Characterization of Lipopolysaccharide Effects on LRRK2 Signaling in RAW Macrophages
Abstract
1. Introduction
2. Results
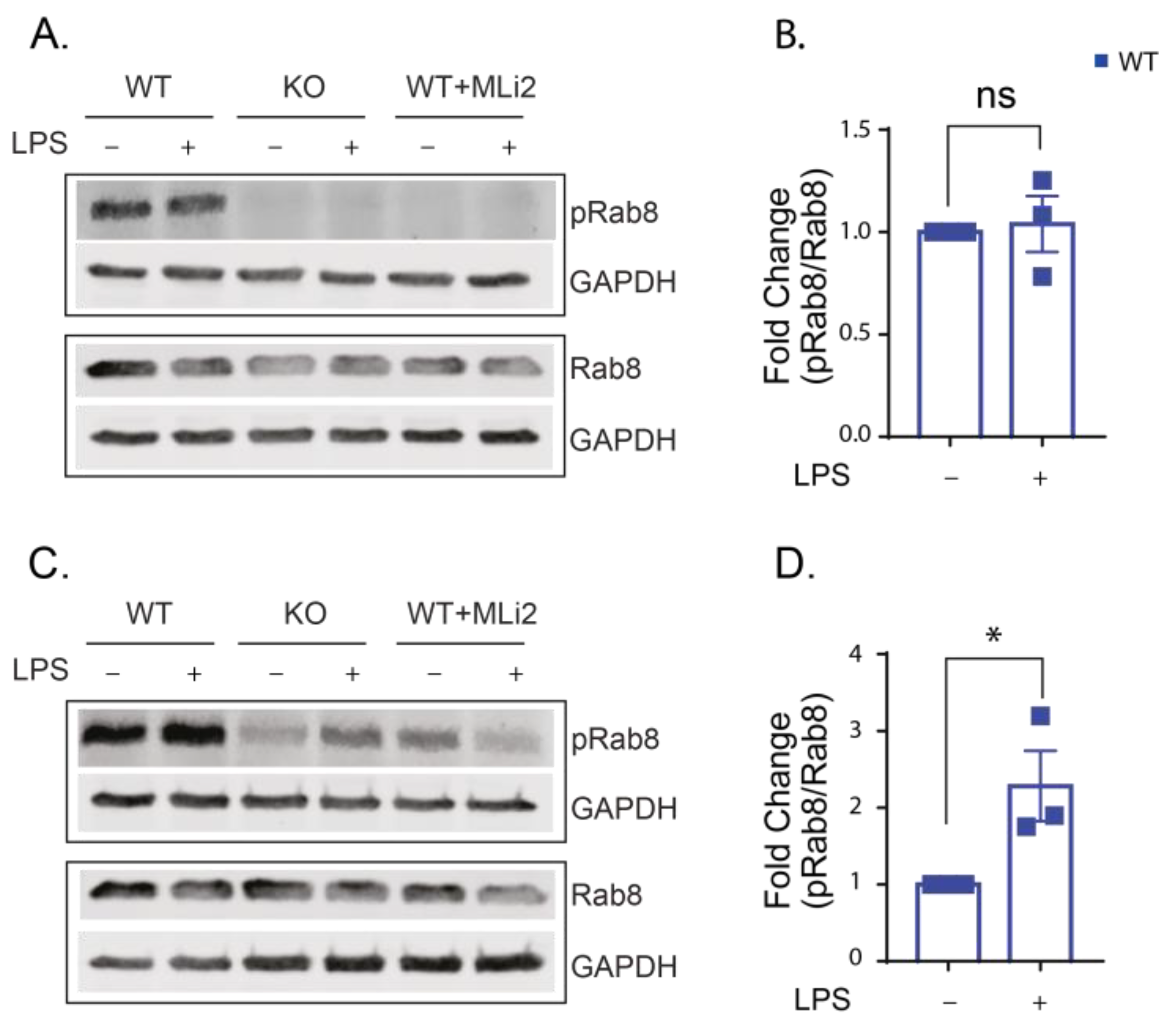
2.1. LPS Increases the Kinase Activity of LRRK2
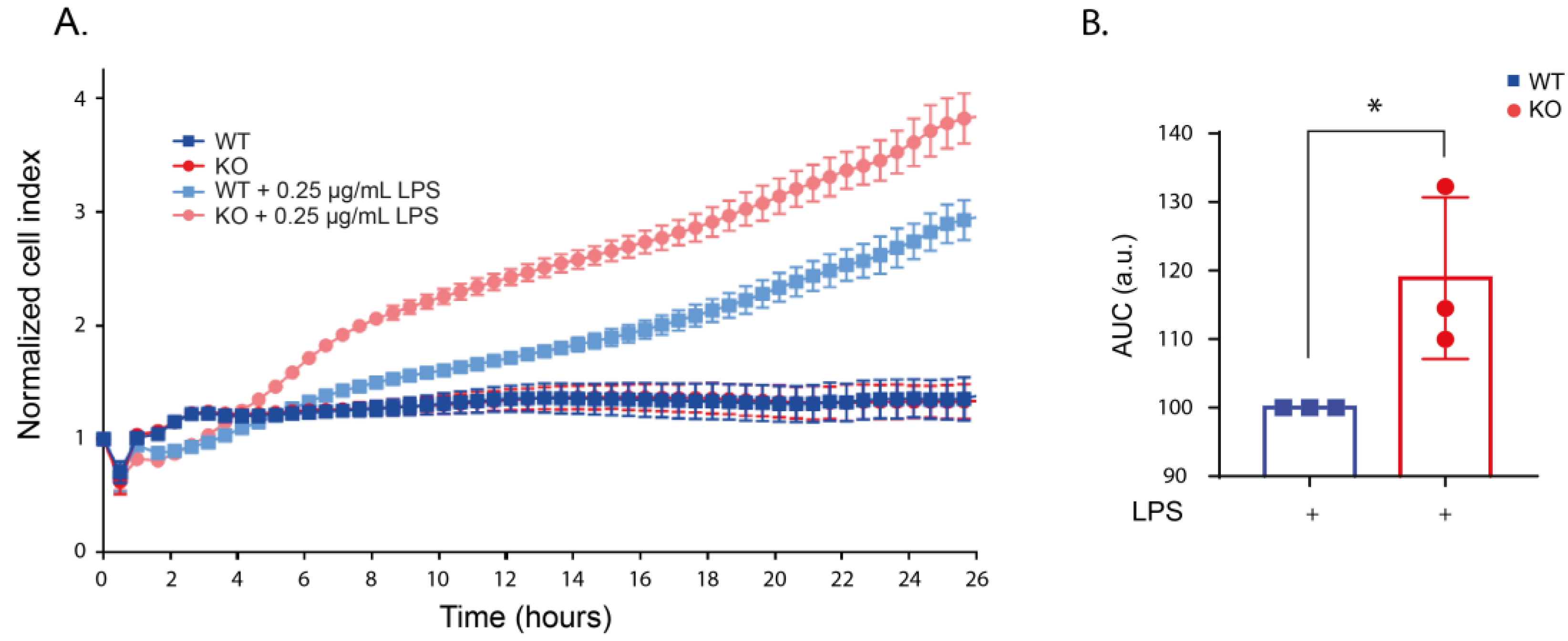
2.2. LRRK2 Deletion Changes the Cell Morphology after LPS Stimulation
2.3. LRRK2 Mediates LPS-Stimulated ROS Production
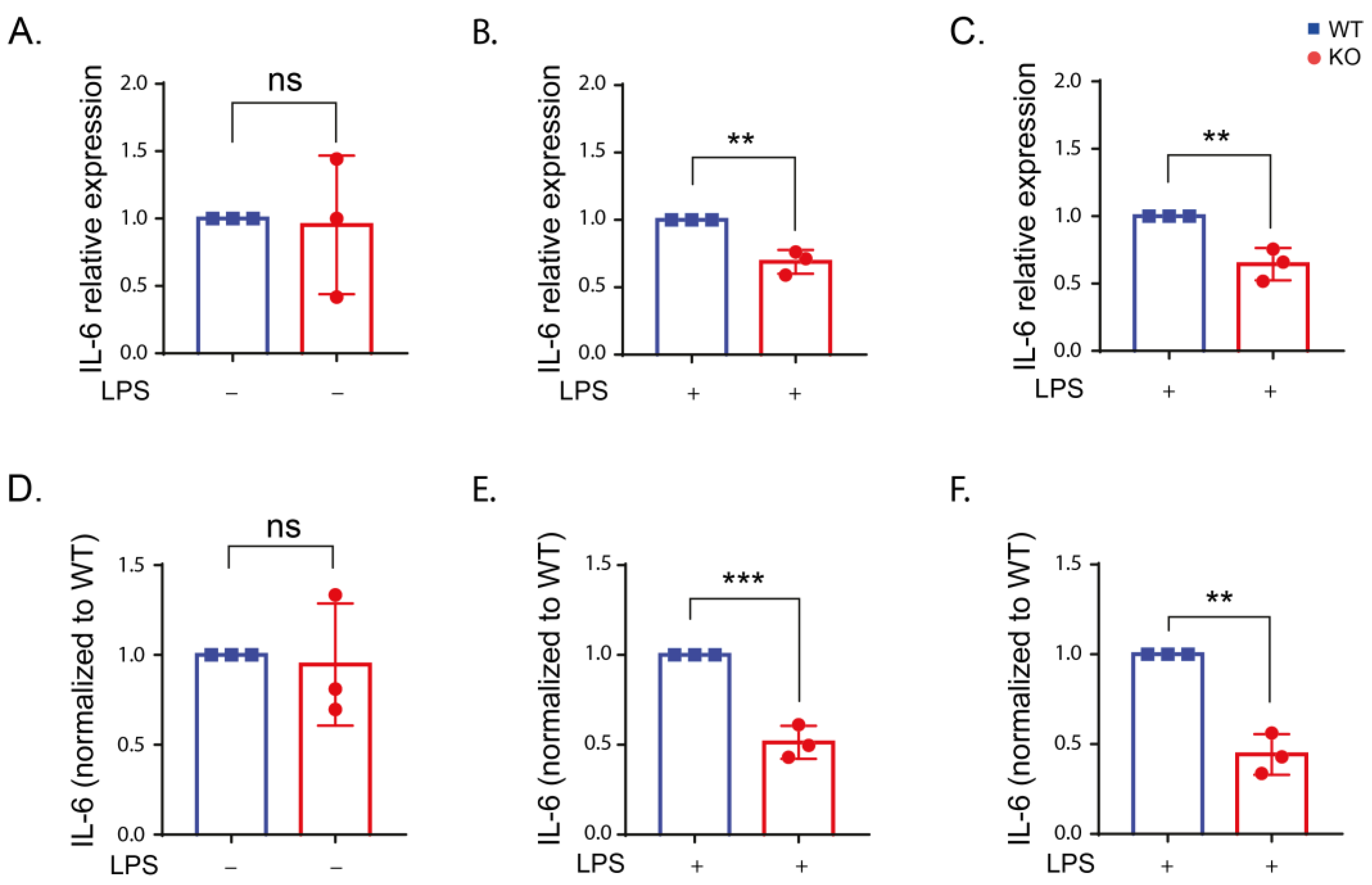
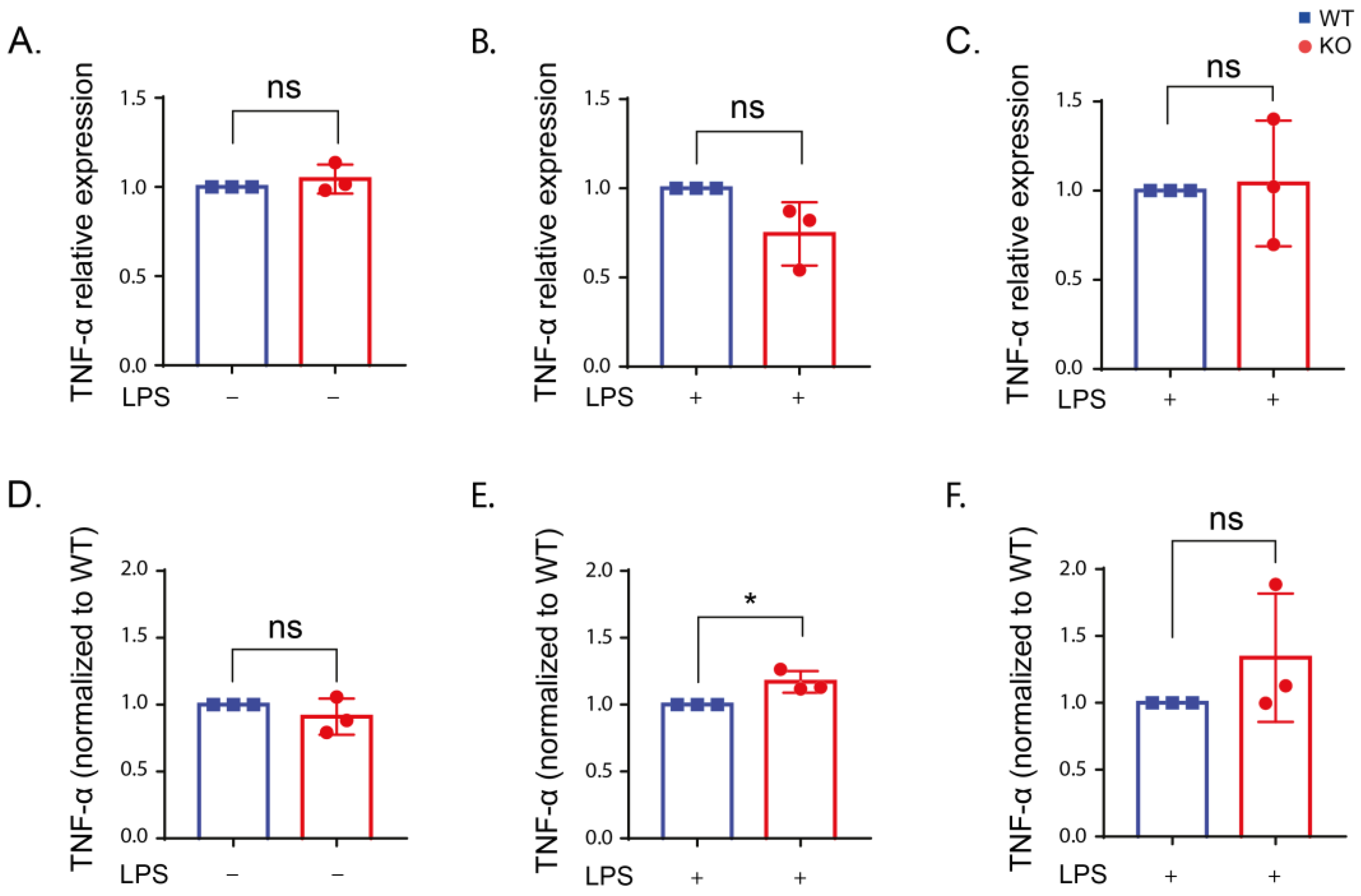
2.4. Inflammatory Cytokine Production in Stimulated RAW Macrophages
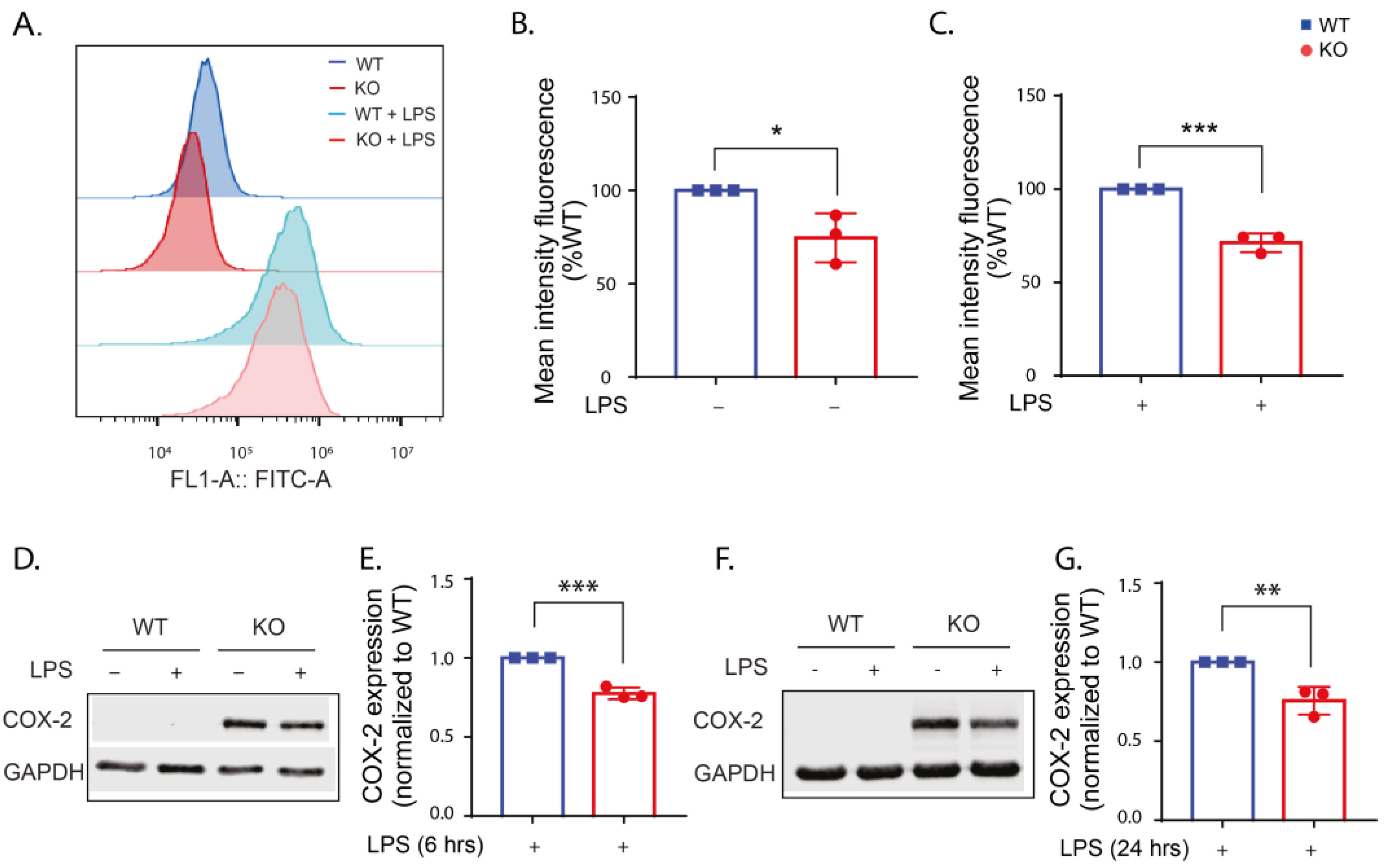
2.5. LRRK2 Deletion Reduces COX-2 Expression
2.6. LRRK2 Influences Glycolysis following LPS Stimulation
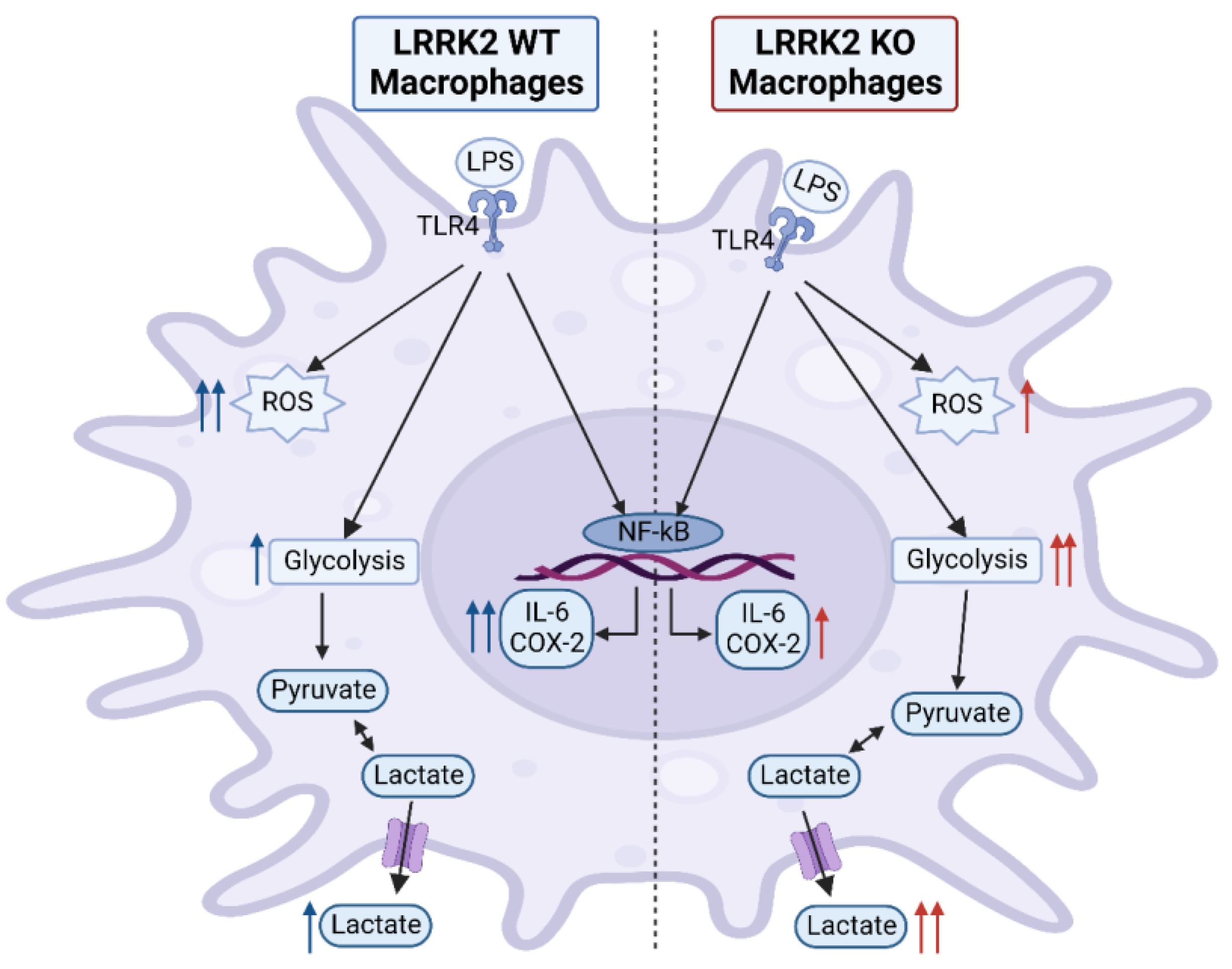
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Treatment
4.3. Western Blotting
4.4. Real-Time Cell Impedance Measurements
4.5. Flow Cytometry Measurement
4.5.1. Reactive Oxygen Species (ROS) Levels
4.5.2. Cyclooxygenase-2 (COX-2) Detection
4.6. RNA Extraction and Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
4.7. Enzyme-Linked Immunosorbent Assay (ELISA) for IL-6 and TNF-α
4.8. Lactate Measurement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 4-HNE | 4-hydroxynonenal |
| ANOVA | analysis of variance |
| AUC | area under curve |
| BMDMs | bone-marrow-derived macrophage |
| COX-2 | cyclooxygenase-2 |
| DCF | dichlorodihydrofluorescein |
| DCFDA | 2′,7′-dichlorodihydrofluorescein diacetate |
| ECAR | extracellular acidification rate |
| ELISA | enzyme-linked immunosorbent assay |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| IL-1β | interleukin-1 beta |
| IL-6 | interleukin 6 |
| INF-γ | interferon-gamma |
| iPSC | induced pluripotent stem cell |
| LLOMe | L-leucyl-L-leucine methyl ester |
| LRRK2 | leucine rich-repeat kinase 2 |
| MAPK | mitogen-activated protein kinase |
| MEFs | mouse embryonic fibroblasts |
| NF-κB | nuclear factor-kappa B |
| NSAIDs | non-steroidal anti-inflammatory drugs |
| PD | Parkinson’s disease |
| PI3Kγ | phosphatidylinositol 3-kinase γ |
| qPCR | reverse transcription quantitative real-time polymerase chain reaction |
| ROS | reactive oxygen species |
| RPL13A | ribosomal protein L13A |
| SD | standard deviation |
| SEM | standard error of the mean |
| SNpc | substantia nigra pars compacta |
| TLR4 | toll-like receptor 4 |
| TNF-α | tumor necrosis factor alpha. |
References
- Tysnes, O.B.; Storstein, A. Epidemiology of Parkinson’s Disease. J. Neural Transm. 2017, 124, 901–905. [Google Scholar] [CrossRef]
- Greffard, S.; Verny, M.; Bonnet, A.M.; Beinis, J.Y.; Gallinari, C.; Meaume, S.; Piette, F.; Hauw, J.J.; Duyckaerts, C. Motor Score of the Unified Parkinson Disease Rating Scale as a Good Predictor of Lewy Body-Associated Neuronal Loss in the Substantia Nigra. Arch. Neurol. 2006, 63, 584–588. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Nalls, M.A.; Singleton, A.B. The Genetic Architecture of Parkinson’s Disease. Lancet Neurol. 2020, 19, 170–178. [Google Scholar] [CrossRef]
- Reed, X.; Bandrés-Ciga, S.; Blauwendraat, C.; Cookson, M.R. The Role of Monogenic Genes in Idiopathic Parkinson’s Disease. Neurobiol. Dis. 2019, 124, 230–239. [Google Scholar] [CrossRef]
- Guadagnolo, D.; Piane, M.; Torrisi, M.R.; Pizzuti, A.; Petrucci, S. Genotype-Phenotype Correlations in Monogenic Parkinson Disease: A Review on Clinical and Molecular Findings. Front. Neurol. 2021, 12, 648588. [Google Scholar] [CrossRef]
- Marín, I. The Parkinson Disease Gene LRRK2: Evolutionary and Structural Insights. Mol. Biol. Evol. 2006, 23, 2423–2433. [Google Scholar] [CrossRef]
- Myasnikov, A.; Zhu, H.; Hixson, P.; Xie, B.; Yu, K.; Pitre, A.; Peng, J.; Sun, J. Structural Analysis of the Full-Length Human LRRK2. Cell 2021, 184, 3519–3527.e10. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Z.; Ye, T.; Mabrouk, O.S.; Maltbie, T.; Aasly, J.; West, A.B. Elevated LRRK2 Autophosphorylation in Brain-Derived and Peripheral Exosomes in LRRK2 Mutation Carriers. Acta Neuropathol. Commun. 2017, 5, 86. [Google Scholar] [CrossRef]
- Di Maio, R.; Hoffman, E.K.; Rocha, E.M.; Keeney, M.T.; Sanders, L.H.; De Miranda, B.R.; Zharikov, A.; Van Laar, A.; Stepan, A.F.; Lanz, T.A.; et al. LRRK2 Activation in Idiopathic Parkinson’s Disease. Sci. Transl. Med. 2018, 10, eaar5429. [Google Scholar] [CrossRef]
- Wallings, R.; Manzoni, C.; Bandopadhyay, R. Cellular Processes Associated with LRRK2 Function and Dysfunction. FEBS J. 2015, 282, 2806–2826. [Google Scholar] [CrossRef]
- Martin, I.; Kim, J.W.; Dawson, V.L.; Dawson, T.M. LRRK2 Pathobiology in Parkinson’s Disease. J. Neurochem. 2014, 131, 554–565. [Google Scholar] [CrossRef]
- Tsafaras, G.; Baekelandt, V. The Role of LRRK2 in the Periphery: Link with Parkinson’s Disease and Inflammatory Diseases. Neurobiol. Dis. 2022, 172, 105806. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Zhou, J. Neuroinflammation in Parkinson’s Disease and Its Potential as Therapeutic Target. Transl. Neurodegener. 2015, 4, 1–9. [Google Scholar] [CrossRef]
- Gardet, A.; Benita, Y.; Li, C.; Sands, B.E.; Ballester, I.; Stevens, C.; Korzenik, J.R.; Rioux, J.D.; Daly, M.J.; Xavier, R.J.; et al. LRRK2 Is Involved in the IFN-γ Response and Host Response to Pathogens. J. Immunol. 2010, 185, 5577–5585. [Google Scholar] [CrossRef]
- Hakimi, M.; Selvanantham, T.; Swinton, E.; Padmore, R.F.; Tong, Y.; Kabbach, G.; Venderova, K.; Girardin, S.E.; Bulman, D.E.; Scherzer, C.R.; et al. Parkinson’s Disease-Linked LRRK2 Is Expressed in Circulating and Tissue Immune Cells and Upregulated Following Recognition of Microbial Structures. J. Neural Transm. 2011, 118, 795–808. [Google Scholar] [CrossRef]
- Miklossy, J.; Arai, T.; Guo, J.P.; Klegeris, A.; Yu, S.; McGeer, E.G.; McGeer, P.L. LRRK2 Expression in Normal and Pathologic Human Brain and in Human Cell Lines. J. Neuropathol. Exp. Neurol. 2006, 65, 953–963. [Google Scholar] [CrossRef]
- Atashrazm, F.; Hammond, D.; Perera, G.; Bolliger, M.F.; Matar, E.; Halliday, G.M.; Schüle, B.; Lewis, S.J.G.; Nichols, R.J.; Dzamko, N. LRRK2-Mediated Rab10 Phosphorylation in Immune Cells from Parkinson’s Disease Patients. Mov. Disord. 2019, 34, 406–415. [Google Scholar] [CrossRef]
- Cook, D.A.; Kannarkat, G.T.; Cintron, A.F.; Butkovich, L.M.; Fraser, K.B.; Chang, J.; Grigoryan, N.; Factor, S.A.; West, A.B.; Boss, J.M.; et al. LRRK2 Levels in Immune Cells Are Increased in Parkinson’s Disease. NPJ Park. Dis. 2017, 3, 1–12. [Google Scholar] [CrossRef]
- Zhang, F.-R.; Huang, W.; Chen, S.-M.; Sun, L.-D.; Liu, H.; Li, Y.; Cui, Y.; Yan, X.-X.; Yang, H.-T.; Yang, R.-D.; et al. Genomewide Association Study of Leprosy. N. Engl. J. Med. 2009, 361, 2609–2618. [Google Scholar] [CrossRef]
- Hui, K.Y.; Fernandez-Hernandez, H.; Hu, J.; Schaffner, A.; Pankratz, N.; Hsu, N.Y.; Chuang, L.S.; Carmi, S.; Villaverde, N.; Li, X.; et al. Functional Variants in the LRRK2 Gene Confer Shared Effects on Risk for Crohn’s Disease and Parkinson’s Disease. Sci. Transl. Med. 2018, 10, eaai7795. [Google Scholar] [CrossRef]
- Deng, I.; Corrigan, F.; Zhai, G.; Zhou, X.-F.; Bobrovskaya, L. Lipopolysaccharide Animal Models of Parkinson’s Disease: Recent Progress and Relevance to Clinical Disease. Brain Behav. Immun.—Health 2020, 4, 100060. [Google Scholar] [CrossRef] [PubMed]
- Beier, E.E.; Neal, M.; Alam, G.; Edler, M.; Wu, L.J.; Richardson, J.R. Alternative Microglial Activation Is Associated with Cessation of Progressive Dopamine Neuron Loss in Mice Systemically Administered Lipopolysaccharide. Neurobiol. Dis. 2017, 108, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Bodea, L.G.; Wang, Y.; Linnartz-Gerlach, B.; Kopatz, J.; Sinkkonen, L.; Musgrove, R.; Kaoma, T.; Muller, A.; Vallar, L.; Di Monte, D.A.; et al. Neurodegeneration by Activation of the Microglial Complement–Phagosome Pathway. J. Neurosci. 2014, 34, 8546–8556. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, C.; Wu, W.Y.; Chen, F.; Wu, Y.Y.; Li, W.Z.; Chen, H.Q.; Yin, Y.Y. Biochanin A Protects Dopaminergic Neurons against Lipopolysaccharide-Induced Damage and Oxidative Stress in a Rat Model of Parkinson’s Disease. Pharmacol. Biochem. Behav. 2015, 138, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Castaño, A.; Herrera, A.J.; Cano, J.; Machado, A. Lipopolysaccharide Intranigral Injection Induces Inflammatory Reaction and Damage in Nigrostriatal Dopaminergic System. J. Neurochem. 1998, 70, 1584–1592. [Google Scholar] [CrossRef]
- Machado, A.; Herrera, A.J.; Venero, J.L.; Santiago, M.; de Pablos, R.M.; Villarán, R.F.; Espinosa-Oliva, A.M.; Argüelles, S.; Sarmiento, M.; Delgado-Cortés, M.J.; et al. Inflammatory Animal Model for Parkinson’s Disease: The Intranigral Injection of LPS Induced the Inflammatory Process along with the Selective Degeneration of Nigrostriatal Dopaminergic Neurons. ISRN Neurol. 2011, 2011, 1–16. [Google Scholar] [CrossRef]
- Kuss, M.; Adamopoulou, E.; Kahle, P.J. Interferon-γ Induces Leucine-Rich Repeat Kinase LRRK2 via Extracellular Signal-Regulated Kinase ERK5 in Macrophages. J. Neurochem. 2014, 129, 980–987. [Google Scholar] [CrossRef]
- Gillardon, F.; Schmid, R.; Draheim, H. Parkinson’s Disease-Linked Leucine-Rich Repeat Kinase 2(R1441G) Mutation Increases Proinflammatory Cytokine Release from Activated Primary Microglial Cells and Resultant Neurotoxicity. Neuroscience 2012, 208, 41–48. [Google Scholar] [CrossRef]
- Moehle, M.S.; Webber, P.J.; Tse, T.; Sukar, N.; Standaert, D.G.; Desilva, T.M.; Cowell, R.M.; West, A.B. LRRK2 Inhibition Attenuates Microglial Inflammatory Responses. J. Neurosci. 2012, 32, 1602–1611. [Google Scholar] [CrossRef]
- Daher, J.P.L.; Volpicelli-Daley, L.A.; Blackburn, J.P.; Moehle, M.S.; West, A.B. Abrogation of α-Synuclein -Mediated Dopaminergic Neurodegeneration in LRRK2-Deficient Rats. Proc. Natl. Acad. Sci. USA 2014, 111, 9289–9294. [Google Scholar] [CrossRef]
- Dzamko, N.; Inesta-Vaquera, F.; Zhang, J.; Xie, C.; Cai, H.; Arthur, S.; Tan, L.; Choi, H.; Gray, N.; Cohen, P.; et al. The IkappaB Kinase Family Phosphorylates the Parkinson’s Disease Kinase LRRK2 at Ser935 and Ser910 during Toll-Like Receptor Signaling. PLoS ONE 2012, 7, e39132. [Google Scholar] [CrossRef]
- Schapansky, J.; Nardozzi, J.D.; Felizia, F.; LaVoie, M.J. Membrane Recruitment of Endogenous LRRK2 Precedes Its Potent Regulation of Autophagy. Hum. Mol. Genet. 2014, 23, 4201–4214. [Google Scholar] [CrossRef]
- Li, T.; Ning, B.; Kong, L.; Dai, B.; He, X.; Thomas, J.M.; Sawa, A.; Ross, C.A.; Smith, W.W. A Lrrk2 Gtp Binding Inhibitor, 68, Reduces Lps-induced Signaling Events and Tnf-α Release in Human Lymphoblasts. Cells 2021, 10, 480. [Google Scholar] [CrossRef]
- Ho, D.H.; Je, A.R.; Lee, H.; Son, I.; Kweon, H.S.; Kim, H.G.; Seol, W. LRRK2 Kinase Activity Induces Mitochondrial Fission in Microglia via Drp1 and Modulates Neuroinflammation. Exp. Neurobiol. 2018, 27, 171–180. [Google Scholar] [CrossRef]
- Russo, I.; Berti, G.; Plotegher, N.; Bernardo, G.; Filograna, R.; Bubacco, L.; Greggio, E. Leucine-Rich Repeat Kinase 2 Positively Regulates Inflammation and down-Regulates NF-ΚB P50 Signaling in Cultured Microglia Cells. J. Neuroinflamm. 2015, 12, 230. [Google Scholar] [CrossRef]
- Kim, B.; Yang, M.S.; Choi, D.; Kim, J.H.; Kim, H.S.; Seol, W.; Choi, S.; Jou, I.; Kim, E.Y.; Joe, E. hye Impaired Inflammatory Responses in Murine Lrrk2-Knockdown Brain Microglia. PLoS ONE 2012, 7, e34693. [Google Scholar] [CrossRef]
- Fan, C.; Wu, L.H.; Zhang, G.F.; Xu, F.; Zhang, S.; Zhang, X.; Sun, L.; Yu, Y.; Zhang, Y.; Ye, R.D. 4’-Hydroxywogonin Suppresses Lipopolysaccharide-Induced Inflammatory Responses in RAW 264.7 Macrophages and Acute Lung Injury Mice. PLoS ONE 2017, 12, e0181191. [Google Scholar] [CrossRef]
- Yauger, Y.J.; Bermudez, S.; Moritz, K.E.; Glaser, E.; Stoica, B.; Byrnes, K.R. Iron Accentuated Reactive Oxygen Species Release by NADPH Oxidase in Activated Microglia Contributes to Oxidative Stress in Vitro. J. Neuroinflamm. 2019, 16, 41. [Google Scholar] [CrossRef]
- More, G.K.; Makola, R.T. In-Vitro Analysis of Free Radical Scavenging Activities and Suppression of LPS-Induced ROS Production in Macrophage Cells by Solanum Sisymbriifolium Extracts. Sci. Rep. 2020, 10, 6493. [Google Scholar] [CrossRef]
- Jakubowski, W.; Bartosz, G. 2,7-Dichlorofluorescin Oxidation and Reactive Oxygen Species: What Does It Measure? Cell Biol. Int. 2000, 24, 757–760. [Google Scholar] [CrossRef]
- Sakai, J.; Cammarota, E.; Wright, J.A.; Cicuta, P.; Gottschalk, R.A.; Li, N.; Fraser, I.D.C.; Bryant, C.E. Lipopolysaccharide-Induced NF-ΚB Nuclear Translocation Is Primarily Dependent on MyD88, but TNFα Expression Requires TRIF and MyD88. Sci. Rep. 2017, 7, 1428. [Google Scholar] [CrossRef]
- Dorrington, M.G.; Fraser, I.D.C. NF-ΚB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef]
- Teismann, P.; Tieu, K.; Choi, D.K.; Wu, D.C.; Naini, A.; Hunot, S.; Vila, M.; Jackson-Lewis, V.; Przedborski, S. Cyclooxygenase-2 Is Instrumental in Parkinson’s Disease Neurodegeneration. Proc. Natl. Acad. Sci. USA 2003, 100, 5473–5478. [Google Scholar] [CrossRef]
- Knott, C.; Stern, G.; Wilkin, G.P. Inflammatory Regulators in Parkinson’s Disease: INOS, Lipocortin-1, and Cyclooxygenases-1 and -2. Mol. Cell. Neurosci. 2000, 16, 724–739. [Google Scholar] [CrossRef]
- Sánchez-Pernaute, R.; Ferree, A.; Cooper, O.; Yu, M.; Brownell, A.L.; Isacson, O. Selective COX-2 Inhibition Prevents Progressive Dopamine Neuron Degeneration in a Rat Model of Parkinson’s Disease. J. Neuroinflamm. 2004, 1, 6. [Google Scholar] [CrossRef]
- Vijitruth, R.; Liu, M.; Choi, D.Y.; Nguyen, X.V.; Hunter, R.L.; Bing, G. Cyclooxygenase-2 Mediates Microglial Activation and Secondary Dopaminergic Cell Death in the Mouse MPTP Model of Parkinson’s Disease. J. Neuroinflamm. 2006, 3, 6. [Google Scholar] [CrossRef]
- Lopez de Maturana, R.; Aguila, J.C.; Sousa, A.; Vazquez, N.; del Rio, P.; Aiastui, A.; Gorostidi, A.; Lopez de Munain, A.; Sanchez-Pernaute, R. Leucine-Rich Repeat Kinase 2 Modulates Cyclooxygenase 2 and the Inflammatory Response in Idiopathic and Genetic Parkinson’s Disease. Neurobiol. Aging 2014, 35, 1116–1124. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic Reprogramming in Macrophage Responses. Biomark. Res. 2021, 9, 1–17. [Google Scholar] [CrossRef]
- Wallings, R.L.; Herrick, M.K.; Tansey, M.G. LRRK2 at the Interface Between Peripheral and Central Immune Function in Parkinson’s. Front. Neurosci. 2020, 14, 443. [Google Scholar] [CrossRef]
- Su, X.; Federoff, H.J. Immune Responses in Parkinson’s Disease: Interplay between Central and Peripheral Immune Systems. Biomed. Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Oun, A.; Sabogal-Guaqueta, A.M.; Galuh, S.; Alexander, A.; Kortholt, A.; Dolga, A. The Multifaceted Role of LRRK2 in Parkinson’s Disease: From Human IPSC to Organoids. Neurobiol. Dis. 2022, 173, 105837. [Google Scholar] [CrossRef]
- Nazish, I.; Arber, C.; Piers, T.M.; Warner, T.T.; Hardy, J.A.; Lewis, P.A.; Pocock, J.M.; Bandopadhyay, R. Abrogation of LRRK2 Dependent Rab10 Phosphorylation with TLR4 Activation and Alterations in Evoked Cytokine Release in Immune Cells. Neurochem. Int. 2021, 147, 105070. [Google Scholar] [CrossRef]
- Herbst, S.; Campbell, P.; Harvey, J.; Bernard, E.M.; Papayannopoulos, V.; Wood, N.W.; Morris, H.R.; Gutierrez, M.G. LRRK 2 Activation Controls the Repair of Damaged Endomembranes in Macrophages. EMBO J. 2020, 39, e104494. [Google Scholar] [CrossRef]
- Wall, A.A.; Luo, L.; Hung, Y.; Tong, S.J.; Condon, N.D.; Blumenthal, A.; Sweet, M.J.; Stow, J.L. Small GTPase Rab8a-Recruited Phosphatidylinositol 3-Kinase γ Regulates Signaling and Cytokine Outputs from Endosomal Toll-like Receptors. J. Biol. Chem. 2017, 292, 4411–4422. [Google Scholar] [CrossRef]
- Dias, V.; Junn, E.; Mouradian, M.M. The Role of Oxidative Stress in Parkinson’s Disease. J. Parkinsons. Dis. 2013, 3, 461–491. [Google Scholar] [CrossRef]
- Kim, J.; Pajarillo, E.; Rizor, A.; Son, D.S.; Lee, J.; Aschner, M.; Lee, E. LRRK2 Kinase Plays a Critical Role in Manganese-Induced Inflammation and Apoptosis in Microglia. PLoS ONE 2019, 14, e0210248. [Google Scholar] [CrossRef]
- Helton, L.G.; Soliman, A.; Von Zweydorf, F.; Kentros, M.; Manschwetus, J.T.; Hall, S.; Gilsbach, B.; Ho, F.Y.; Athanasopoulos, P.S.; Singh, R.K.; et al. Allosteric Inhibition of Parkinson’s-Linked LRRK2 by Constrained Peptides. ACS Chem. Biol. 2021, 16, 2326–2338. [Google Scholar] [CrossRef]
- Saez-Atienzar, S.; Bonet-Ponce, L.; Blesa, J.R.J.; Romero, F.J.F.J.; Murphy, M.P.M.; Jordan, J.; Galindo, M.F.M. The LRRK2 Inhibitor GSK2578215A Induces Protective Autophagy in SH-SY5Y Cells: Involvement of Drp-1-Mediated Mitochondrial Fission and Mitochondrial-Derived ROS Signaling. Cell Death Dis. 2014, 5, e1368-10. [Google Scholar] [CrossRef]
- Reale, M.; Iarlori, C.; Thomas, A.; Gambi, D.; Perfetti, B.; Di Nicola, M.; Onofrj, M. Peripheral Cytokines Profile in Parkinson’s Disease. Brain. Behav. Immun. 2009, 23, 55–63. [Google Scholar] [CrossRef]
- Nagatsu, T.; Sawada, M. Inflammatory Process in Parkinsons Disease: Role for Cytokines. Curr. Pharm. Des. 2005, 11, 999–1016. [Google Scholar] [CrossRef]
- Tansey, M.G.; Frank-Cannon, T.C.; McCoy, M.K.; Jae, K.L.; Martinez, T.N.; McAlpine, F.E.; Ruhn, K.A.; Tran, T.A. Neuroinflammation in Parkinson’s Disease: Is There Sufficient Evidence for Mechanism-Based Interventional Therapy? Front. Biosci. 2008, 13, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, K.; Apel, A.; Schulte, C.; Schneiderhan-Marra, N.; Pont-Sunyer, C.; Vilas, D.; Ruiz-Martinez, J.; Langkamp, M.; Corvol, J.C.; Cormier, F.; et al. Inflammatory Profile in LRRK2-Associated Prodromal and Clinical PD. J. Neuroinflamm. 2016, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, K.; Schulte, C.; Schneiderhan-Marra, N.; Apel, A.; Pont-Sunyer, C.; Vilas, D.; Ruiz-Martinez, J.; Langkamp, M.; Corvol, J.-C.; Cormier, F.; et al. Inflammatory Profile Discriminates Clinical Subtypes in LRRK2-Associated Parkinson’s Disease. Eur. J. Neurol. 2017, 24, 427.e6. [Google Scholar] [CrossRef] [PubMed]
- Panagiotakopoulou, V.; Ivanyuk, D.; De Cicco, S.; Haq, W.; Arsić, A.; Yu, C.; Messelodi, D.; Oldrati, M.; Schöndorf, D.C.; Perez, M.J.; et al. Interferon-γ Signaling Synergizes with LRRK2 in Neurons and Microglia Derived from Human Induced Pluripotent Stem Cells. Nat. Commun. 2020, 11, 5163. [Google Scholar] [CrossRef]
- Ahmadi Rastegar, D.; Hughes, L.P.; Perera, G.; Keshiya, S.; Zhong, S.; Gao, J.; Halliday, G.M.; Schüle, B.; Dzamko, N. Effect of LRRK2 Protein and Activity on Stimulated Cytokines in Human Monocytes and Macrophages. NPJ Park. Dis. 2022, 8, 34. [Google Scholar] [CrossRef]
- Dwyer, Z.; Rudyk, C.; Thompson, A.; Farmer, K.; Fenner, B.; Fortin, T.; Derksen, A.; Sun, H.; Hayley, S. Leucine-Rich Repeat Kinase-2 (LRRK2) Modulates Microglial Phenotype and Dopaminergic Neurodegeneration. Neurobiol. Aging 2020, 91, 45–55. [Google Scholar] [CrossRef]
- Kubo, M.; Nagashima, R.; Kurihara, M.; Kawakami, F.; Maekawa, T.; Eshima, K.; Ohta, E.; Kato, H.; Obata, F. Leucine-Rich Repeat Kinase 2 Controls Inflammatory Cytokines Production through NF-ΚB Phosphorylation and Antigen Presentation in Bone Marrow-Derived Dendritic Cells. Int. J. Mol. Sci. 2020, 21, 1890. [Google Scholar] [CrossRef]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The Pro- and Anti-Inflammatory Properties of the Cytokine Interleukin-6. Biochim. Biophys. Acta Mol. Cell Res. 2011, 1813, 878–888. [Google Scholar] [CrossRef]
- San Luciano, M.; Tanner, C.M.; Meng, C.; Marras, C.; Goldman, S.M.; Lang, A.E.; Tolosa, E.; Schüle, B.; Langston, J.W.; Brice, A.; et al. Nonsteroidal Anti-Inflammatory Use and LRRK2 Parkinson’s Disease Penetrance. Mov. Disord. 2020, 35, 1755–1764. [Google Scholar] [CrossRef]
- Cai, R.; Zhang, Y.; Simmering, J.E.; Schultz, J.L.; Li, Y.; Fernandez-Carasa, I.; Consiglio, A.; Raya, A.; Polgreen, P.M.; Narayanan, N.S.; et al. Enhancing Glycolysis Attenuates Parkinson’s Disease Progression in Models and Clinical Databases. J. Clin. Investig. 2019, 129, 4539–4549. [Google Scholar] [CrossRef]
- Nikonova, E.V.; Xiong, Y.; Tanis, K.Q.; Dawson, V.L.; Vogel, R.L.; Finney, E.M.; Stone, D.J.; Reynolds, I.J.; Kern, J.T.; Dawson, T.M. Transcriptional Responses to Loss or Gain of Function of the Leucine-Rich Repeat Kinase 2 (LRRK2) Gene Uncover Biological Processes Modulated by LRRK2 Activity. Hum. Mol. Genet. 2012, 21, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Nabar, N.R.; Heijjer, C.N.; Shi, C.S.; Hwang, I.Y.; Ganesan, S.; Karlsson, M.C.I.; Kehrl, J.H. LRRK2 Is Required for CD38-Mediated NAADP-Ca2+ Signaling and the Downstream Activation of TFEB (Transcription Factor EB) in Immune Cells. Autophagy 2022, 18, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Teslaa, T.; Teitell, M.A. Techniques to Monitor Glycolysis. Methods Enzymol. 2014, 542, 91–114. [Google Scholar] [CrossRef]
- Toyofuku, T.; Okamoto, Y.; Ishikawa, T.; Sasawatari, S.; Kumanogoh, A. LRRK2 Regulates Endoplasmic Reticulum–Mitochondrial Tethering through the PERK -mediated Ubiquitination Pathway. EMBO J. 2020, 39, e100875. [Google Scholar] [CrossRef]
- Weindel, C.G.; Bell, S.L.; Vail, K.J.; West, K.O.; Patrick, K.L.; Watson, R.O. LRRK2 Maintains Mitochondrial Homeostasis and Regulates Innate Immune Responses to Mycobacterium Tuberculosis. Elife 2020, 9, e51071. [Google Scholar] [CrossRef]
- Diemert, S.; Dolga, A.M.M.; Tobaben, S.; Grohm, J.; Pfeifer, S.; Oexler, E.; Culmsee, C. Impedance Measurement for Real Time Detection of Neuronal Cell Death. J. Neurosci. Methods 2012, 203, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Krabbendam, I.E.; Honrath, B.; Dilberger, B.; Iannetti, E.F.; Branicky, R.S.; Meyer, T.; Evers, B.; Dekker, F.J.; Koopman, W.J.H.; Beyrath, J.; et al. SK Channel-Mediated Metabolic Escape to Glycolysis Inhibits Ferroptosis and Supports Stress Resistance in C. Elegans. Cell Death Dis. 2020, 11, 263. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oun, A.; Hoeksema, E.; Soliman, A.; Brouwer, F.; García-Reyes, F.; Pots, H.; Trombetta-Lima, M.; Kortholt, A.; Dolga, A.M. Characterization of Lipopolysaccharide Effects on LRRK2 Signaling in RAW Macrophages. Int. J. Mol. Sci. 2023, 24, 1644. https://doi.org/10.3390/ijms24021644
Oun A, Hoeksema E, Soliman A, Brouwer F, García-Reyes F, Pots H, Trombetta-Lima M, Kortholt A, Dolga AM. Characterization of Lipopolysaccharide Effects on LRRK2 Signaling in RAW Macrophages. International Journal of Molecular Sciences. 2023; 24(2):1644. https://doi.org/10.3390/ijms24021644
Chicago/Turabian StyleOun, Asmaa, Emmy Hoeksema, Ahmed Soliman, Famke Brouwer, Fabiola García-Reyes, Henderikus Pots, Marina Trombetta-Lima, Arjan Kortholt, and Amalia M. Dolga. 2023. "Characterization of Lipopolysaccharide Effects on LRRK2 Signaling in RAW Macrophages" International Journal of Molecular Sciences 24, no. 2: 1644. https://doi.org/10.3390/ijms24021644
APA StyleOun, A., Hoeksema, E., Soliman, A., Brouwer, F., García-Reyes, F., Pots, H., Trombetta-Lima, M., Kortholt, A., & Dolga, A. M. (2023). Characterization of Lipopolysaccharide Effects on LRRK2 Signaling in RAW Macrophages. International Journal of Molecular Sciences, 24(2), 1644. https://doi.org/10.3390/ijms24021644






