; Kenney, L.J.; Yan, J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci. Rep. 2012, 2, 509. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.S.; Singh, N.; Bonocora, R.P.; Fitzgerald, D.M.; Wade, J.T.; Grainger, D.C. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014, 28, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Kotlajich, M.V.; Hron, D.R.; Boudreau, B.A.; Sun, Z.; Lyubchenko, Y.L.; Landick, R. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. eLife 2015, 4, e04970. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C.D. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef]
- Ali, S.S.; Whitney, J.C.; Stevenson, J.; Robinson, H.; Howell, P.L.; Navarre, W.W. Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex. J. Biol. Chem. 2013, 288, 13356–13369. [Google Scholar] [CrossRef]
- Boudreau, B.A.; Hron, D.R.; Qin, L.; van der Valk, R.A.; Kotlajich, M.V.; Dame, R.T.; Landick, R. StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments. Nucleic Acids Res. 2018, 46, 5525–5546. [Google Scholar] [CrossRef]
- Liu, X.; Lin, S.; Liu, T.; Zhou, Y.; Wang, W.; Yao, J.; Guo, Y.; Tang, K.; Chen, R.; Benedik, M.J.; et al. Xenogeneic silencing relies on temperature-dependent phosphorylation of the host H-NS protein in Shewanella. Nucleic Acids Res. 2021, 49, 3427–3440. [Google Scholar] [CrossRef]
- Will, W.R.; Bale, D.H.; Reid, P.J.; Libby, S.J.; Fang, F.C. Evolutionary expansion of a regulatory network by counter-silencing. Nat. Commun. 2014, 5, 5270. [Google Scholar] [CrossRef]
- Banda, M.M.; Zavala-Alvarado, C.; Pérez-Morales, D.; Bustamante, V.H. SlyA and HilD counteract H-NS-mediated repression on the ssrAB virulence operon of Salmonella enterica Serovar Typhimurium and thus promote its activation by OmpR. J. Bacteriol. 2019, 201, e00530-18. [Google Scholar] [CrossRef] [PubMed]
- Hustmyer, C.M.; Wolfe, M.B.; Welch, R.A.; Landick, R. RfaH counter-silences inhibition of transcript elongation by H-NS-StpA nucleoprotein filaments in pathogenic Escherichia coli. mBio 2022, 13, e0266222. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Groisman, E.A. Salmonella expresses foreign genes during infection by degrading their silencer. Proc. Natl. Acad. Sci. USA 2020, 117, 8074–8082. [Google Scholar] [CrossRef] [PubMed]
- Oberto, J.; Nabti, S.; Jooste, V.; Mignot, H.; Rouviere-Yaniv, J. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS ONE 2009, 4, e4367. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.I.; Kahramanoglou, C.; Ali, R.M.; Fraser, G.M.; Seshasayee, A.S.N.; Luscombe, N.M. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012, 40, 3524–3537. [Google Scholar] [CrossRef] [PubMed]
- Pinson, V.; Takahashi, M.; Rouviere-Yaniv, J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA J. Mol. Biol. 1999, 287, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Kamashev, D.; Balandina, A.; Rouviere-Yaniv, J. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999, 18, 5434–5444. [Google Scholar] [CrossRef]
- Kamashev, D.; Rouviere-Yaniv, J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000, 19, 6527–6535. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S.; Aki, T.; Adhya, S. Atomic force microscopic demonstration of DNA looping by GalR and HU. Nucleic Acids Res. 1997, 25, 873–876. [Google Scholar] [CrossRef]
- Ferrándiz, M.-J.; Carreño, D.; Ayora, S.; de la Campa, A.G. HU of Streptococcus pneumoniae is essential for the preservation of DNA supercoiling. Front. Microbiol. 2018, 9, 493. [Google Scholar] [CrossRef]
- Verma, S.C.; Harned, A.; Narayan, K.; Adhya, S. Non-specific and specific DNA binding modes of bacterial histone, HU, separately regulate distinct physiological processes through different mechanisms. Mol. Microbiol. 2023, 119, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Ribardo, D.A.; McIver, K.S. Defining the Mga regulon: Comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 2006, 62, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Hondorp, E.R.; McIver, K.S. The Mga virulence regulon: Infection where the grass is greener. Mol. Microbiol. 2007, 66, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Bourgogne, A.; Drysdale, M.; Hilsenbeck, S.G.; Peterson, S.N.; Koehler, T.M. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 2003, 71, 2736–2743. [Google Scholar] [CrossRef] [PubMed]
- Hemsley, C.; Joyce, E.; Hava, D.L.; Kawale, A.; Camilli, A. MgrA, an orthologue of Mga, acts as a transcriptional repressor of the genes within the rlrA pathogenicity islet in Streptococcus pneumoniae. J. Bacteriol. 2003, 185, 6640–6647. [Google Scholar] [CrossRef]
- Solano-Collado, V.; Espinosa, M.; Bravo, A. Activator role of the pneumococcal Mga-like virulence transcriptional regulator. J. Bacteriol. 2012, 194, 4197–4207. [Google Scholar] [CrossRef]
- Ruiz-Cruz, S.; Espinosa, M.; Goldmann, O.; Bravo, A. Global regulation of gene expression by the MafR protein of Enterococcus faecalis. Front. Microbiol. 2016, 6, 1521. [Google Scholar] [CrossRef]
- Ruiz-Cruz, S.; Moreno-Blanco, A.; Espinosa, M.; Bravo, A. Transcriptional activation by MafR, a global regulator of Enterococcus faecalis. Sci. Rep. 2019, 9, 6146. [Google Scholar] [CrossRef]
- Furuta, Y.; Cheng, C.; Zorigt, T.; Paudel, A.; Izumi, S.; Tsujinouchi, M.; Shimizu, T.; Meijer, W.G.; Higashi, H. Direct regulons of AtxA, the master virulence regulator of Bacillus anthracis. mSystems 2021, 6, e0029121. [Google Scholar] [CrossRef]
- Hammerstrom, T.G.; Horton, L.B.; Swick, M.C.; Joachimiak, A.; Osipiuk, J.; Koehler, T.M. Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity. Mol. Microbiol. 2015, 95, 426–441. [Google Scholar] [CrossRef] [PubMed]
- Solano-Collado, V.; Lurz, R.; Espinosa, M.; Bravo, A. The pneumococcal MgaSpn virulence transcriptional regulator generates multimeric complexes on linear double-stranded DNA. Nucleic Acids Res. 2013, 41, 6975–6991. [Google Scholar] [CrossRef] [PubMed]
- Tsvetanova, B.; Wilson, A.C.; Bongiorni, C.; Chiang, C.; Hoch, J.A.; Perego, M. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol. Microbiol. 2007, 63, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Hondorp, E.R.; Hou, S.C.; Hause, L.L.; Gera, K.; Lee, C.-E.; McIver, K.S. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol. Microbiol. 2013, 88, 1176–1193. [Google Scholar] [CrossRef] [PubMed]
- Sanson, M.; Makthal, N.; Gavagan, M.; Cantu, C.; Olsen, R.J.; Musser, J.M.; Kumaraswami, M. Phosphorylation events in the multiple gene regulator of group A Streptococcus significantly influence global gene expression and virulence. Infect. Immun. 2015, 83, 2382–2395. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Cruz, S.; Moreno-Blanco, A.; Espinosa, M.; Bravo, A. DNA-binding properties of MafR, a global regulator of Enterococcus faecalis. FEBS Lett. 2018, 592, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Solano-Collado, V.; Hüttener, M.; Espinosa, M.; Juárez, A.; Bravo, A. MgaSpn and H-NS: Two unrelated global regulators with similar DNA-binding properties. Front. Mol. Biosci. 2016, 3, 60. [Google Scholar] [CrossRef] [PubMed]
- Hause, L.L.; McIver, K.S. Nucleotides critical for the interaction of the Streptococcus pyogenes Mga virulence regulator with Mga-regulated promoter sequences. J. Bacteriol. 2012, 194, 4904–4919. [Google Scholar] [CrossRef]
- Hadjifrangiskou, M.; Koehler, T.M. Intrinsic curvature associated with the coordinately regulated anthrax toxin gene promoters. Microbiology 2008, 154, 2501–2512. [Google Scholar] [CrossRef]
- Moreno-Blanco, A.; Solano-Collado, V.; Ortuno-Camuñas, A.; Espinosa, M.; Ruiz-Cruz, S.; Bravo, A. PclR is a transcriptional activator of the gene that encodes the pneumococcal collagen-like protein PclA. Sci. Rep. 2022, 12, 11827. [Google Scholar] [CrossRef]
- Paterson, G.K.; Nieminen, L.; Jefferies, J.M.; Mitchell, T.J. PclA, a pneumococcal collagen-like protein with selected strain distribution, contributes to adherence and invasion of host cells. FEMS Microbiol. Lett. 2008, 285, 170–176. [Google Scholar] [CrossRef]
- Cernansky, R. Biodiversity moves beyond counting species. Nature 2017, 546, 22–24. [Google Scholar] [CrossRef] [PubMed]
- May, M. How to fight antibiotic resistance. Nat. Med. 2023, 29, 1583–1586. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, F.; Trujillo, M.; Dennehy, J.J. Why do antibiotics exist? mBio 2021, 12, e01966-21. [Google Scholar] [CrossRef]
- Averill, C.; Anthony, M.A.; Baldrian, P.; Finkbeiner, F.; van den Hoogen, J.; Kiers, T.; Kohout, P.; Hirt, E.; Smith, G.R.; Crowther, T.W. Defending Earth’s terrestrial microbiome. Nat. Microbiol. 2022, 7, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Hoyles, L. Human microbiome myths and misconceptions. Nat. Microbiol. 2023, 8, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Choudoir, M.J.; Eggleston, E.M. Reciprocal inclusion of microbiomes and environmental justice contributes solutions to global environmental health challenges. mSystems 2022, 7, e0146221. [Google Scholar] [CrossRef]
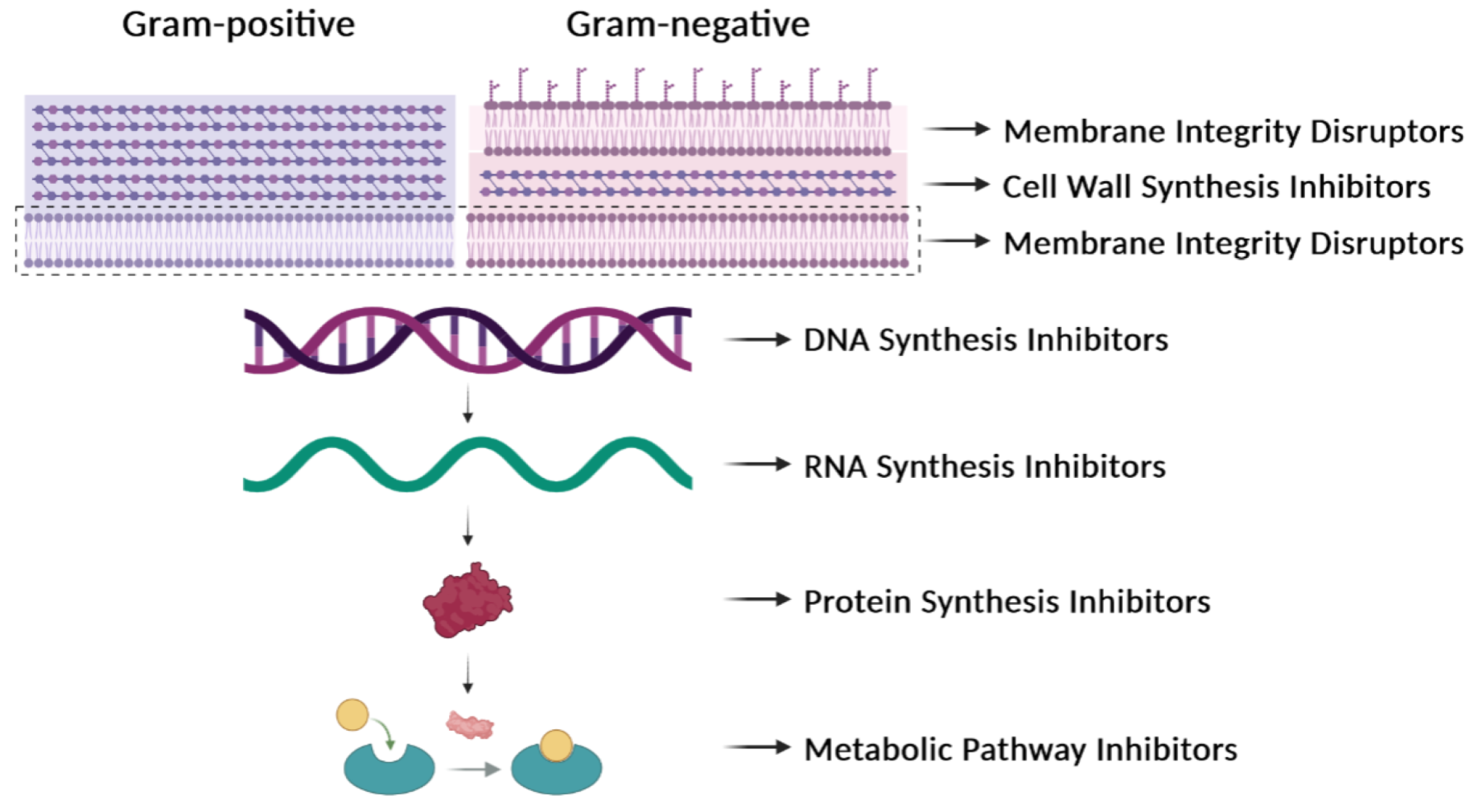
| Antibiotic Type: Examples | Targets | Resistance |
|---|---|---|
| β-Lactams: penicillins, cephalosporins | Enzymes involved in cell-wall synthesis | Breakage of the β-lactam ring by β-lactamases. Decreased uptake or increased efflux of the β-lactam. Changes in the active site of the target. |
| Cationic peptides: colistin, polymyxin B | Bacterial membrane | Lipopolysaccharide modifications. Efflux pumps. Capsular polysaccharide production. |
| Fluoroquinolones: ciprofloxacin | DNA gyrase and topoisomerase IV | Mutations in target enzymes. |
| Rifamycins: rifampin, rifabutin, rifapentine | RNA polymerase | Mutations in the RNA polymerase (β subunit). Drug inactivation. Impeded uptake of the drug. |
| Aminoglycosides: streptomycin, gentamicin | 30S ribosomal subunit | Enzymatic modification of the drug. Enzymatic modification of the target site. Efflux pumps. |
| Tetracyclines: tetracycline, doxycycline, minocycline | 30S ribosomal subunit | Efflux pumps. Ribosomal protection proteins. Enzymatic inactivation of tetracycline. |
| Phenicols: chloramphenicol, thiamphenicol, florfenicol | 50S ribosomal subunit | Ribosomal mutations. Enzymatic modification (methylation) of the target site. Enzymatic inactivation (acetylation) of the drug. Efflux pumps. |
| Macrolides: erythromycin, azithromycin, clarithromycin | 50S ribosomal subunit | Ribosomal modification by methylation or mutation. Efflux pumps. Drug inactivation. |
| Lincosamides: clindamycin | 50S ribosomal subunit | Ribosomal modification by methylation or mutation. Efflux pumps. Drug inactivation. |
| Sulfonamides: sulfamethazine, sulfadiazine | Dihydropteroate synthase in the folic acid pathway | Mutations in the target enzyme. Sulfonamide resistance mediated by HGT (genes encoding variants of the target enzyme). Drug degradation. |
| Element | Main Features |
|---|---|
| Plasmids | DNA molecules that replicate autonomously. |
| Conjugative Plasmids Mobilizable Plasmids | Encode all elements needed for conjugation. Encode a relaxase and an origin of transfer. Need the conjugative machinery provided by ‘auxiliary’ plasmids. |
| Integrative and Conjugative Elements (ICEs) | Integrated elements that can excise, replicate, and transfer by conjugation. Previously termed ‘Conjugative Transposons’. |
| Integrative and Mobilizable Elements (IMEs) | Integrated elements that can excise and replicate but need conjugative machinery provided by ‘auxiliary’ elements for their transfer. Previously termed ‘Mobilizable Transposons’. |
| Insertion Sequences (ISs) | Integrated elements that encode only transposition machinery. |
| Prophages | Bacteriophage genomes totally or partially inserted into the host chromosome. |
| Phage satellites | Elements that exploit phages for transfer between bacteria and can encode ARGs. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravo, A.; Moreno-Blanco, A.; Espinosa, M. One Earth: The Equilibrium between the Human and the Bacterial Worlds. Int. J. Mol. Sci. 2023, 24, 15047. https://doi.org/10.3390/ijms242015047
Bravo A, Moreno-Blanco A, Espinosa M. One Earth: The Equilibrium between the Human and the Bacterial Worlds. International Journal of Molecular Sciences. 2023; 24(20):15047. https://doi.org/10.3390/ijms242015047
Chicago/Turabian StyleBravo, Alicia, Ana Moreno-Blanco, and Manuel Espinosa. 2023. "One Earth: The Equilibrium between the Human and the Bacterial Worlds" International Journal of Molecular Sciences 24, no. 20: 15047. https://doi.org/10.3390/ijms242015047
APA StyleBravo, A., Moreno-Blanco, A., & Espinosa, M. (2023). One Earth: The Equilibrium between the Human and the Bacterial Worlds. International Journal of Molecular Sciences, 24(20), 15047. https://doi.org/10.3390/ijms242015047





