Monitoring the Conformational Changes of the Aβ(25−35) Peptide in SDS Micelles: A Matter of Time
Abstract
1. Introduction
2. Results
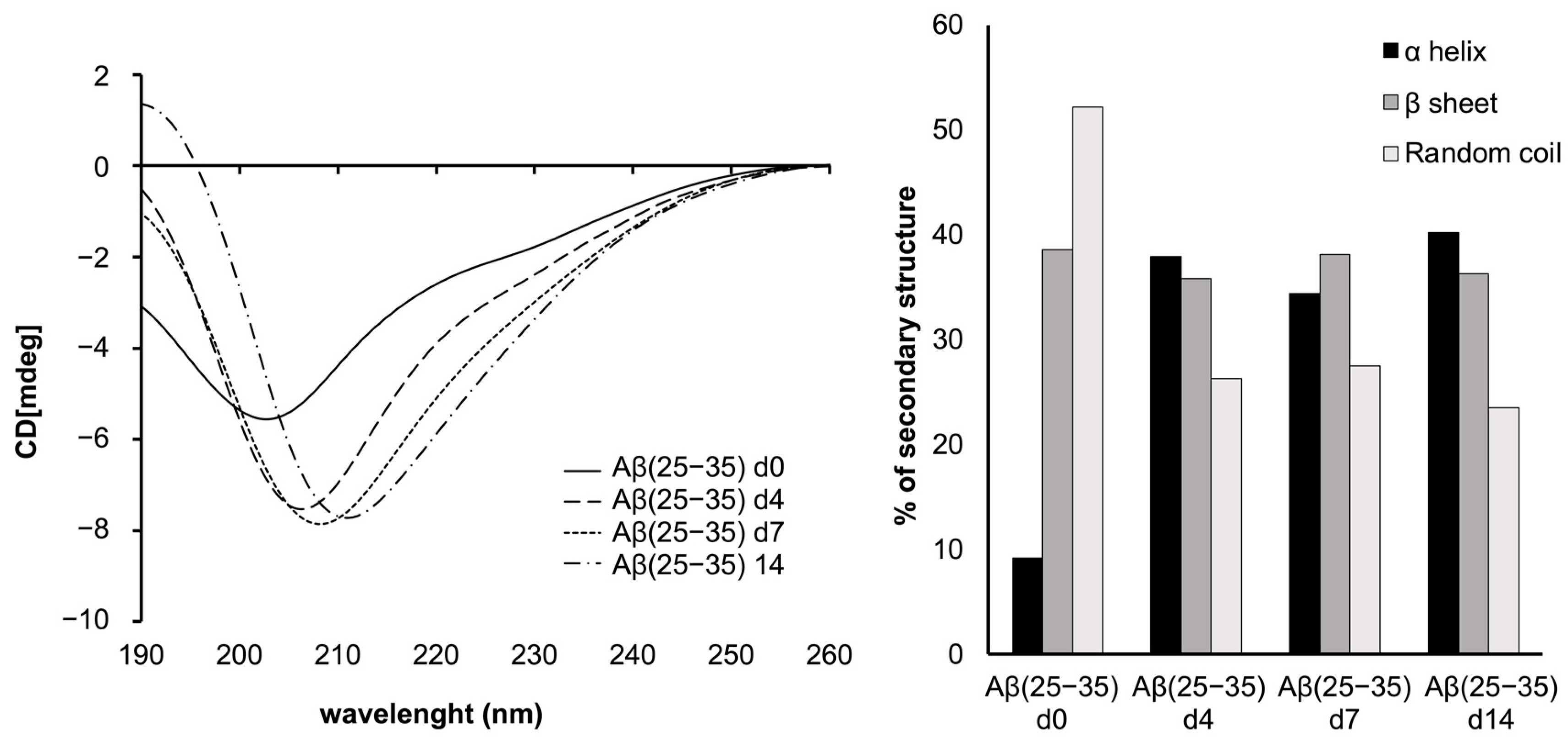
2.1. Circular Dichroism Experiments
2.2. NMR Spectroscopy
2.2.1. DOSY Experiments
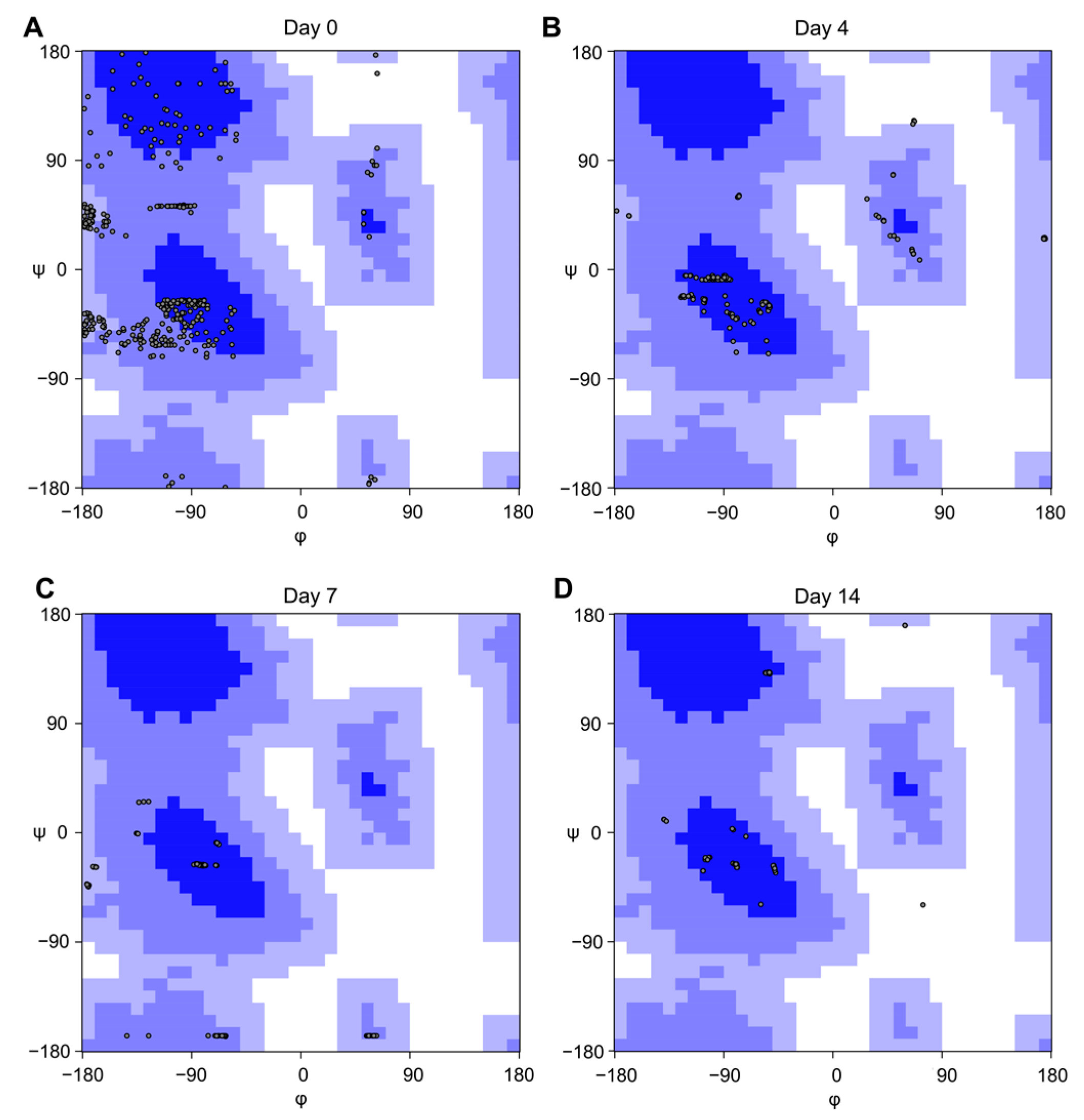
2.2.2. Analysis of Aβ(25−35) Structures
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.1.1. Aβ(25−35) Peptide Synthesis
4.1.2. Sample Preparation for Analyses
4.2. CD Experiments
4.3. NMR Experiments
4.3.1. NMR Data Recording and Processing
4.3.2. Structure Calculations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimer’s Dement. 2013, 9, 63–75.e2. [Google Scholar] [CrossRef] [PubMed]
- Fiandaca, M.S.; Mapstone, M.E.; Cheema, A.K.; Federoff, H.J. The critical need for defining preclinical biomarkers in Alzheimer’s disease. Alzheimer’s Dement. 2014, 10, S196–S212. [Google Scholar] [CrossRef] [PubMed]
- Goyal, D.; Shuaib, S.; Mann, S.; Goyal, B. Rationally designed peptides and peptidomimetics as inhibitors of amyloid-β (Aβ) aggregation: Potential therapeutics of Alzheimer’s disease. ACS Comb. Sci. 2017, 19, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Meyers, E.A.; Amouyel, P.; Bovenkamp, D.E.; Carrillo, M.C.; De Buchy, G.D.; Dumont, M.; Fillit, H.; Friedman, L.; Henderson-Begg, G.; Hort, J. Commentary: Global Alzheimer’s disease and Alzheimer’s disease related dementia research funding organizations support and engage the research community throughout the COVID-19 pandemic. Alzheimer’s Dement. 2022, 18, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Dickson, D.W. The pathogenesis of senile plaques. J. Neuropathol. Exp. Neurol. 1997, 56, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Translating cell biology into therapeutic advances in Alzheimer’s disease. Nature 1999, 399, A23–A31. [Google Scholar] [CrossRef]
- Hoyer, W.; Grönwall, C.; Jonsson, A.; Ståhl, S.; Härd, T. Stabilization of a β-hairpin in monomeric Alzheimer’s amyloid-β peptide inhibits amyloid formation. Proc. Natl. Acad. Sci. USA 2008, 105, 5099–5104. [Google Scholar] [CrossRef]
- Abelein, A.; Abrahams, J.P.; Danielsson, J.; Gräslund, A.; Jarvet, J.; Luo, J.; Tiiman, A.; Wärmländer, S.K. The hairpin conformation of the amyloid β peptide is an important structural motif along the aggregation pathway. JBIC J. Biol. Inorg. Chem. 2014, 19, 623–634. [Google Scholar] [CrossRef]
- Ahmed, M.; Davis, J.; Aucoin, D.; Sato, T.; Ahuja, S.; Aimoto, S.; Elliott, J.I.; Van Nostrand, W.E.; Smith, S.O. Structural conversion of neurotoxic amyloid-β 1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 2010, 17, 561. [Google Scholar] [CrossRef]
- Wärmländer, S.; Tiiman, A.; Abelein, A.; Luo, J.; Jarvet, J.; Söderberg, K.L.; Danielsson, J.; Gräslund, A. Biophysical Studies of the Amyloid β-Peptide: Interactions with Metal Ions and Small Molecules. ChemBioChem 2013, 14, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Wallin, C.; Luo, J.; Jarvet, J.; Wärmländer, S.K.; Gräslund, A. The Amyloid-β Peptide in Amyloid Formation Processes: Interactions with Blood Proteins and Naturally Occurring Metal Ions. Isr. J. Chem. 2017, 57, 674–685. [Google Scholar] [CrossRef]
- Scheidt, H.A.; Morgado, I.; Rothemund, S.; Huster, D.; Fändrich, M. Solid-state NMR spectroscopic investigation of Aβ protofibrils: Implication of a β-sheet remodeling upon maturation into terminal amyloid fibrils. Angew. Chem. Int. Ed. 2011, 50, 2837–2840. [Google Scholar] [CrossRef]
- Scheidt, H.A.; Morgado, I.; Rothemund, S.; Huster, D. Dynamics of amyloid β fibrils revealed by solid-state NMR. J. Biol. Chem. 2012, 287, 2017–2021. [Google Scholar] [CrossRef]
- Scheidt, H.A.; Morgado, I.; Huster, D. Solid-state NMR reveals a close structural relationship between amyloid-β protofibrils and oligomers. J. Biol. Chem. 2012, 287, 22822–22826. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.; Luheshi, L.M.; Söllvander, S.; de Barros, T.P.; Macao, B.; Knowles, T.P.; Biverstål, H.; Lendel, C.; Ekholm-Petterson, F.; Dubnovitsky, A. Stabilization of neurotoxic Alzheimer amyloid-β oligomers by protein engineering. Proc. Natl. Acad. Sci. USA 2010, 107, 15595–15600. [Google Scholar] [CrossRef] [PubMed]
- Kotler, S.A.; Walsh, P.; Brender, J.R.; Ramamoorthy, A. Differences between amyloid-β aggregation in solution and on the membrane: Insights into elucidation of the mechanistic details of Alzheimer’s disease. Chem. Soc. Rev. 2014, 43, 6692–6700. [Google Scholar] [CrossRef] [PubMed]
- Talafous, J.; Marcinowski, K.J.; Klopman, G.; Zagorski, M.G. Solution Structure of Residues 1–28 of the Amyloid. beta.-Peptide. Biochemistry 1994, 33, 7788–7796. [Google Scholar] [CrossRef] [PubMed]
- Sticht, H.; Bayer, P.; Willbold, D.; Dames, S.; Hilbich, C.; Beyreuther, K.; Frank, R.W.; Rösch, P. Structure of amyloid A4-(1–40)-peptide of Alzheimer’s disease. Eur. J. Biochem. 1995, 233, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Crescenzi, O.; Tomaselli, S.; Guerrini, R.; Salvadori, S.; D’Ursi, A.M.; Temussi, P.A.; Picone, D. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment: Similarity with a virus fusion domain. Eur. J. Biochem. 2002, 269, 5642–5648. [Google Scholar] [CrossRef]
- D’Ursi, A.M.; Armenante, M.R.; Guerrini, R.; Salvadori, S.; Sorrentino, G.; Picone, D. Solution structure of amyloid β-peptide (25−35) in different media. J. Med. Chem. 2004, 47, 4231–4238. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Esposito, V.; Vangone, P.; van Nuland, N.A.; Bonvin, A.M.; Guerrini, R.; Tancredi, T.; Temussi, P.A.; Picone, D. The α-to-β conformational transition of Alzheimer’s Aβ-(1–42) peptide in aqueous media is reversible: A step by step conformational analysis suggests the location of β conformation seeding. ChemBioChem 2006, 7, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Zirah, S.; Kozin, S.A.; Mazur, A.K.; Blond, A.; Cheminant, M.; Ségalas-Milazzo, I.; Debey, P.; Rebuffat, S. Structural changes of region 1-16 of the Alzheimer disease amyloid β-peptide upon zinc binding and in vitro aging. J. Biol. Chem. 2006, 281, 2151–2161. [Google Scholar] [CrossRef]
- Österlund, N.; Luo, J.; Wärmländer, S.K.; Gräslund, A. Membrane-mimetic systems for biophysical studies of the amyloid-β peptide. Biochim. Biophys. Acta-Proteins Proteom. 2019, 1867, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Grimaldi, M.; Buonocore, M.; Stillitano, I.; D’Ursi, A.M. Exploring the early stages of the amyloid Aβ (1–42) peptide aggregation process: An NMR study. Pharmaceuticals 2021, 14, 732. [Google Scholar] [CrossRef]
- Santoro, A.; Grimaldi, M.; Buonocore, M.; Stillitano, I.; Gloria, A.; Santin, M.; Bobba, F.; Saponetti, M.S.; Ciaglia, E.; D’Ursi, A.M. New Aβ (1–42) ligands from anti-amyloid antibodies: Design, synthesis, and structural interaction. Eur. J. Med. Chem. 2022, 237, 114400. [Google Scholar] [CrossRef] [PubMed]
- Nadezhdin, K.; Bocharova, O.; Bocharov, E.; Arseniev, A. Structural and dynamic study of the transmembrane domain of the amyloid precursor protein. Acta Nat. 2011, 3, 69–76. [Google Scholar] [CrossRef]
- Serra-Batiste, M.; Ninot-Pedrosa, M.; Bayoumi, M.; Gairí, M.; Maglia, G.; Carulla, N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. USA 2016, 113, 10866–10871. [Google Scholar] [CrossRef]
- Coles, M.; Bicknell, W.; Watson, A.A.; Fairlie, D.P.; Craik, D.J. Solution structure of amyloid β-peptide (1− 40) in a water− micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry 1998, 37, 11064–11077. [Google Scholar] [CrossRef]
- Shao, H.; Jao, S.C.; Ma, K.; Zagorski, M.G. Solution structures of micelle-bound amyloid β-(1–40) and β-(1–42) peptides of Alzheimer’s disease. J. Mol. Biol. 1999, 285, 755–773. [Google Scholar] [CrossRef]
- Rangachari, V.; Reed, D.K.; Moore, B.D.; Rosenberry, T.L. Secondary structure and interfacial aggregation of amyloid-β (1− 40) on sodium dodecyl sulfate micelles. Biochemistry 2006, 45, 8639–8648. [Google Scholar] [CrossRef] [PubMed]
- Usachev, K.S.; Filippov, A.; Khairutdinov, B.; Antzutkin, O.; Klochkov, V. NMR structure of the Arctic mutation of the Alzheimer’s Aβ (1–40) peptide docked to SDS micelles. J. Mol. Struct. 2014, 1076, 518–523. [Google Scholar] [CrossRef]
- Eichler, J. Peptides as protein binding site mimetics. Curr. Opin. Chem. Biol. 2008, 12, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Groß, A.; Hashimoto, C.; Sticht, H.; Eichler, J. Synthetic peptides as protein mimics. Front. Bioeng. Biotechnol. 2016, 3, 211. [Google Scholar] [CrossRef]
- Marin, O.; Meggio, F.; Boldyreff, B.; Issinger, O.-G.; Pinna, L.A. Dissection of the dual function of the β-subunit of protein kinase CK2 (‘casein kinase-2′): A synthetic peptide reproducing the carboxyl-terminal domain mimicks the positive but not the negative effects of the whole protein. FEBS Lett. 1995, 363, 111–114. [Google Scholar] [CrossRef]
- Grimaldi, M.; Stillitano, I.; Amodio, G.; Santoro, A.; Buonocore, M.; Moltedo, O.; Remondelli, P.; D’Ursi, A.M. Structural basis of antiviral activity of peptides from MPER of FIV gp36. PLoS ONE 2018, 13, e0204042. [Google Scholar] [CrossRef]
- Buonocore, M.; Santoro, A.; Grimaldi, M.; Covelli, V.; Firoznezhad, M.; Rodriquez, M.; Santin, M.; D’Ursi, A.M. Structural analysis of a simplified model reproducing SARS-CoV-2 S RBD/ACE2 binding site. Heliyon 2022, 8, e11568. [Google Scholar] [CrossRef]
- Usachev, K.; Filippov, A.; Filippova, E.; Antzutkin, O.; Klochkov, V. Solution structures of Alzheimer’s amyloid Aβ13–23 peptide: NMR studies in solution and in SDS. J. Mol. Struct. 2013, 1049, 436–440. [Google Scholar] [CrossRef]
- Rodziewicz-Motowidło, S.; Czaplewska, P.; Sikorska, E.; Spodzieja, M.; Kołodziejczyk, A.S. The Arctic mutation alters helix length and type in the 11–28 β-amyloid peptide monomer—CD, NMR and MD studies in an SDS micelle. J. Struct. Biol. 2008, 164, 199–209. [Google Scholar] [CrossRef]
- Usachev, K.S.; Filippov, A.V.; Antzutkin, O.N.; Klochkov, V.V. Use of a combination of the RDC method and NOESY NMR spectroscopy to determine the structure of Alzheimer’s amyloid Aβ10–35 peptide in solution and in SDS micelles. Eur. Biophys. J. 2013, 42, 803–810. [Google Scholar] [CrossRef]
- Grimaldi, M.; Scrima, M.; Esposito, C.; Vitiello, G.; Ramunno, A.; Limongelli, V.; D’Errico, G.; Novellino, E.; D’Ursi, A.M. Membrane charge dependent states of the β-amyloid fragment Aβ (16–35) with differently charged micelle aggregates. Biochim. Biophys. Acta (BBA)-Biomembr. 2010, 1798, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, M.; Marino, S.D.; Florenzano, F.; Ciotta, M.T.; Nori, S.L.; Rodriquez, M.; Sorrentino, G.; D’Ursi, A.M.; Scrima, M. β-Amyloid-acetylcholine molecular interaction: New role of cholinergic mediators in anti-Alzheimer therapy? Future Med. Chem. 2016, 8, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Randino, R.; Grimaldi, M.; Persico, M.; De Santis, A.; Cini, E.; Cabri, W.; Riva, A.; D’Errico, G.; Fattorusso, C.; D’Ursi, A.M. Investigating the neuroprotective effects of turmeric extract: Structural interactions of β-amyloid peptide with single curcuminoids. Sci. Rep. 2016, 6, 38846. [Google Scholar] [CrossRef]
- Sublimi Saponetti, M.; Grimaldi, M.; Scrima, M.; Albonetti, C.; Nori, S.L.; Cucolo, A.; Bobba, F.; D’Ursi, A.M. Aggregation of ass (25–35) on dopc and dopc/dha bilayers: An atomic force microscopy study. PLoS ONE 2014, 9, e115780. [Google Scholar] [CrossRef] [PubMed]
- Pike, C.J.; Walencewicz-Wasserman, A.J.; Kosmoski, J.; Cribbs, D.H.; Glabe, C.G.; Cotman, C.W. Structure-activity analyses of β-amyloid peptides: Contributions of the β25–35 region to aggregation and neurotoxicity. J. Neurochem. 1995, 64, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Terzi, E.; Hoelzemann, G.; Seelig, J. Alzheimer. Beta.-amyloid peptide 25−35: Electrostatic interactions with phospholipid membranes. Biochemistry 1994, 33, 7434–7441. [Google Scholar] [CrossRef]
- Kohno, T.; Kobayashi, K.; Maeda, T.; Sato, K.; Takashima, A. Three-dimensional structures of the amyloid β peptide (25− 35) in membrane-mimicking environment. Biochemistry 1996, 35, 16094–16104. [Google Scholar] [CrossRef] [PubMed]
- Francis, F.; Koulakoff, A.; Boucher, D.; Chafey, P.; Schaar, B.; Vinet, M.-C.; Friocourt, G.; McDonnell, N.; Reiner, O.; Kahn, A. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron 1999, 23, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P. Pathways towards and away from Alzheimer’s disease. Nature 2004, 430, 631–639. [Google Scholar] [CrossRef]
- Aleksis, R.; Oleskovs, F.; Jaudzems, K.; Pahnke, J.; Biverstål, H. Structural studies of amyloid-β peptides: Unlocking the mechanism of aggregation and the associated toxicity. Biochimie 2017, 140, 176–192. [Google Scholar] [CrossRef]
- Wilkins, D.K.; Grimshaw, S.B.; Receveur, V.; Dobson, C.M.; Jones, J.A.; Smith, L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 1999, 38, 16424–16431. [Google Scholar] [CrossRef] [PubMed]
- Mazer, N.A.; Benedek, G.B.; Carey, M.C. An investigation of the micellar phase of sodium dodecyl sulfate in aqueous sodium chloride solutions using quasielastic light scattering spectroscopy. J. Phys. Chem. 1976, 80, 1075–1085. [Google Scholar] [CrossRef]
- Shastry, T.A.; Morris-Cohen, A.J.; Weiss, E.A.; Hersam, M.C. Probing carbon nanotube–surfactant interactions with two-dimensional DOSY NMR. J. Am. Chem. Soc. 2013, 135, 6750–6753. [Google Scholar] [CrossRef]
- Arkhipov, V.P.; Arkhipov, R.V.; Kuzina, N.A.; Filippov, A. Study of the premicellar state in aqueous solutions of sodium dodecyl sulfate by nuclear magnetic resonance diffusion. Magn. Reson. Chem. 2021, 59, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.; Kneller, D. SPARKY 3.114; University of California: San Francisco, CA, USA, 2007. [Google Scholar]
- Kneller, D.; Kuntz, I. UCSF Sparky-an NMR display, annotation and assignment tool. Proc. J. Cell. Biochem. 1993, 53, 254. [Google Scholar]
- Wüthrich, K. NMR with proteins and nucleic acids. Eur. News 1986, 17, 11–13. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR structure calculation with CYANA. In Protein NMR Techniques; Springer: Berlin/Heidelberg, Germany, 2004; pp. 353–378. [Google Scholar]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Grover, S.; Jain, S. Aducanumab: A review of the first approved amyloid-targeting antibody for Alzheimer’s disease. Drugs Ther. Perspect. 2022, 38, 443–454. [Google Scholar] [CrossRef]
- Marcinowski, K.J.; Shao, H.; Clancy, E.L.; Zagorski, M.G. Solution structure model of residues 1−28 of the Amyloid β-peptide when bound to micelles. J. Am. Chem. Soc. 1998, 120, 11082–11091. [Google Scholar] [CrossRef]
- Thimons, K.L.; Brazdil, L.C.; Harrison, D.; Fisch, M.R. Effects of pentanol isomers on the growth of SDS micelles in 0.5 M NaCl. J. Phys. Chem. B 1997, 101, 11087–11091. [Google Scholar] [CrossRef]
- Javadian, S.; Gharibi, H.; Sohrabi, B.; Bijanzadeh, H.; Safarpour, M.; Behjatmanesh-Ardakani, R. Determination of the physico-chemical parameters and aggregation number of surfactant in micelles in binary alcohol–water mixtures. J. Mol. Liq. 2008, 137, 74–79. [Google Scholar] [CrossRef]
- Moore, P.N.; Puvvada, S.; Blankschtein, D. Challenging the surfactant monomer skin penetration model: Penetration of sodium dodecyl sulfate micelles into the epidermis. J. Cosmet. Sci. 2003, 54, 29–46. [Google Scholar] [PubMed]
- Merrifield, R.B. Solid-phase peptide synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1969, 32, 221–296. [Google Scholar]
- Jao, S.-C.; Ma, K.; Talafous, J.; Orlando, R.; Zagorski, M.G. Trifluoroacetic acid pretreatment reproducibly disaggregates the amyloid β-peptide. Amyloid 1997, 4, 240–252. [Google Scholar] [CrossRef]
- Pellegrini, M.; Mierke, D.F. Structural characterization of peptide hormone/receptor interactions by NMR spectroscopy. Pept. Sci. 1999, 51, 208–220. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolym. Orig. Res. Biomol. 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Parella, T.; Adell, P.; Sánchez-Ferrando, F.; Virgili, A. Effective multiple-solvent suppression scheme using the excitation sculpting principle. Magn. Reson. Chem. 1998, 36, 245–249. [Google Scholar] [CrossRef]
- Stilbs, P. Molecular self-diffusion coefficients in Fourier transform nuclear magnetic resonance spectrometric analysis of complex mixtures. Anal. Chem. 1981, 53, 2135–2137. [Google Scholar] [CrossRef]
- Vasenkov, S.; Galvosas, P.; Geier, O.; Nestle, N.; Stallmach, F.; Kärger, J. Determination of genuine diffusivities in heterogeneous media using stimulated echo pulsed field gradient NMR. J. Magn. Reson. 2001, 149, 228–233. [Google Scholar] [CrossRef]
- Stejskal, E.O.; Tanner, J.E. Spin diffusion measurements: Spin echoes in the presence of a time-dependent field gradient. J. Chem. Phys. 1965, 42, 288–292. [Google Scholar] [CrossRef]
- Skaanning, L.K.; Santoro, A.; Skamris, T.; Martinsen, J.H.; D’Ursi, A.M.; Bucciarelli, S.; Vestergaard, B.; Bugge, K.; Langkilde, A.E.; Kragelund, B.B. The non-fibrillating N-terminal of α-synuclein binds and co-fibrillates with heparin. Biomolecules 2020, 10, 1192. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L. Maestro 12.5.139; Schrödinger LLC: New York, NY, USA, 2018. [Google Scholar]

| D (m2/s) SDS | D (m2/s) Aβ(25−35) | |
|---|---|---|
| Day 0 | 6.98 ± 0.06 × 10−11 | 6.78 ± 0.07 × 10−11 |
| Day 4 | 6.67 ± 0.40 × 10−11 | 6.45 ± 0.17 × 10−11 |
| Day 7 | 6.45± 0.04 × 10−11 | 6.43 ± 0.19 × 10−11 |
| Day 14 | 6.58 ± 0.19 × 10−11 | 6.60 ± 0.14 × 10−11 |
| Day 0 | Day 4 | Day 7 | Day 14 | |
| Number of Experimental Restraints after CYANA | ||||
| Total NOEs | 169 | 203 | 217 | 217 |
| Intra residual | 112 | 118 | 123 | 121 |
| Short-range | 53 | 56 | 60 | 60 |
| Medium-range | 4 | 29 | 34 | 36 |
| Long-range | 0 | 0 | 0 | 0 |
| RMSD | ||||
| bb/heavy Å | 2.15/3.09 | 0.63/1.21 | 0.58/1.21 | 0.25/0.94 |
| Ramachandran analysis | ||||
| Favorable regions | 40.0% | 40.6% | 41.7% | 84.3% |
| Additional allowed regions | 41.7% | 43.1% | 29.7% | 14.3% |
| Generously allowed regions | 18.0% | 14.9% | 28.6% | 1.1% |
| Disallowed regions | 0.3% | 1.4% | 0.0% | 0.3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santoro, A.; Buonocore, M.; Grimaldi, M.; Napolitano, E.; D’Ursi, A.M. Monitoring the Conformational Changes of the Aβ(25−35) Peptide in SDS Micelles: A Matter of Time. Int. J. Mol. Sci. 2023, 24, 971. https://doi.org/10.3390/ijms24020971
Santoro A, Buonocore M, Grimaldi M, Napolitano E, D’Ursi AM. Monitoring the Conformational Changes of the Aβ(25−35) Peptide in SDS Micelles: A Matter of Time. International Journal of Molecular Sciences. 2023; 24(2):971. https://doi.org/10.3390/ijms24020971
Chicago/Turabian StyleSantoro, Angelo, Michela Buonocore, Manuela Grimaldi, Enza Napolitano, and Anna Maria D’Ursi. 2023. "Monitoring the Conformational Changes of the Aβ(25−35) Peptide in SDS Micelles: A Matter of Time" International Journal of Molecular Sciences 24, no. 2: 971. https://doi.org/10.3390/ijms24020971
APA StyleSantoro, A., Buonocore, M., Grimaldi, M., Napolitano, E., & D’Ursi, A. M. (2023). Monitoring the Conformational Changes of the Aβ(25−35) Peptide in SDS Micelles: A Matter of Time. International Journal of Molecular Sciences, 24(2), 971. https://doi.org/10.3390/ijms24020971











