Downregulation of CDC25C in NPCs Disturbed Cortical Neurogenesis
Abstract
1. Introduction
2. Results
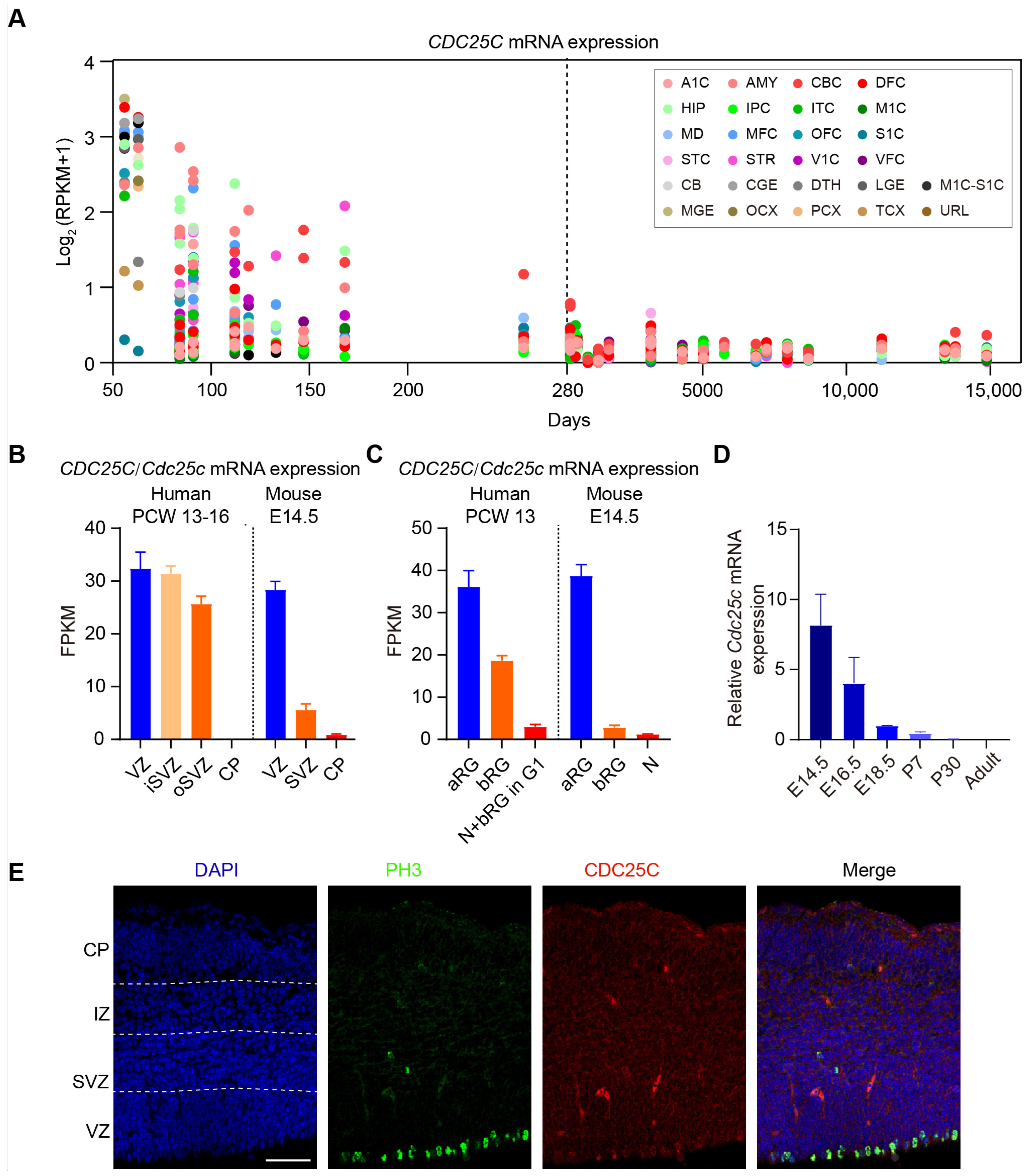
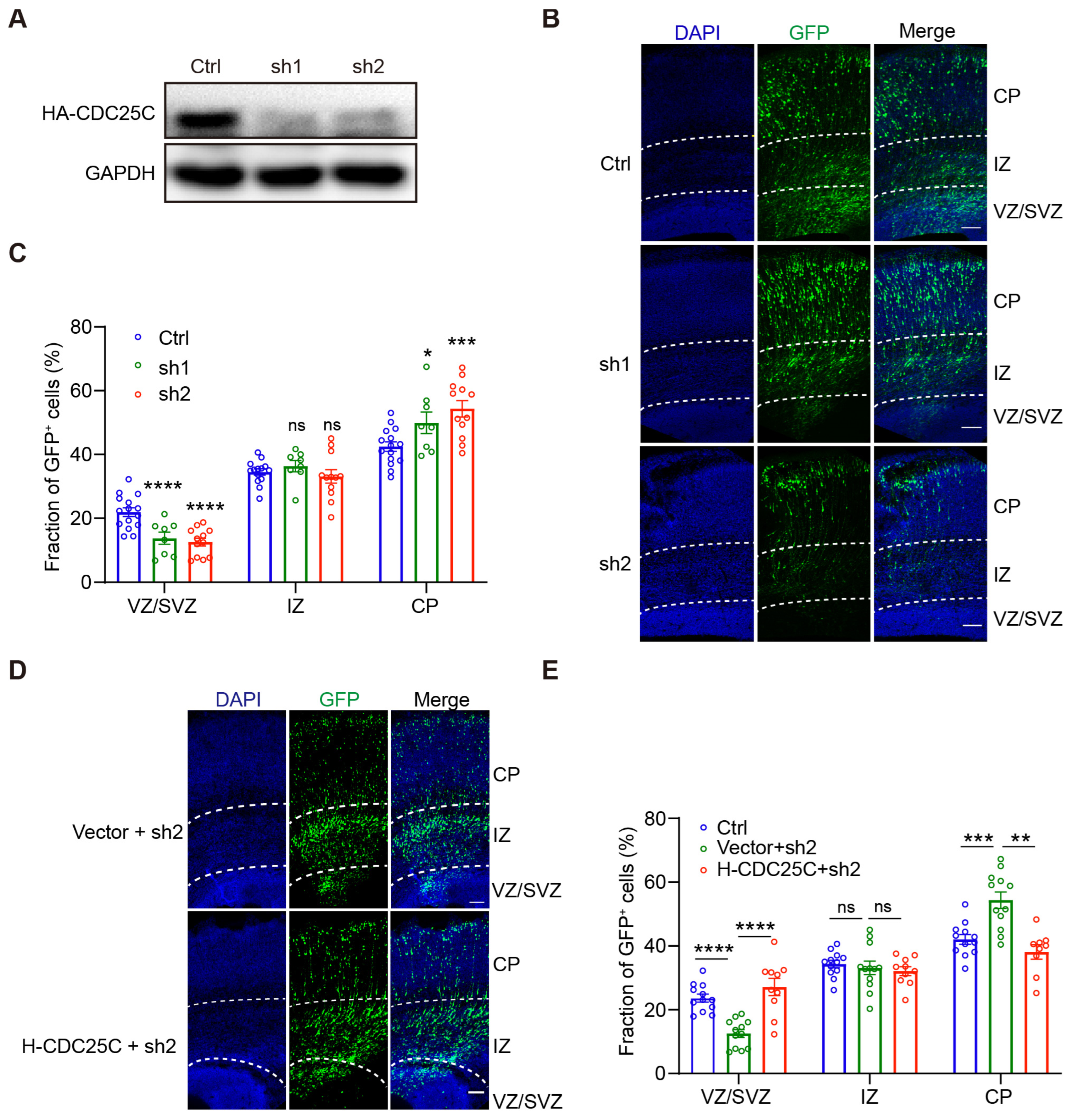
2.1. Silencing of CDC25C Altered Cell Distribution in Developing Neocortex
2.2. Cdc25c Expression Is Essential for Normal Proliferation and Differentiation of NPCs
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Plasmids
4.3. Antibodies
4.4. Cell Culture, Transfection and Western Blotting
4.5. RNA Extraction and QPCR Analysis
4.6. In Utero Electroporation and BrdU Labeling
4.7. Immunostaining
4.8. FACS Analysis
4.9. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huttner, W.B.; Brand, M. Asymmetric division and polarity of neuroepithelial cells. Curr. Opin. Neurobiol. 1997, 7, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Götz, M.; Barde, Y.A. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron 2005, 46, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Knoblich, J.A. Mechanisms of asymmetric stem cell division. Cell 2008, 132, 583–597. [Google Scholar] [CrossRef]
- Franco, S.J.; Müller, U. Shaping our minds: Stem and progenitor cell diversity in the mammalian neocortex. Neuron 2013, 77, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Schlusche, A.K.; Vay, S.U.; Huttner, W.; Isbrandt, D.; Jakovcevski, I.; Kleinenkuhnen, N.; Sandke, S.; Campos-Martín, R.; Florio, M.; Roeper, J.; et al. Developmental HCN channelopathy results in decreased neural progenitor proliferation and microcephaly in mice. Proc. Natl. Acad. Sci. USA 2021, 118, e2009393118. [Google Scholar] [CrossRef] [PubMed]
- Tsampoula, M.; Tarampoulous, I.; Manolakou, T.; Ninou, E.; Politis, P.K. The Neurodevelopmental Disorders Associated Gene Rnf113a Regulates Survival and Differentiation Properties of Neural Stem Cells. Stem. Cells Dev. 2022, 40, 678–690. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Sievert, D.; Smith, D.E.C.; Mendes, M.I.; Chen, D.Y.; Stanley, V.; Ghosh, S.; Wang, Y.; Kara, M.; et al. Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat. Commun. 2020, 11, 4038. [Google Scholar] [CrossRef]
- Dehay, C.; Kennedy, H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007, 8, 438–450. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef]
- Wickramasinghe, D.; Becker, S.; Ernst, M.K.; Resnick, J.L.; Centanni, J.M.; Tessarollo, L.; Grabel, L.B.; Donovan, P.J. Two CDC25 homologues are differentially expressed during mouse development. Development 1995, 121, 2047–2056. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, D.; Li, Z.; Huang, Y.; Wu, Q.; Ru, G.; Chen, M.; Wang, B. Oxidative stress-induced DNA damage of mouse zygotes triggers G2/M checkpoint and phosphorylates Cdc25 and Cdc2. Cell Stress Chaperones 2016, 21, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Al-Matouq, J.; Holmes, T.R.; Hansen, L.A. CDC25B and CDC25C overexpression in nonmelanoma skin cancer suppresses cell death. Mol. Carcinog. 2019, 58, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.C.; Park, J.E.; Park, B.C.; Kim, J.H.; Jeong, D.G.; Park, S.G.; Cho, S. Cell cycle-dependent Cdc25C phosphatase determines cell survival by regulating apoptosis signal-regulating kinase 1. Cell Death Differ. 2015, 22, 1605–1617. [Google Scholar] [CrossRef]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tian, C.; Xu, Y.; Xie, W.L.; Zhang, J.; Zhang, B.Y.; Ren, K.; Wang, K.; Chen, C.; Wang, S.B.; et al. Abortive cell cycle events in the brains of scrapie-infected hamsters with remarkable decreases of PLK3/Cdc25C and increases of PLK1/cyclin B1. Mol. Neurobiol. 2013, 48, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Vincent, F.; Shah, K. Deregulated Cdk5 triggers aberrant activation of cell cycle kinases and phosphatases inducing neuronal death. J. Cell Sci. 2012, 125, 5124–5137. [Google Scholar] [CrossRef] [PubMed]
- Wezyk, M.; Szybinska, A.; Wojsiat, J.; Szczerba, M.; Day, K.; Ronnholm, H.; Kele, M.; Berdynski, M.; Peplonska, B.; Fichna, J.P.; et al. Overactive BRCA1 Affects Presenilin 1 in Induced Pluripotent Stem Cell-Derived Neurons in Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 62, 175–202. [Google Scholar] [CrossRef]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Holper, S.; Kruger, M.; Stainier, D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Gunther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Chen, M.S.; Hurov, J.; White, L.S.; Woodford-Thomas, T.; Piwnica-Worms, H. Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol. Cell. Biol. 2001, 21, 3853–3861. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.M.; White, L.S.; Donovan, P.J.; Piwnica-Worms, H. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol. Cell. Biol. 2005, 25, 2853–2860. [Google Scholar] [CrossRef] [PubMed]
- Fietz, S.A.; Lachmann, R.; Brandl, H.; Kircher, M.; Samusik, N.; Schroder, R.; Lakshmanaperumal, N.; Henry, I.; Vogt, J.; Riehn, A.; et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. USA 2012, 109, 11836–11841. [Google Scholar] [CrossRef]
- Florio, M.; Albert, M.; Taverna, E.; Namba, T.; Brandl, H.; Lewitus, E.; Haffner, C.; Sykes, A.; Wong, F.K.; Peters, J.; et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 2015, 347, 1465–1470. [Google Scholar] [CrossRef]
- Zhou, X.; Zhi, Y.; Yu, J.; Xu, D. The Yin and Yang of Autosomal Recessive Primary Microcephaly Genes: Insights from Neurogenesis and Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 1691. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Yo, M.; Komatsu, N.; Hiratsuka, T.; Kogure, T.; Hoshida, T.; Goshima, N.; Matsuda, M.; Miyoshi, H.; Miyawaki, A. Genetically Encoded Tools for Optical Dissection of the Mammalian Cell Cycle. Mol. Cell 2017, 68, 626–640. [Google Scholar] [CrossRef] [PubMed]
- Godin, J.D.; Nguyen, L. Novel Functions of Core Cell Cycle Regulators in Neuronal Migration. In Cellular and Molecular Control of Neuronal Migration; Springer: Dordrecht, The Netherlands, 2014; pp. 59–74. [Google Scholar] [CrossRef]
- Liu, X.; Zong, W.; Li, T.; Wang, Y.; Xu, X.; Zhou, Z.W.; Wang, Z.Q. The E3 ubiquitin ligase APC/C(C)(dh1) degrades MCPH1 after MCPH1-betaTrCP2-Cdc25A-mediated mitotic entry to ensure neurogenesis. EMBO J. 2017, 36, 3666–3681. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Bonnet, F.; Pignolet, J.; Lobjois, V.; Bel-Vialar, S.; Gautrais, J.; Pituello, F.; Agius, E. Single-cell imaging of the cell cycle reveals CDC25B-induced heterogeneity of G1 phase length in neural progenitor cells. Development 2022, 149, dev199660. [Google Scholar] [CrossRef]
- Gruber, R.; Zhou, Z.; Sukchev, M.; Joerss, T.; Frappart, P.O.; Wang, Z.Q. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat. Cell Biol. 2011, 13, 1325–1334. [Google Scholar] [CrossRef]
- Prasad Tharanga Jayasooriya, R.G.; Dilshara, M.G.; Neelaka Molagoda, I.M.; Park, C.; Park, S.R.; Lee, S.; Choi, Y.H.; Kim, G.Y. Camptothecin induces G(2)/M phase arrest through the ATM-Chk2-Cdc25C axis as a result of autophagy-induced cytoprotection: Implications of reactive oxygen species. Oncotarget 2018, 9, 21744–21757. [Google Scholar] [CrossRef]
- Xu, Z.; Kukekov, N.V.; Greene, L.A. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 2003, 22, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, F.; Wang, Y.; Sun, Y.; Xu, Z. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Rep. 2014, 6, 104–116. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Lu, D.; Yi, W.; Xu, D. Downregulation of CDC25C in NPCs Disturbed Cortical Neurogenesis. Int. J. Mol. Sci. 2023, 24, 1505. https://doi.org/10.3390/ijms24021505
Zhou X, Lu D, Yi W, Xu D. Downregulation of CDC25C in NPCs Disturbed Cortical Neurogenesis. International Journal of Molecular Sciences. 2023; 24(2):1505. https://doi.org/10.3390/ijms24021505
Chicago/Turabian StyleZhou, Xiaokun, Danping Lu, Wenxiang Yi, and Dan Xu. 2023. "Downregulation of CDC25C in NPCs Disturbed Cortical Neurogenesis" International Journal of Molecular Sciences 24, no. 2: 1505. https://doi.org/10.3390/ijms24021505
APA StyleZhou, X., Lu, D., Yi, W., & Xu, D. (2023). Downregulation of CDC25C in NPCs Disturbed Cortical Neurogenesis. International Journal of Molecular Sciences, 24(2), 1505. https://doi.org/10.3390/ijms24021505



