A Joint Transcriptomic and Metabolomic Analysis Reveals the Regulation of Shading on Lignin Biosynthesis in Asparagus
Abstract
1. Introduction
2. Results
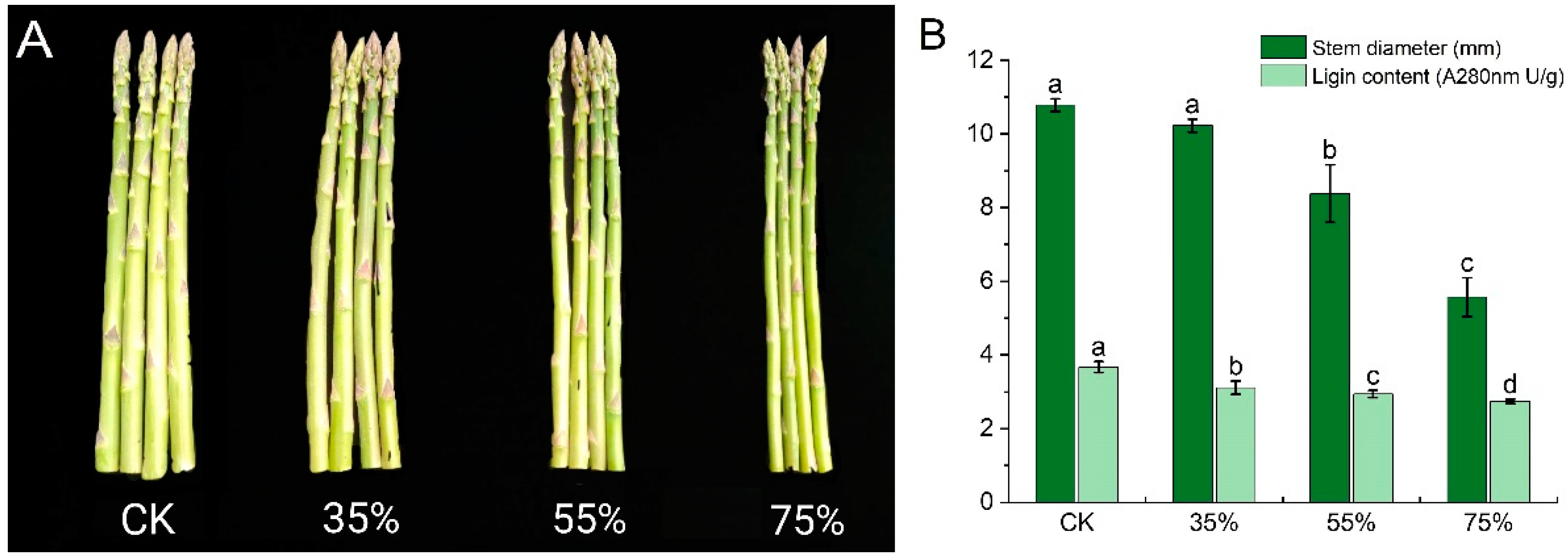
2.1. Influence of Shading on the Morphological Characteristics of Asparagus Tender Stem
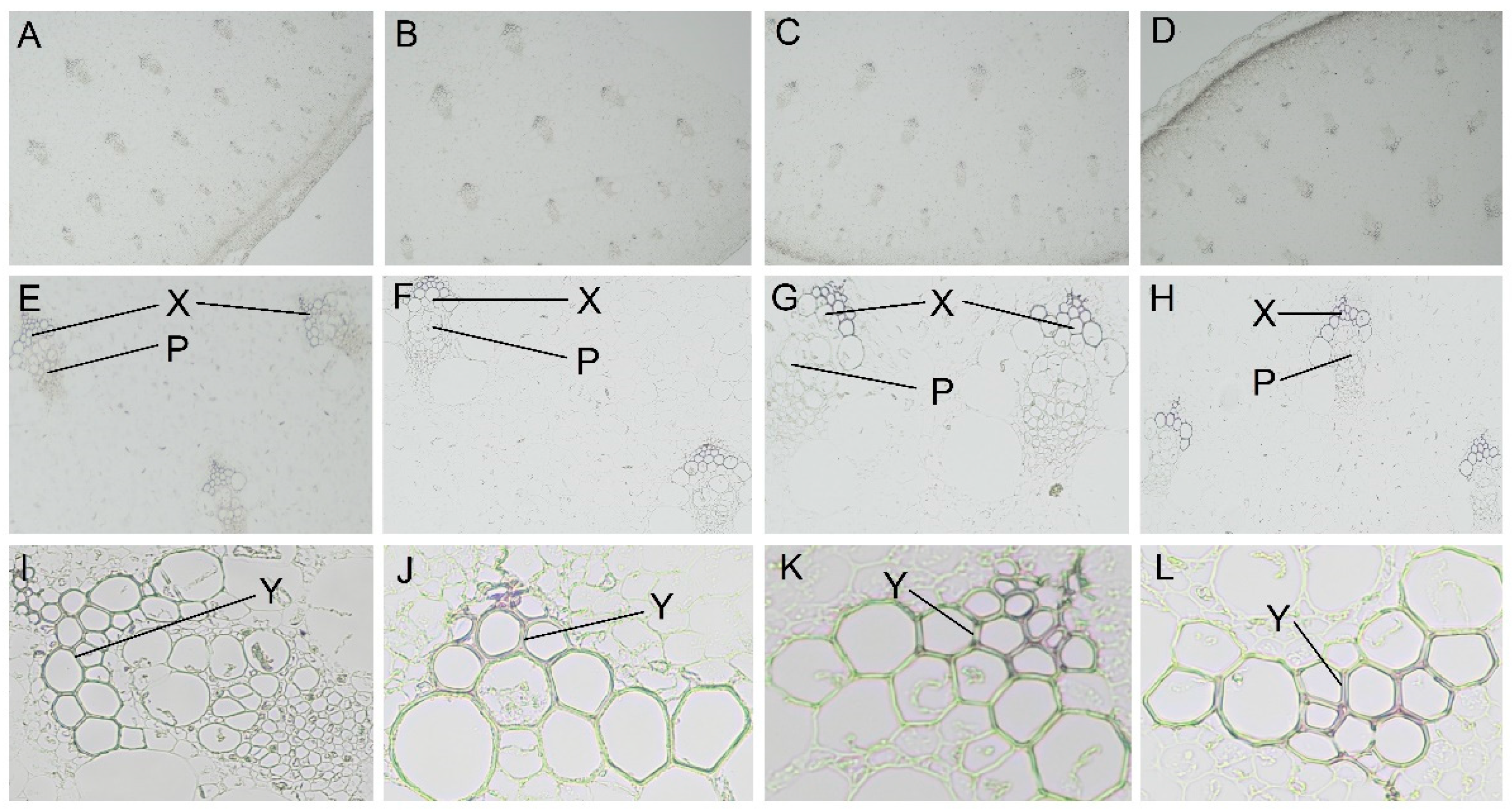
2.2. Histochemical Staining Analysis of Lignin Morphological Distribution in Asparagus under Shading
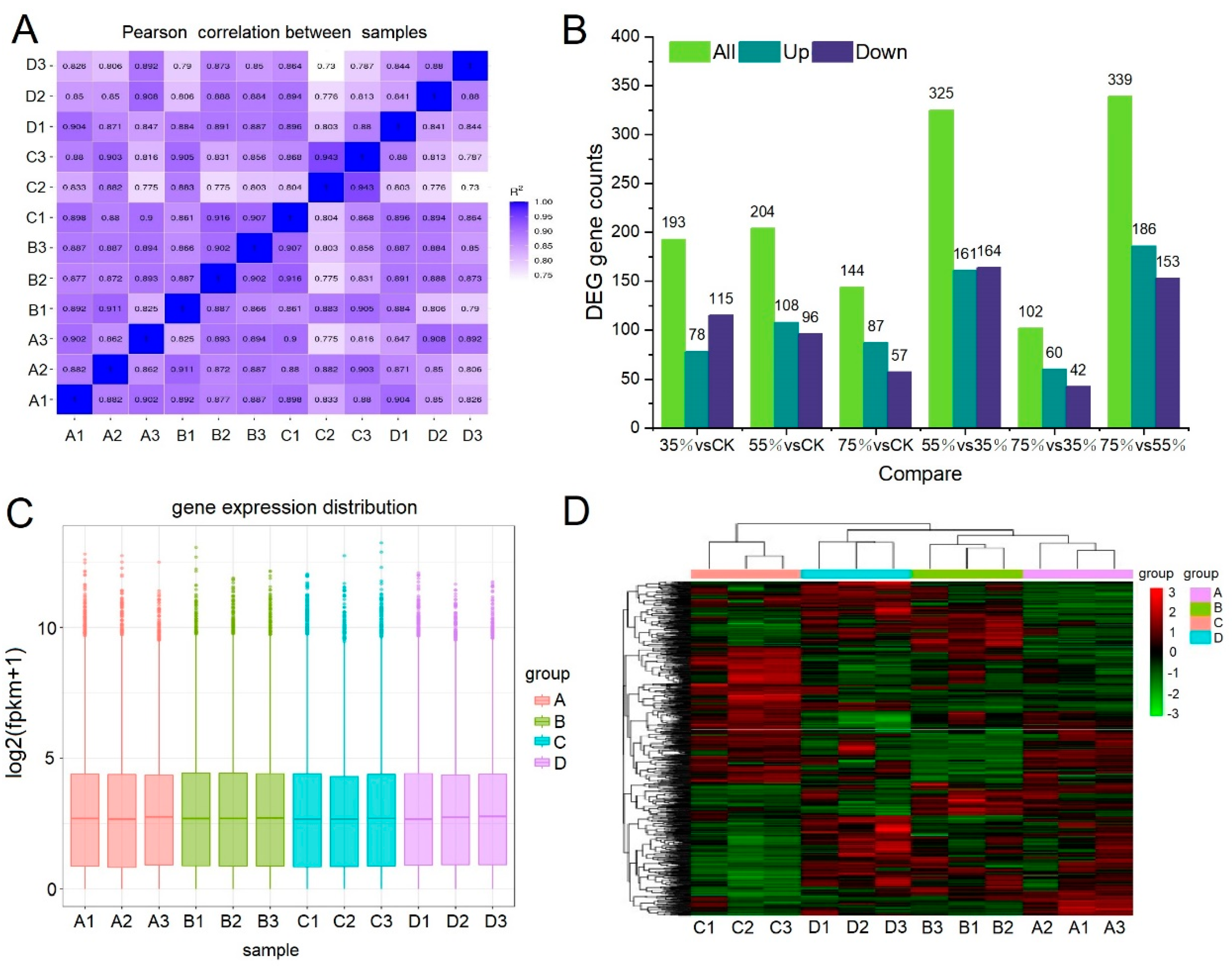
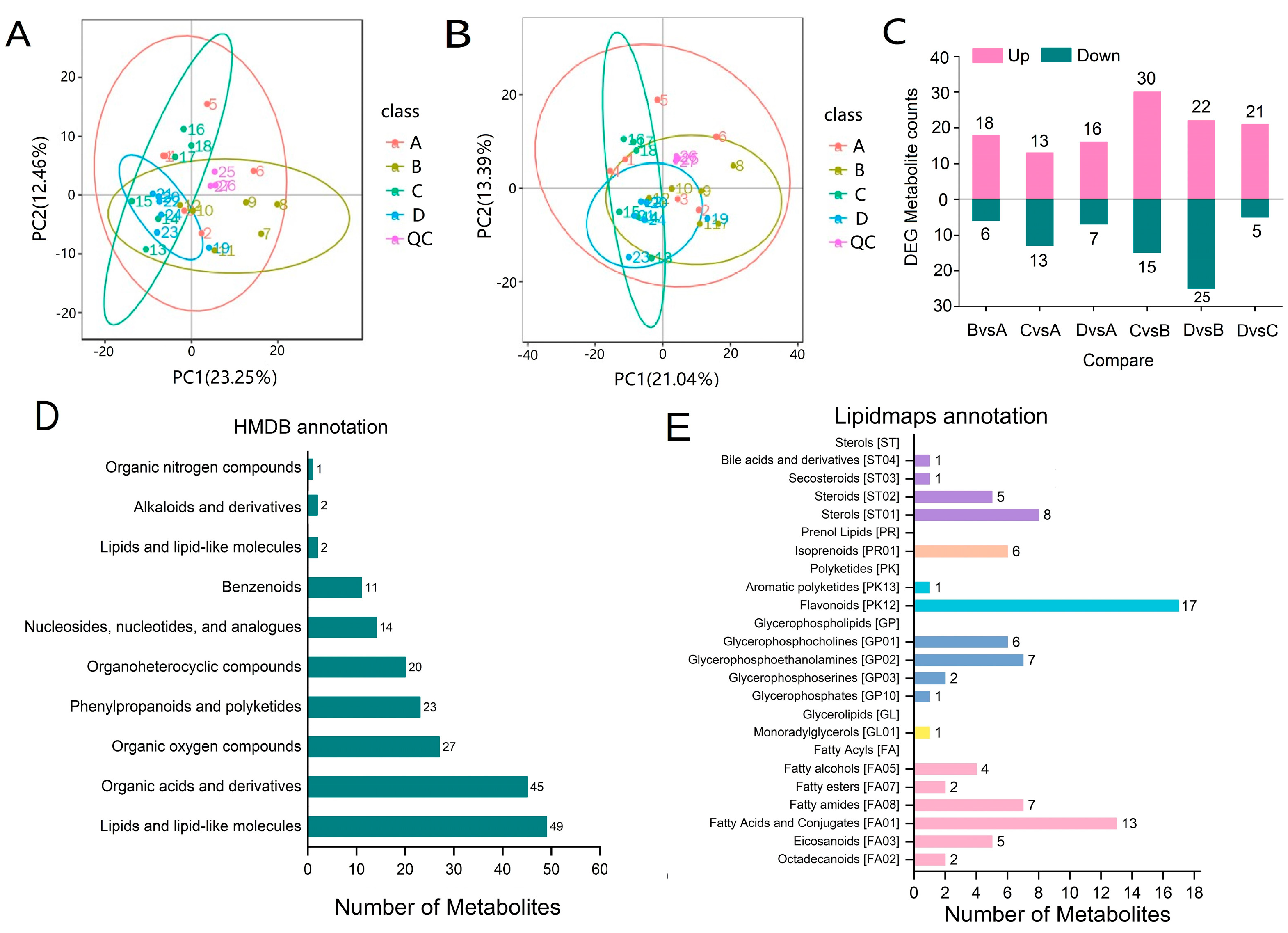
2.3. Transcriptome Sequencing and Whole Gene Expression Analysis
2.4. DEGs Analysis
2.5. Metabolome Analysis
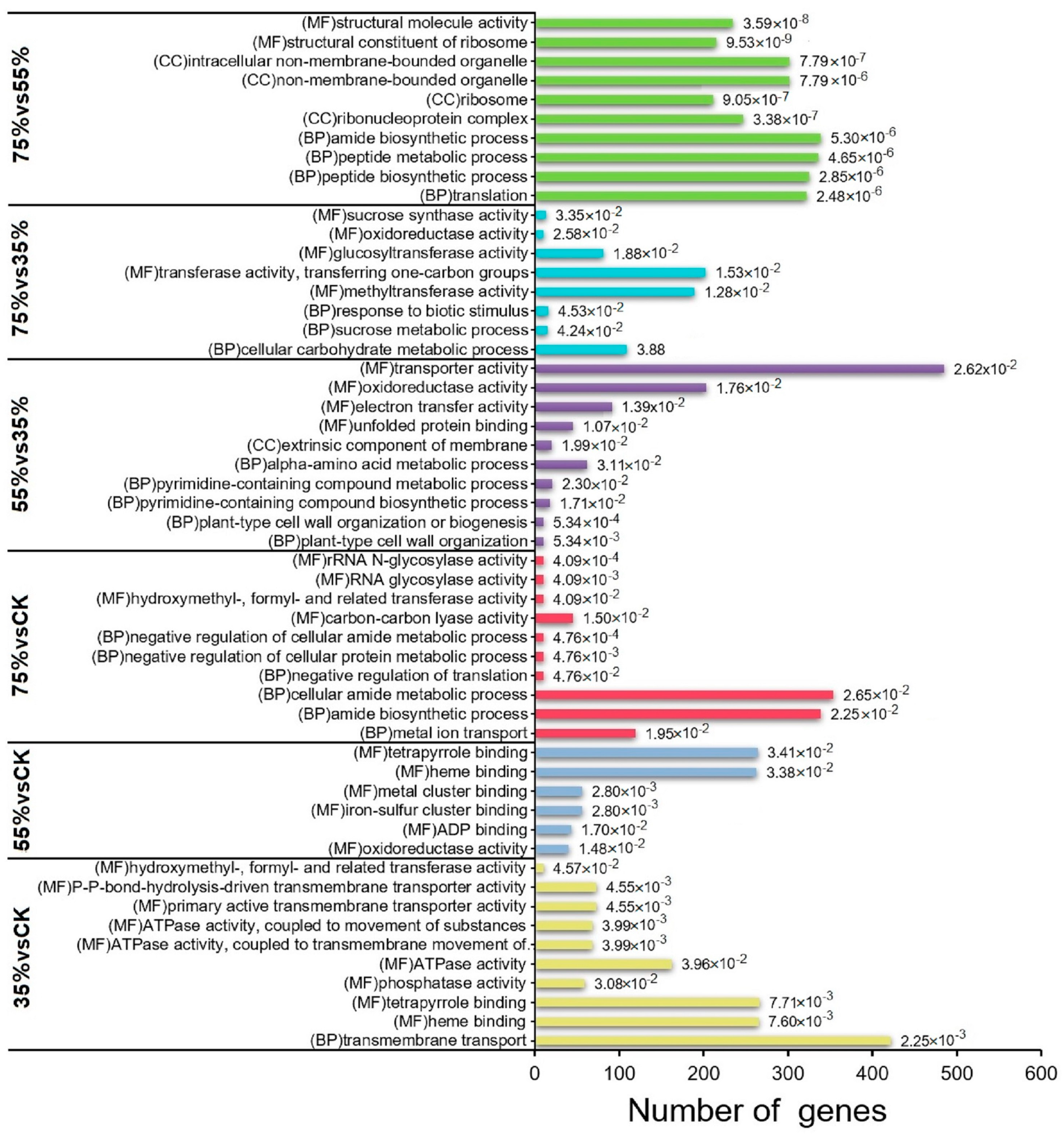
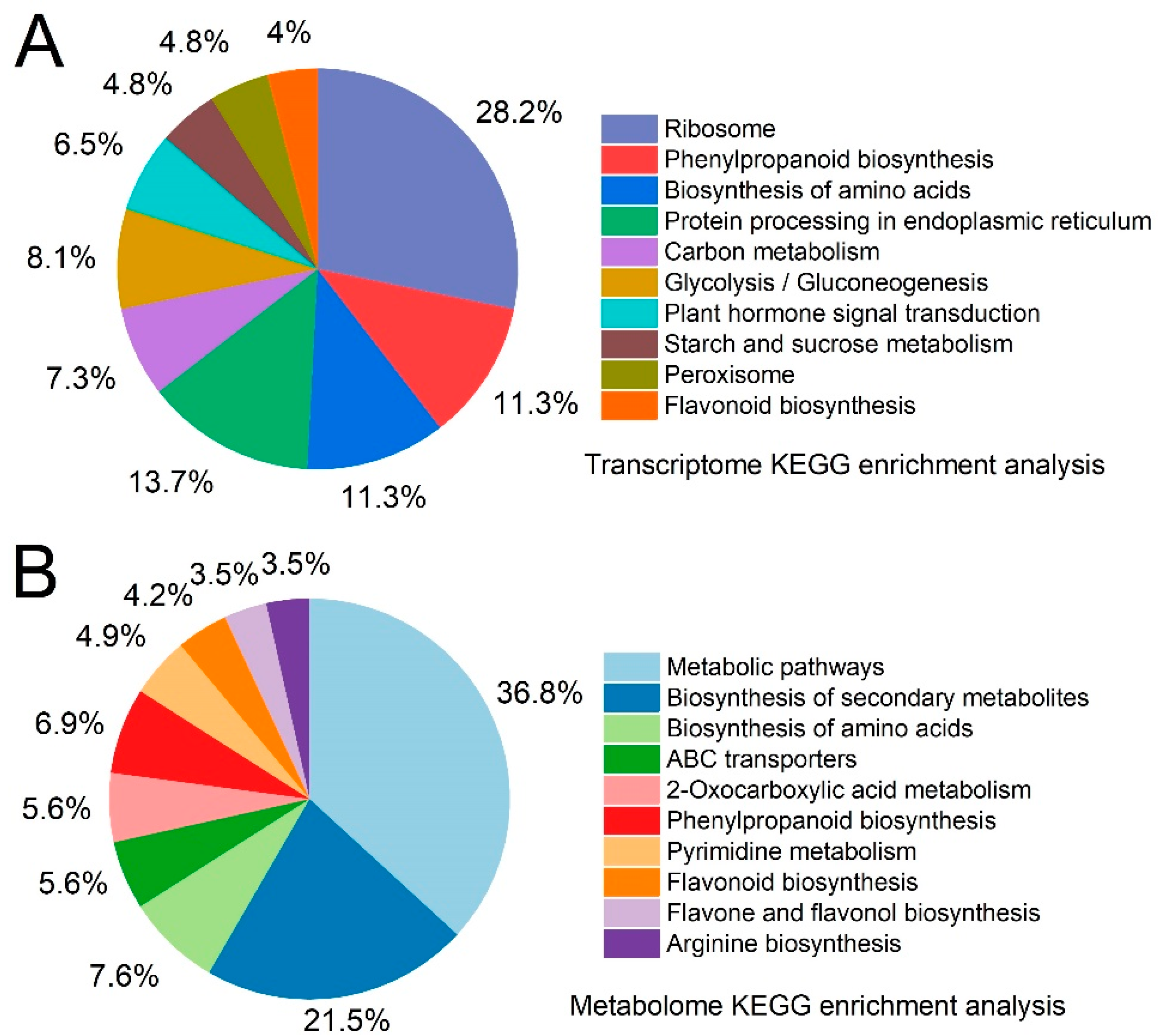
2.6. Transcriptome and Metabolome Pathway Enrichment Analysis
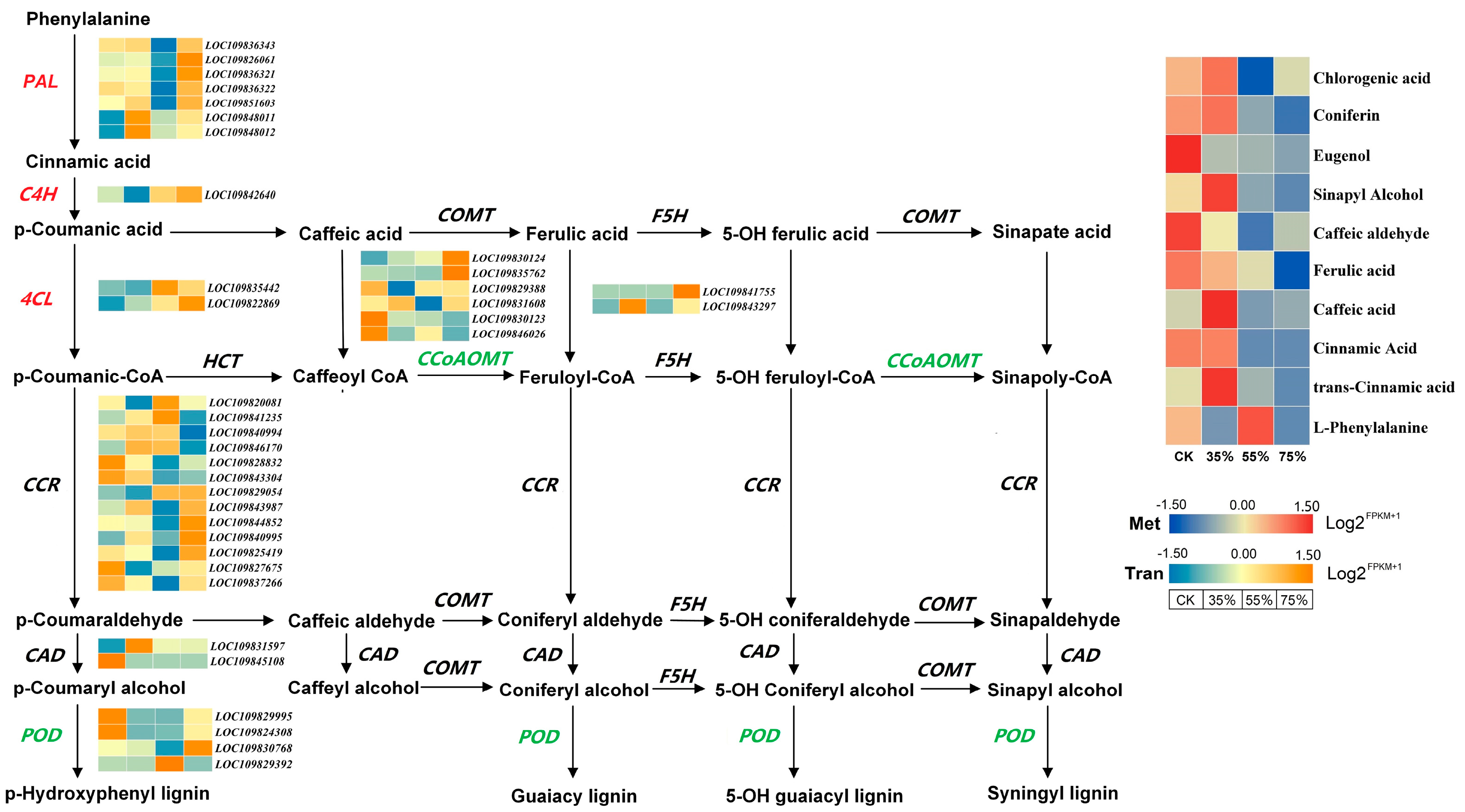
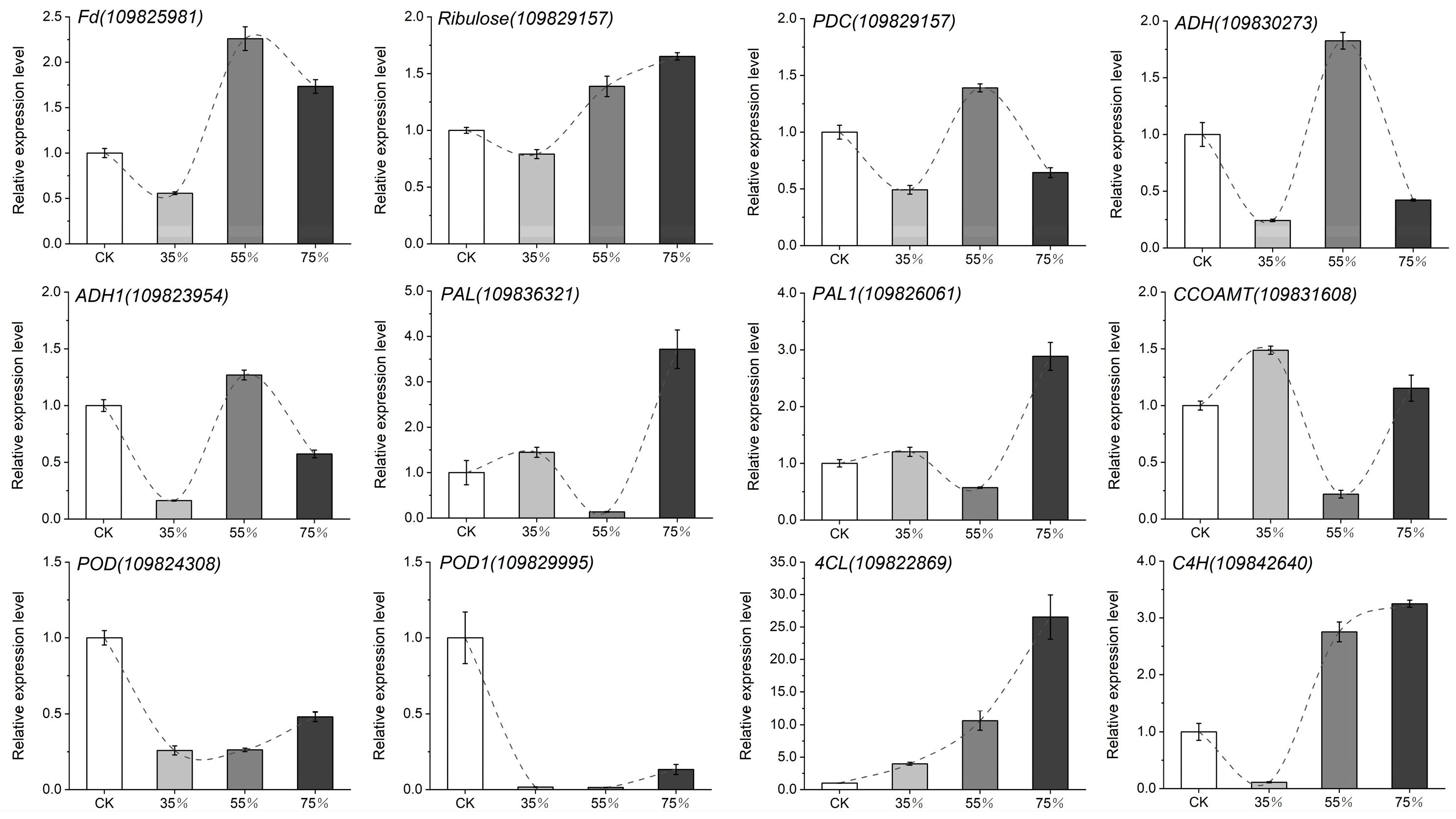
2.7. Transcriptome and Metabolome Reveal Shading Analysis of Lignin Accumulation in Asparagus
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Asparagus Material
5.2. Determination of Stem Diameter and Lignin Content
5.3. Histochemical Staining of Lignin Morphology Distribution
5.4. Transcriptome Sequencing and Annotation
5.5. Extraction and Detection of Metabolites
5.6. RT-qPCR Validation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, P.; Keller, M.; et al. Lignin Valorization: Improving Lignin Processing in the Biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin Biosynthesis and Structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Boudet, A.M.; Kajita, S.; Grima-Pettenati, J.; Goffner, D. Lignins and lignocellulosics: A better control of synthesis for new and improved uses. Trends Plant Sci. 2003, 8, 576–581. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, W.; Ding, Y.; Zhang, J.; Cambula, E.D.; Weng, F.; Liu, Z.; Ding, C.; Tang, S.; Chen, L.; et al. Shading Contributes to the Reduction of Stem Mechanical Strength by Decreasing Cell Wall Synthesis in Japonica Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 881. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-G.; Ren, M.-L.; Liu, T.; Du, Y.-L.; Zhou, T.; Liu, X.-M.; Liu, J.; Hussain, S.; Yang, W.-Y. Effect of shade stress on lignin biosynthesis in soybean stems. J. Integr. Agric. 2018, 17, 1594–1604. [Google Scholar] [CrossRef]
- Mottiar, Y.; Vanholme, R.; Boerjan, W.; Ralph, J.; Mansfield, S.D. Designer lignins: Harnessing the plasticity of lignification. Curr. Opin Biotechnol. 2016, 37, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2004, 22, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.-C.; Liu, C.-J. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chapple, C. The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Liu, C.-J.; Miao, Y.-C.; Zhang, K.-W. Sequestration and Transport of Lignin Monomeric Precursors. Molecules 2011, 16, 710–727. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Chen, Z.; Yu, J.-Q. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Thévenin, J.; Pollet, B.; Letarnec, B.; Saulnier, L.; Gissot, L.; Maia-Grondard, A.; Lapierre, C.; Jouanin, L. The Simultaneous Repression of CCR and CAD, Two Enzymes of the Lignin Biosynthetic Pathway, Results in Sterility and Dwarfism in Arabidopsis thaliana. Mol. Plant 2011, 4, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Rochfort, S.; Liu, Z.; Ran, Y.; Griffith, M.; Badenhorst, P.; Spangenberg, G.; Smith, K.F.; Noel, J.P.; Mouradov, A.; et al. Functional analyses of caffeic acid O-Methyltransferase and Cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell. 2010, 22, 3357–3373. [Google Scholar] [CrossRef] [PubMed]
- Shadle, G.; Chen, F.; Srinivasa Reddy, M.S.; Jackson, L.; Nakashima, J.; Dixon, R.A. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 2007, 68, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, W.C.; Xu, Y.Q.; Li, G.J.; Liao, Y.; Fu, F.L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Linker, R.; Gepstein, S.; Tanimoto, E.; Yamamoto, R.; Neumann, P.M. Progressive inhibition by water deficit of cell wall extensibility and growth along the elongation zone of maize roots is related to increased lignin metabolism and progressive stelar accumulation of wall phenolics. Plant Physiol. 2006, 140, 603–612. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Valliyodan, B.; Zhang, J.; Lenoble, M.E.; Yu, O.; Rogers, E.E.; Sharp, R.E.; Nguyen, H.T. Regulation of growth response to water stress in the soybean primary root. I. Proteomic analysis reveals region-specific regulation of phenylpropanoid metabolism and control of free iron in the elongation zone. Plant Cell Environ. 2010, 33, 223–243. [Google Scholar] [CrossRef]
- Chitrakar, B.; Zhang, M.; Adhikari, B. Asparagus (Asparagus officinalis): Processing effect on nutritional and phytochemical composition of spear and hard-stem byproducts. Trends Food Sci. Technol. 2019, 93, 1–11. [Google Scholar] [CrossRef]
- Kobus-Cisowska, J.; Szymanowska, D.; Szczepaniak, O.M.; Gramza-Michałowska, A.; Kmiecik, D.; Kulczyński, B.; Szulc, P.; Górnaś, P. Composition of polyphenols of asparagus spears (Asparagus officinalis) and their antioxidant potential. Cienc. Rural 2019, 49, e20180863–e20180871. [Google Scholar] [CrossRef]
- Lee, E.J.; Yoo, K.S.; Patil, B.S. Development of a rapid HPLC-UV method for simultaneous quantification of protodioscin and rutin in white and green asparagus spears. J. Food Sci. 2010, 75, C703–C709. [Google Scholar] [CrossRef]
- Ferrara, L.; Dosi, R.; Di Maro, A.; Guida, V.; Cefarelli, G.; Pacifico, S.; Mastellone, C.; Fiorentino, A.; Rosati, A.; Parente, A. Nutritional values, metabolic profile and radical scavenging capacities of wild asparagus (A. acutifolius L.). J. Food Compos. Anal. 2011, 24, 326–333. [Google Scholar] [CrossRef]
- Desoukey, S.F.; El-Nahas, S.E.; Sabh, A.Z.; Taha, Z.K.; El-Shabrawi, H.M. Antimicrobial effect of Asparagus officinalis L. extracts. Plant Arch. 2020, 20, 9253–9264. Available online: http://plantarchives.org/20-2/9253-9264%20(7132) (accessed on 20 December 2022).
- Zhao, Q.; Xie, B.; Yan, J.; Zhao, F.; Xiao, J.; Yao, L.; Zhao, B.; Huang, Y. In vitro antioxidant and antitumor activities of polysaccharides extracted from Asparagus officinalis. Carbohydr. Polym. 2012, 87, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.H.; Shi, W.B.; Xia, X.; Zhao, D.Q.; Wu, Y.Q.; Tao, J. Morphological, microstructural and lignin-related responses of herbaceous peony stem to shading. Sci. Hortic. 2022, 293, 110734. [Google Scholar] [CrossRef]
- Casal, J.J. Shade avoidance. Arab. Book 2012, 10, e0157. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-Wide Characterization of the Lignification Toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Van Acker, R.; Vanholme, R.; Storme, V.; Mortimer, J.C.; Dupree, P.; Boerjan, W. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 2013, 6, 46. [Google Scholar] [CrossRef]
- Zhao, H.; Wei, J.; Jin, Z.; Liu, H.; Tai, W.; Song, Y. Lignin biosynthesis by suppression of two O-methyl-transferases. Chin. Sci. Bull. 2002, 47, 1092–1095. [Google Scholar] [CrossRef]
- Zhong, R.; Iii, W.H.M.; Himmelsbach, D.S.; Ii, F.L.P.; Ye, Z.-H. Essential Role of Caffeoyl Coenzyme A O-Methyltransferase in Lignin Biosynthesis in Woody Poplar Plants. Plant Physiol. 2000, 124, 563–578. [Google Scholar] [CrossRef]
- Mir Derikvand, M.; Sierra, J.B.; Ruel, K.; Pollet, B.; Do, C.T.; Thévenin, J.; Buffard, D.; Jouanin, L.; Lapierre, C. Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 2008, 227, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Ipekci, Z.; Ogras, T.; Altinkut, A. Reduced leaf peroxidase activity is associated with reduced lignin content intransgenic poplar. Plant Biotechnol. 1999, 16, 381–387. [Google Scholar] [CrossRef]
- Hussain, S.; Iqbal, N.; Rahman, T.; Liu, T.; Brestic, M.; Safdar, M.E.; Asghar, M.A.; Farooq, M.U.; Shafiq, I.; Ali, A.; et al. Shade effect on carbohydrates dynamics and stem strength of soybean genotypes. Environ. Exp. Bot. 2019, 162, 374–382. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, H.; Zeng, R.; Wang, X.; Huang, L.; Wang, L.; Wang, X.; Zhang, L. Shade Effects on Peanut Yield Associate with Physiological and Expressional Regulation on Photosynthesis and Sucrose Metabolism. Int. J. Mol. Sci. 2020, 21, 5284. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, X.; Hou, P.; Xue, J.; Liu, G.; Liu, W.; Li, S.; Wang, K.; Xie, R.; Ming, B.; et al. Quantitative effects of solar radiation on maize lodging resistance mechanical properties. Field Crops Res. 2020, 255, 107906. [Google Scholar] [CrossRef]
- Liu, W.-G.; Wen, B.-X.; Zhou, T.; Wang, L.; Gao, Y.; Li, S.-X.; Qin, S.-S.; Liu, J.; Yang, W.-Y. iTRAQ protein profile analysis of soybean stems reveals new aspects critical for lodging in intercropping systems. J. Integr. Agric. 2019, 18, 2029–2040. [Google Scholar] [CrossRef]
- Wen, B.; Zhang, Y.; Hussain, S.; Wang, S.; Zhang, X.; Yang, J.; Xu, M.; Qin, S.; Yang, W.; Liu, W. Slight Shading Stress at Seedling Stage Does not Reduce Lignin Biosynthesis or Affect Lodging Resistance of Soybean Stems. Agronomy 2020, 10, 544. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Oliveira, D.M.; Andreotti, G.C.; De Souza, L.A.; Dos Santos, W.D.; Bonato, C.M.; Antunes, W.C. Increased Gibberellins and Light Levels Promotes Cell Wall Thickness and Enhance Lignin Deposition in Xylem Fibers. Front. Plant Sci. 2018, 9, 1391. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 2009, 60, 509–521. [Google Scholar] [CrossRef]
- Lillo, C.; Lea, U.S.; Ruoff, P. Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 2008, 31, 587–601. [Google Scholar] [CrossRef]
- Elkind, Y.; Edwards, R.; Mavandad, M.; Hedrick, S.A.; Ribak, O.; Dixon, R.A.; Lamb, C.J. Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc. Natl. Acad. Sci. USA 1990, 87, 9057–9061. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Jourdes, M.; Patten, A.M.; Kim, K.-W.; Vassão, D.G.; Lewis, N.G. Dissection of lignin macromolecular configuration and assembly: Comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 2008, 25, 1015–1090. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, G.; Nie, J.; Lin, Y.; Nie, F.; Zhang, J.; Xu, Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Abdulrazzak, N.; Pollet, B.; Ehlting, J.; Larsen, K.; Asnaghi, C.; Ronseau, S.; Proux, C.; Erhardt, M.; Seltzer, V.; Renou, J.-P.; et al. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 2006, 140, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Schoch, G.A.; Morant, M.; Abdulrazzak, N.; Asnaghi, C.; Goepfert, S.; Petersen, M.; Ullmann, P.; Werck-Reichhart, D. The meta-hydroxylation step in the phenylpropanoid pathway: A new level of complexity in the pathway and its regulation. Environ. Chem. Lett. 2006, 4, 127–136. [Google Scholar] [CrossRef]
- Morrison, I.M. A semi-micro method for the determination of lignin and its use in predicting the digestibility of forage crops. J. Sci. Food Agric. 1972, 23, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2012, 8, 17–32. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Li, X.; He, M.; Li, Y.; Lu, W.; Li, M.; Sun, B.; Zheng, Y. A Joint Transcriptomic and Metabolomic Analysis Reveals the Regulation of Shading on Lignin Biosynthesis in Asparagus. Int. J. Mol. Sci. 2023, 24, 1539. https://doi.org/10.3390/ijms24021539
Ma J, Li X, He M, Li Y, Lu W, Li M, Sun B, Zheng Y. A Joint Transcriptomic and Metabolomic Analysis Reveals the Regulation of Shading on Lignin Biosynthesis in Asparagus. International Journal of Molecular Sciences. 2023; 24(2):1539. https://doi.org/10.3390/ijms24021539
Chicago/Turabian StyleMa, Junying, Xiaoyan Li, Maolin He, Yanwen Li, Wei Lu, Mengyao Li, Bo Sun, and Yangxia Zheng. 2023. "A Joint Transcriptomic and Metabolomic Analysis Reveals the Regulation of Shading on Lignin Biosynthesis in Asparagus" International Journal of Molecular Sciences 24, no. 2: 1539. https://doi.org/10.3390/ijms24021539
APA StyleMa, J., Li, X., He, M., Li, Y., Lu, W., Li, M., Sun, B., & Zheng, Y. (2023). A Joint Transcriptomic and Metabolomic Analysis Reveals the Regulation of Shading on Lignin Biosynthesis in Asparagus. International Journal of Molecular Sciences, 24(2), 1539. https://doi.org/10.3390/ijms24021539








