Curcumin Reduces Pathological Endoplasmic Reticulum Stress through Increasing Proteolysis of Mutant Matrilin-3
Abstract
1. Introduction
2. Results
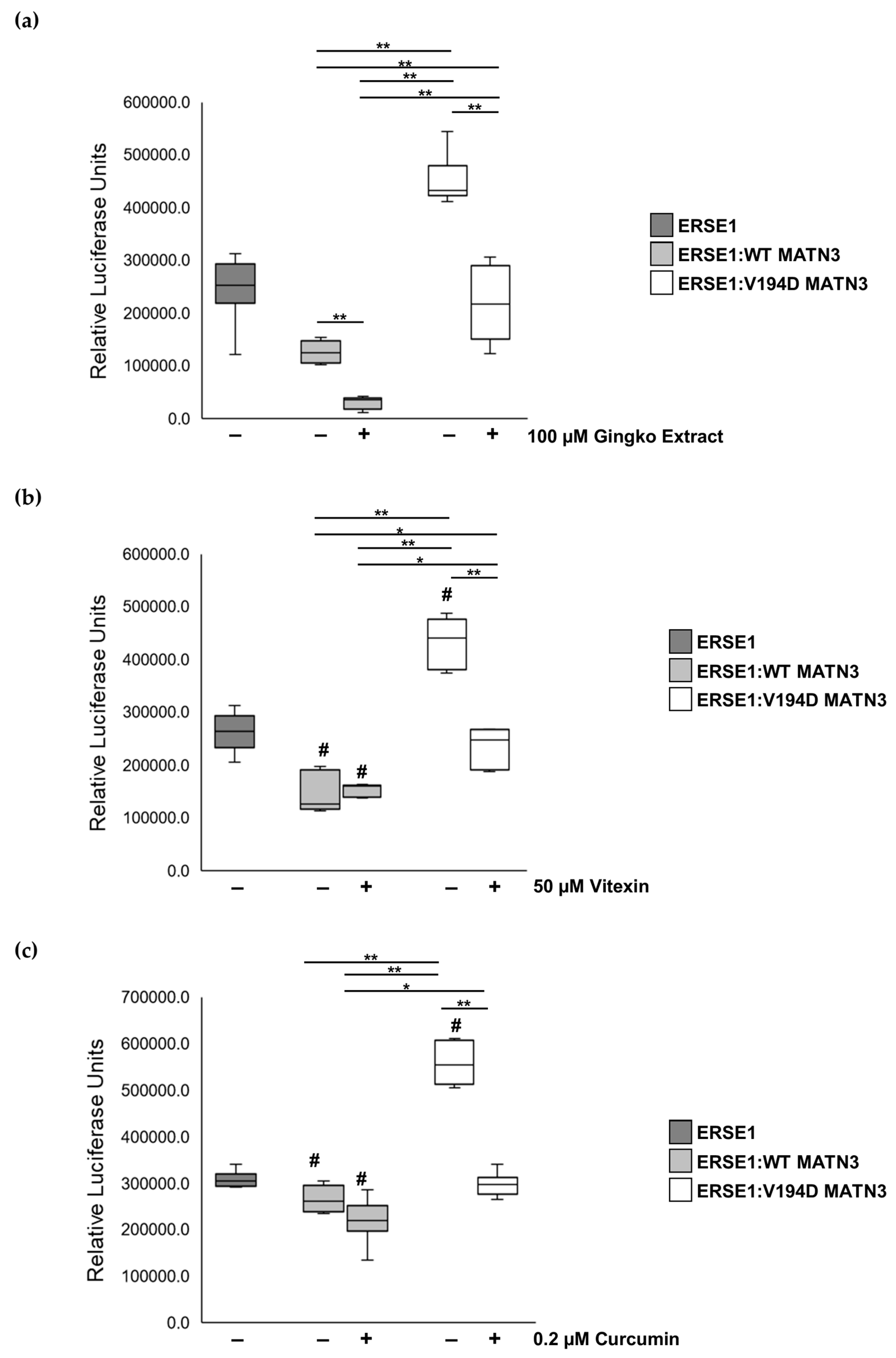
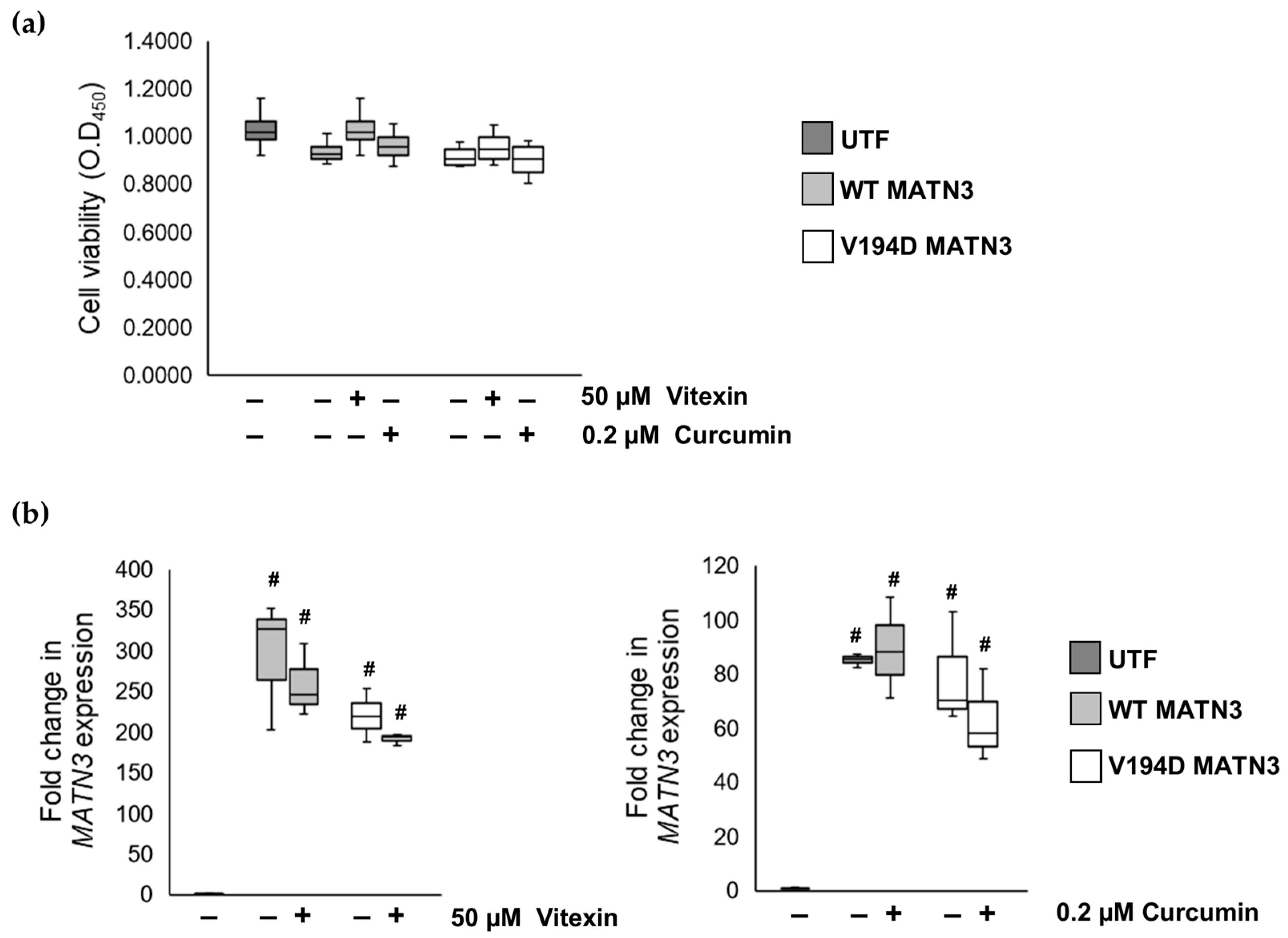
2.1. The Naturally Occurring Chemicals, Vitexin and Curcumin, Significantly Reduce Pathological ER Stress Caused by the Retention of Mutant Matrilin-3 without Affecting Wild-Type Cells
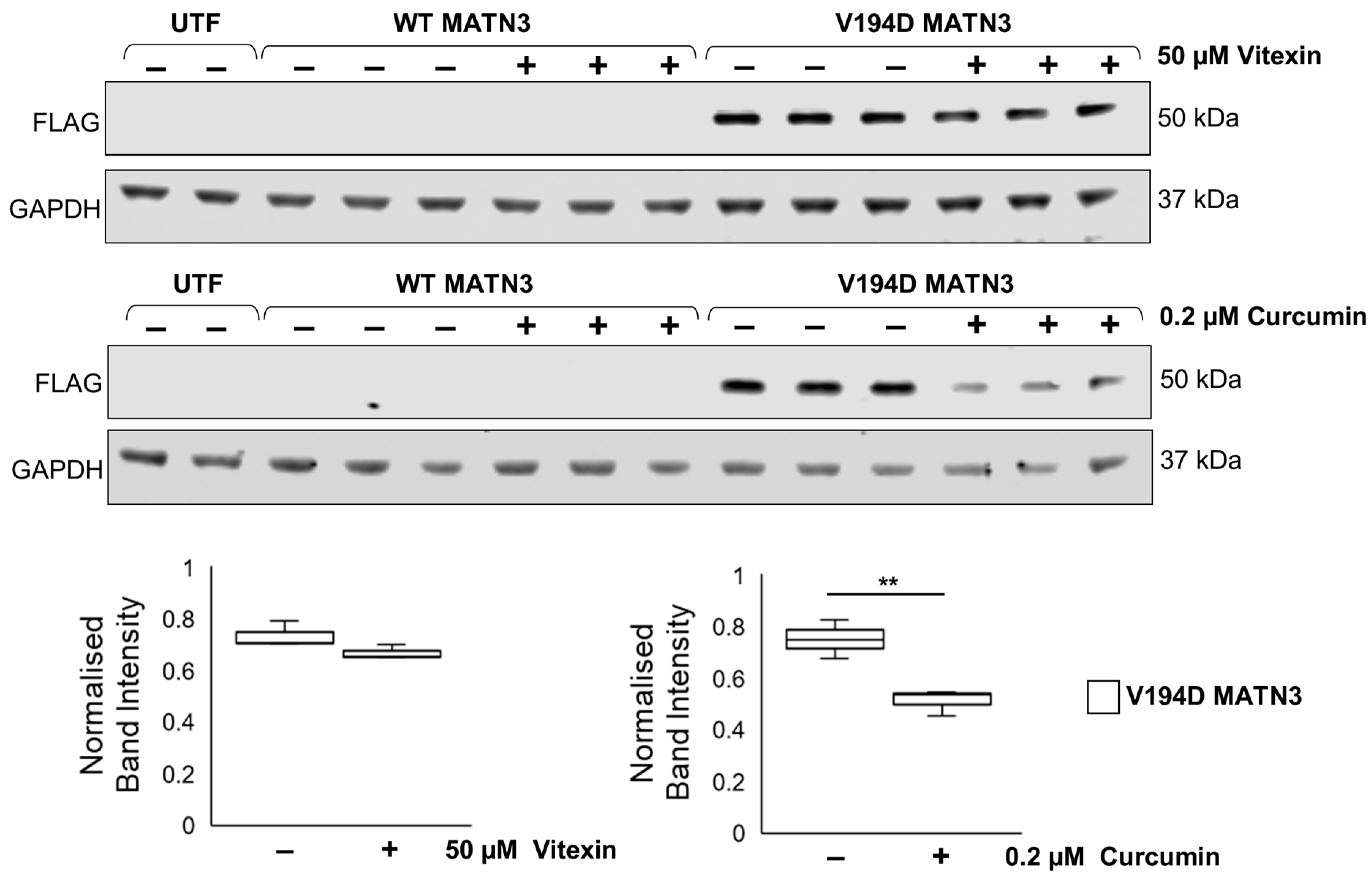
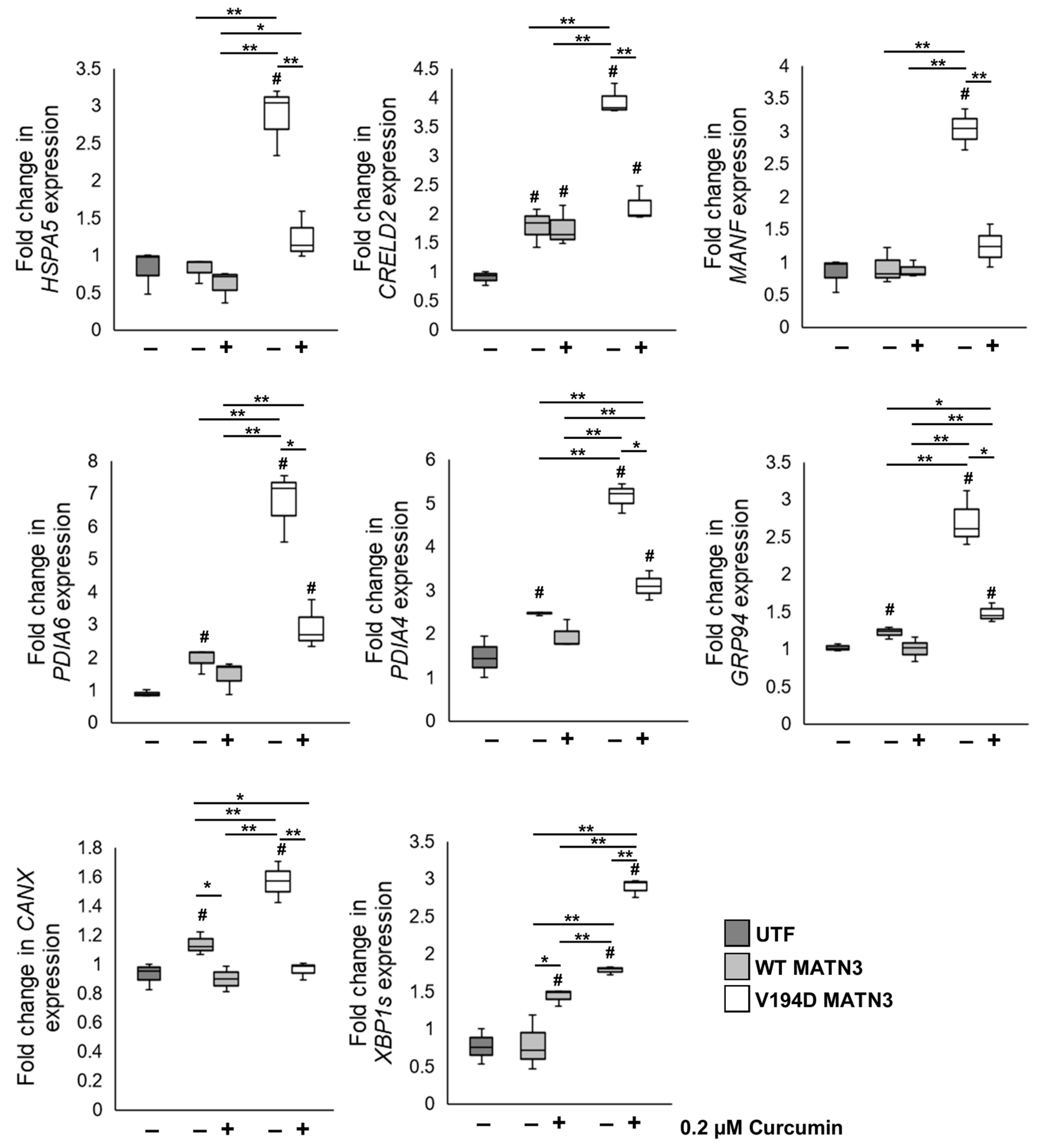
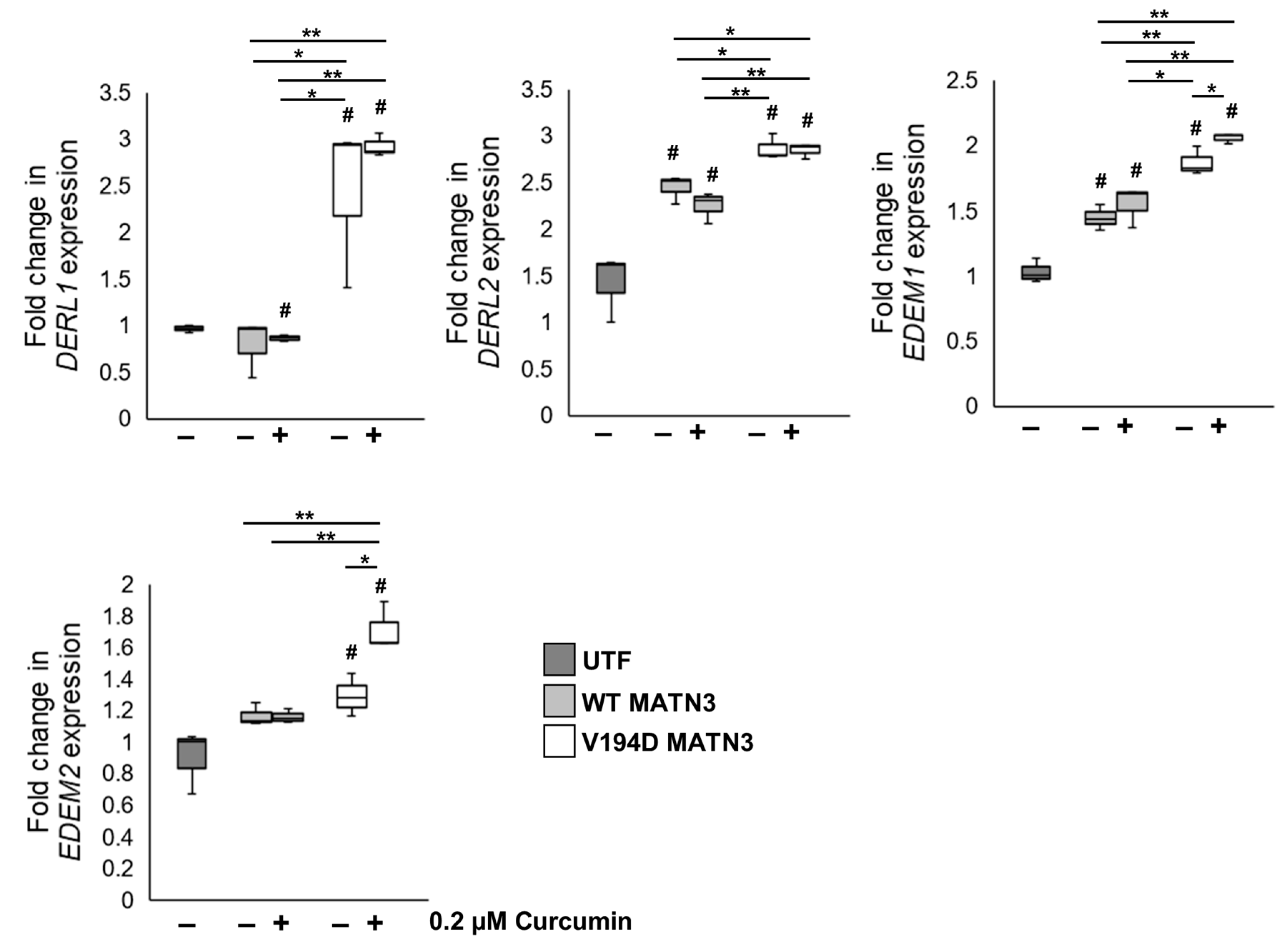
2.2. Curcumin Treatment Reduces the Intracellular Accumulation of Mutant Matrilin-3 and Ameliorates Pathological ER Stress Signalling in a Cell Model of EDM5
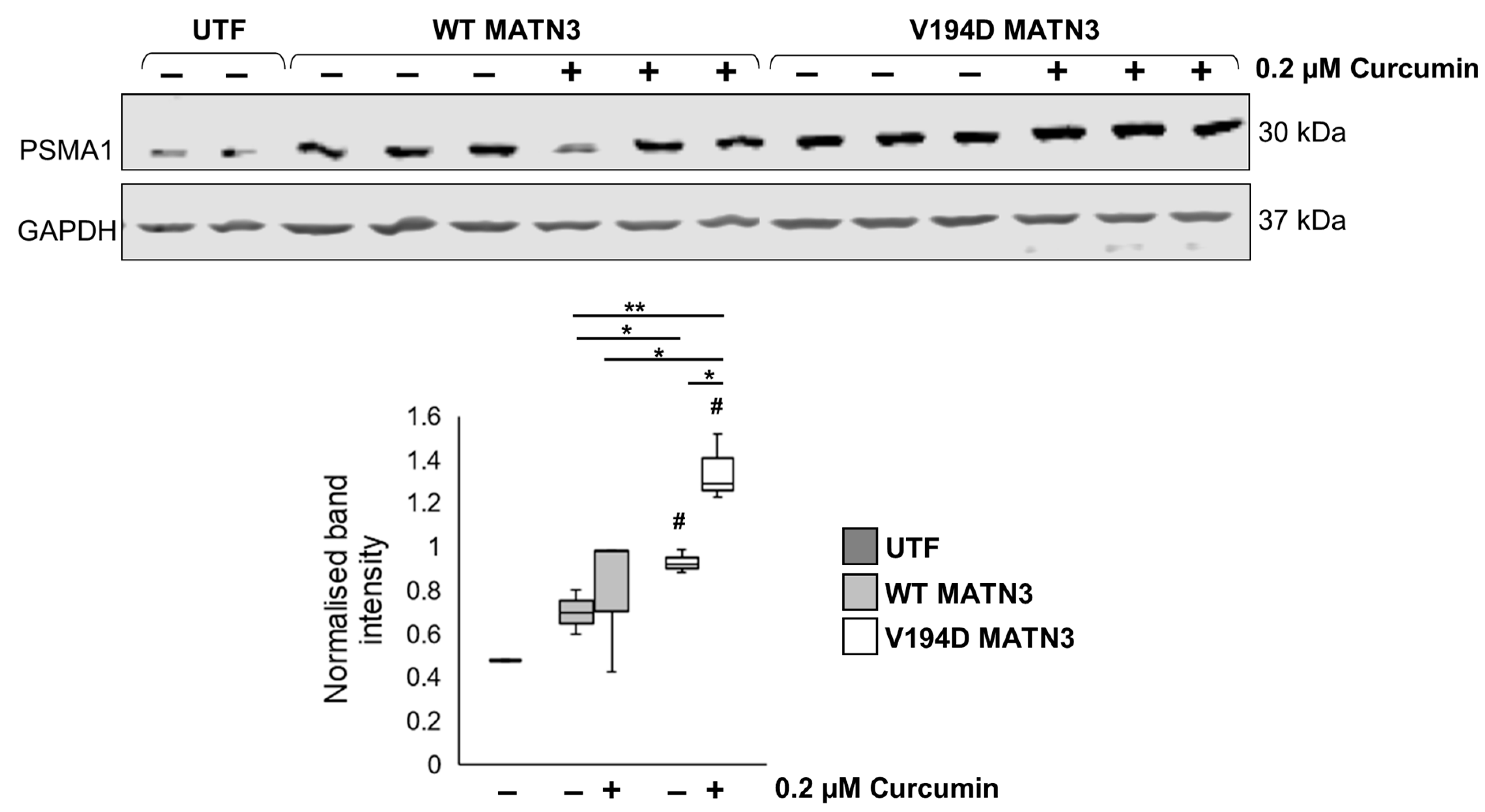
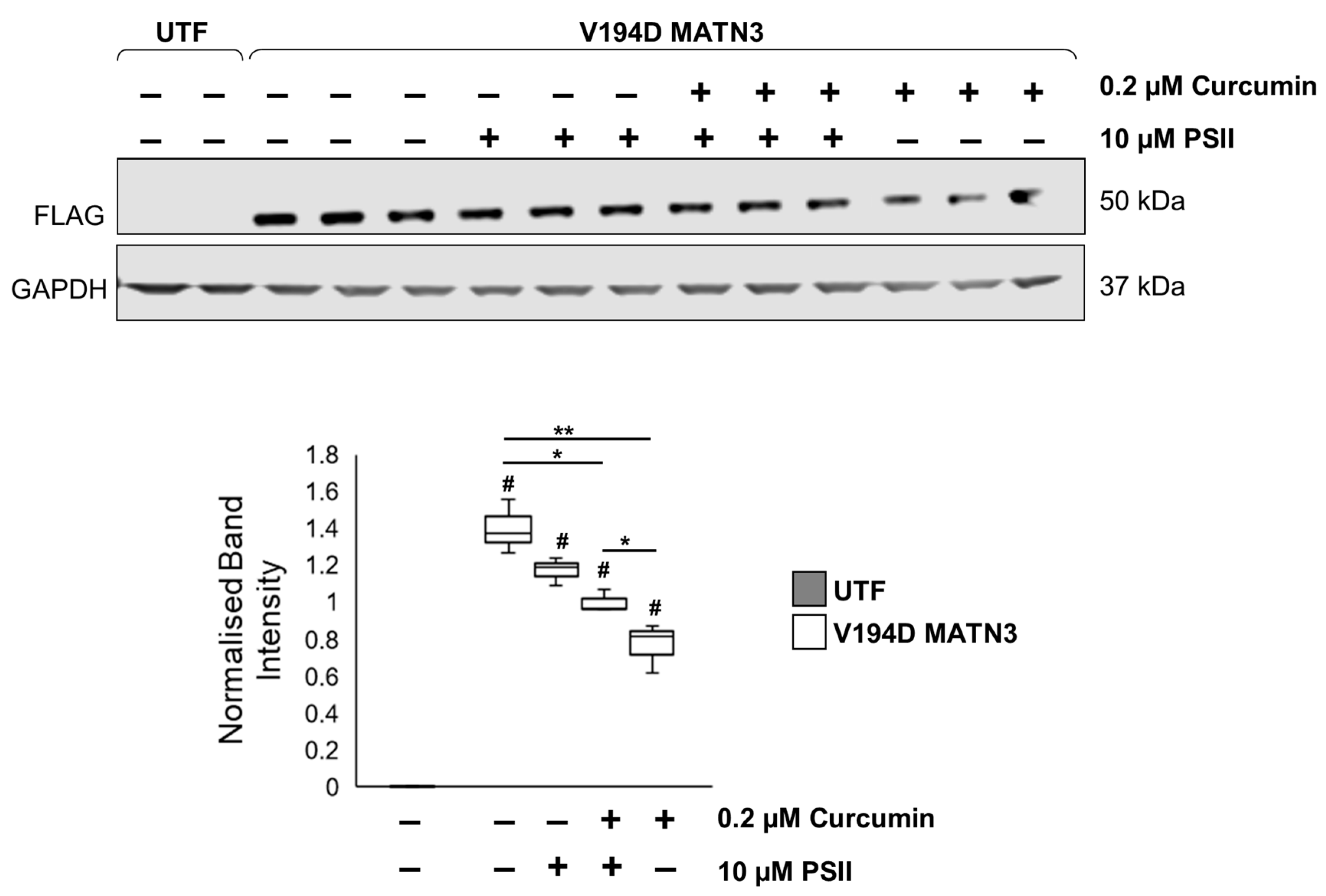
2.3. Curcumin Promotes Proteolysis of Mutant Matrilin-3
3. Discussion and Conclusions
4. Materials and Methods
4.1. Cell Culture
4.2. Luciferase Assay
4.3. Cell Viability
4.4. Protein Extraction and Immunoblotting
4.5. RNA Extraction and Quantitative PCR (qPCR)
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chapman, K.L.; Mortier, G.R.; Chapman, K.; Loughlin, J.; Grant, M.E.; Briggs, M.D. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. Nat. Genet. 2001, 28, 393–396. [Google Scholar] [CrossRef]
- Jackson, G.C.; Barker, F.S.; Jakkula, E.; Czarny-Ratajczak, M.; Mäkitie, O.; Cole, W.G.; Wright, M.J.; Smithson, S.F.; Suri, M.; Rogala, P.; et al. Missense mutations in the beta strands of the single A-domain of matrilin-3 result in multiple epiphyseal dysplasia. J. Med. Genet. 2004, 41, 52–59. [Google Scholar] [CrossRef]
- Dennis, E.P.; Greenhalgh-Maychell, P.L.; Briggs, M.D. Multiple epiphyseal dysplasia and related disorders: Molecular genetics, disease mechanisms, and therapeutic avenues. Dev. Dyn. 2021, 250, 345–359. [Google Scholar] [CrossRef]
- Cotterill, S.L.; Jackson, G.C.; Leighton, M.P.; Wagener, R.; Mäkitie, O.; Cole, W.G.; Briggs, M.D. Multiple epiphyseal dysplasia mutations in MATN3 cause misfolding of the A-domain and prevent secretion of mutant matrilin-3. Hum. Mutat. 2005, 26, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Leighton, M.P.; Nundlall, S.; Starborg, T.; Meadows, R.S.; Suleman, F.; Knowles, L.; Wagener, R.; Thornton, D.J.; Kadler, K.E.; Boot-Handford, R.P.; et al. Decreased chondrocyte proliferation and dysregulated apoptosis in the cartilage growth plate are key features of a murine model of epiphyseal dysplasia caused by a matn3 mutation. Hum. Mol. Genet. 2007, 16, 1728–1741. [Google Scholar] [CrossRef]
- Nundlall, S.; Rajpar, M.H.; Bell, P.A.; Clowes, C.; Zeeff, L.A.; Gardner, B.; Thornton, D.J.; Boot-Handford, R.P.; Briggs, M.D. An unfolded protein response is the initial cellular response to the expression of mutant matrilin-3 in a mouse model of multiple epiphyseal dysplasia. Cell Stress Chaperones 2010, 15, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Hartley, C.L.; Edwards, S.; Mullan, L.; Bell, P.A.; Fresquet, M.; Boot-Handford, R.P.; Briggs, M.D. Armet/Manf and Creld2 are components of a specialized ER stress response provoked by inappropriate formation of disulphide bonds: Implications for genetic skeletal diseases. Hum. Mol. Genet. 2013, 22, 5262–5275. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Freddi, S.; Chan, D.; Cheah, K.S.; Bateman, J.F. Misfolding of collagen X chains harboring Schmid metaphyseal chondrodysplasia mutations results in aberrant disulfide bond formation, intracellular retention, and activation of the unfolded protein response. J. Biol. Chem. 2005, 280, 15544–15552. [Google Scholar] [CrossRef] [PubMed]
- Mullan, L.A.; Mularczyk, E.J.; Kung, L.H.; Forouhan, M.; Wragg, J.M.; Goodacre, R.; Bateman, J.F.; Swanton, E.; Briggs, M.D.; Boot-Handford, R.P. Increased intracellular proteolysis reduces disease severity in an ER stress-associated dwarfism. J. Clin. Invest. 2017, 127, 3861–3865. [Google Scholar] [CrossRef] [PubMed]
- Forouhan, M.; Sonntag, S.; Boot-Handford, R.P. Carbamazepine reduces disease severity in a mouse model of metaphyseal chondrodysplasia type Schmid caused by a premature stop codon (Y632X) in the Col10a1 gene. Hum. Mol. Genet. 2018, 27, 3840–3853. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.G.; Lai, L.Y.; Wen, X.J.; Zheng, T.; Cheung, C.W.; Zhou, S.Q.; Xu, S.Y. Neuroprotective effect of ginkgolide B on bupivacaine-induced apoptosis in SH-SY5Y cells. Oxid. Med. Cell Longev. 2013, 2013, 159864. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wang, S.; Fu, L.; Liu, D.; Zhu, Y.; Xu, A. Bilobalide protection of normal human melanocytes from hydrogen peroxide-induced oxidative damage via promotion of antioxidase expression and inhibition of endoplasmic reticulum stress. Clin. Exp. Dermatol. 2016, 41, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, B.; Huang, F.; Liu, B.; Xie, Y. Curcumin inhibits lipolysis via suppression of ER stress in adipose tissue and prevents hepatic insulin resistance. J. Lipid. Res. 2016, 57, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.L.; Li, J.L.; Xue, E.X.; Dou, H.C.; Lin, J.T.; Chen, K.; Wu, H.Q.; Wu, L.; Xuan, J.; Huang, Q.S. Vitexin alleviates ER-stress-activated apoptosis and the related inflammation in chondrocytes and inhibits the degeneration of cartilage in rats. Food Funct. 2018, 9, 5740–5749. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, J.; Zhang, P.; Si, A.; Zhang, Z.; Zhao, L.; Lv, F.; Zhao, G. Ginkgolide B ameliorates myocardial ischemia reperfusion injury in rats via inhibiting endoplasmic reticulum stress. Drug Des. Devel. Ther. 2019, 13, 767–774. [Google Scholar] [CrossRef]
- Guan, G.; Lei, L.; Lv, Q.; Gong, Y.; Yang, L. Curcumin attenuates palmitic acid-induced cell apoptosis by inhibiting endoplasmic reticulum stress in H9C2 cardiomyocytes. Hum. Exp. Toxicol. 2019, 38, 655–664. [Google Scholar] [CrossRef]
- Feng, K.; Ge, Y.; Chen, Z.; Li, X.; Liu, Z.; Li, X.; Li, H.; Tang, T.; Yang, F.; Wang, X. Curcumin Inhibits the PERK-eIF2α-CHOP Pathway through Promoting SIRT1 Expression in Oxidative Stress-induced Rat Chondrocytes and Ameliorates Osteoarthritis Progression in a Rat Model. Oxid. Med. Cell Longev. 2019, 2019, 8574386. [Google Scholar] [CrossRef] [PubMed]
- Malar, D.S.; Prasanth, M.I.; Jeyakumar, M.; Balamurugan, K.; Devi, K.P. Vitexin prevents Aβ proteotoxicity in transgenic Caenorhabditis elegans model of Alzheimer’s disease by modulating unfolded protein response. J. Biochem. Mol. Toxicol. 2021, 35, e22632. [Google Scholar] [CrossRef] [PubMed]
- Kou, J.; Wang, M.; Shi, J.; Zhang, H.; Pu, X.; Song, S.; Yang, C.; Yan, Y.; Döring, Y.; Xie, X.; et al. Curcumin Reduces Cognitive Deficits by Inhibiting Neuroinflammation through the Endoplasmic Reticulum Stress Pathway in Apolipoprotein E4 Transgenic Mice. ACS Omega 2021, 6, 6654–6662. [Google Scholar] [CrossRef]
- Jiang, Y.; Gong, Q.; Gong, Y.; Zhuo, C.; Huang, J.; Tang, Q. Vitexin Attenuates Non-alcoholic Fatty Liver Disease Lipid Accumulation in High Fat-Diet Fed Mice by Activating Autophagy and Reducing Endoplasmic Reticulum Stress in Liver. Biol. Pharm. Bull. 2022, 45, 260–267. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Weng, Y.; Wang, D.; Wang, R.; Wang, H.; Wang, L.; Shen, S.; Wang, H.; Li, Y.; et al. Curcumin Inhibits Hyperandrogen-Induced IRE1α-XBP1 Pathway Activation by Activating the PI3K/AKT Signaling in Ovarian Granulosa Cells of PCOS Model Rats. Oxid. Med. Cell Longev. 2022, 2022, 2113293. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shi, Q.; Xu, S.; Du, C.; Liang, L.; Wu, K.; Wang, K.; Wang, X.; Chang, L.S.; He, D.; et al. Curcumin promotes KLF5 proteasome degradation through downregulating YAP/TAZ in bladder cancer cells. Int. J. Mol. Sci. 2014, 15, 15173–15187. [Google Scholar] [CrossRef]
- Ali, A.; Banerjea, A.C. Curcumin inhibits HIV-1 by promoting Tat protein degradation. Sci. Rep. 2016, 6, 27539. [Google Scholar] [CrossRef]
- Piróg, K.A.; Dennis, E.P.; Hartley, C.L.; Jackson, R.M.; Soul, J.; Schwartz, J.M.; Bateman, J.F.; Boot-Handford, R.P.; Briggs, M.D. XBP1 signalling is essential for alleviating mutant protein aggregation in ER-stress related skeletal disease. PLoS Genet. 2019, 15, e1008215. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Kang, T.I.; So, J.S. Roles of XBP1s in Transcriptional Regulation of Target Genes. Biomedicines 2021, 9, 791. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.; Calanca, V.; Galli, C.; Lucca, P.; Paganetti, P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 2003, 299, 1397–1400. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Hosokawa, N.; Wada, I.; Nagata, K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 2003, 299, 1394–1397. [Google Scholar] [CrossRef]
- Olivari, S.; Cali, T.; Salo, K.E.; Paganetti, P.; Ruddock, L.W.; Molinari, M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun 2006, 349, 1278–1284. [Google Scholar] [CrossRef]
- Olivari, S.; Molinari, M. Glycoprotein folding and the role of EDEM1, EDEM2 and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 2007, 581, 3658–3664. [Google Scholar] [CrossRef]
- Lilley, B.N.; Ploegh, H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 2004, 429, 834–840. [Google Scholar] [CrossRef]
- Ye, Y.; Shibata, Y.; Yun, C.; Ron, D.; Rapoport, T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 2004, 429, 841–847. [Google Scholar] [CrossRef]
- Oda, Y.; Okada, T.; Yoshida, H.; Kaufman, R.J.; Nagata, K.; Mori, K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006, 172, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Van Erk, M.J.; Teuling, E.; Staal, Y.C.; Huybers, S.; Van Bladeren, P.J.; Aarts, J.M.; Van Ommen, B. Time- and dose-dependent effects of curcumin on gene expression in human colon cancer cells. J. Carcinog. 2004, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Besio, R.; Iula, G.; Garibaldi, N.; Cipolla, L.; Sabbioneda, S.; Biggiogera, M.; Marini, J.C.; Rossi, A.; Forlino, A. 4-PBA ameliorates cellular homeostasis in fibroblasts from osteogenesis imperfecta patients by enhancing autophagy and stimulating protein secretion. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1642–1652. [Google Scholar] [CrossRef]
- Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef] [PubMed]
- Bell, P.A.; Wagener, R.; Zaucke, F.; Koch, M.; Selley, J.; Warwood, S.; Knight, D.; Boot-Handford, R.P.; Thornton, D.J.; Briggs, M.D. Analysis of the cartilage proteome from three different mouse models of genetic skeletal diseases reveals common and discrete disease signatures. Biol. Open 2013, 2, 802–811. [Google Scholar] [CrossRef]
- Helms, K.M.; Wilson, R.C.; Ogungbe, I.V.; Setzer, W.N.; Twigg, P.D. Vitexin inhibits polyubiquitin synthesis by the ubiquitin-conjugating enzyme E2-25K. Nat. Prod. Commun. 2011, 6, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Wada, I.; Hasegawa, K.; Yorihuzi, T.; Tremblay, L.O.; Herscovics, A.; Nagata, K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001, 2, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Ninagawa, S.; Okada, T.; Sumitomo, Y.; Kamiya, Y.; Kato, K.; Horimoto, S.; Ishikawa, T.; Takeda, S.; Sakuma, T.; Yamamoto, T.; et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 2014, 206, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Hebert, D.N.; Foellmer, B.; Helenius, A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell 1995, 81, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Wagener, R.; Kobbe, B.; Paulsson, M. Primary structure of matrilin-3, a new member of a family of extracellular matrix proteins related to cartilage matrix protein (matrilin-1) and von Willebrand factor. FEBS Lett. 1997, 413, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Jana, N.R.; Dikshit, P.; Goswami, A.; Nukina, N. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J. Biol. Chem. 2004, 279, 11680–11685. [Google Scholar] [CrossRef] [PubMed]
- Dikshit, P.; Goswami, A.; Mishra, A.; Chatterjee, M.; Jana, N.R. Curcumin induces stress response, neurite outgrowth and prevent NF-kappaB activation by inhibiting the proteasome function. Neurotox. Res. 2006, 9, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Milacic, V.; Banerjee, S.; Landis-Piwowar, K.R.; Sarkar, F.H.; Majumdar, A.P.; Dou, Q.P. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008, 68, 7283–7292. [Google Scholar] [CrossRef]
- Hasima, N.; Aggarwal, B.B. Targeting proteasomal pathways by dietary curcumin for cancer prevention and treatment. Curr. Med. Chem. 2014, 21, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Kunnumakkara, A.B.; Bordoloi, D.; Padmavathi, G.; Monisha, J.; Roy, N.K.; Prasad, S.; Aggarwal, B.B. Curcumin, the golden nutraceutical: Multitargeting for multiple chronic diseases. Br. J. Pharmacol. 2017, 174, 1325–1348. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Wang, P.; Ye, Y.; Yuan, W.; Tan, Y.; Zhang, S.; Meng, Q. Curcumin exerts a protective effect on murine knee chondrocytes treated with IL-1β through blocking the NF-κB/HIF-2α signaling pathway. Ann. Transl. Med. 2021, 9, 940. [Google Scholar] [CrossRef]
- Dennis, E.P.; Edwards, S.M.; Jackson, R.M.; Hartley, C.L.; Tsompani, D.; Capulli, M.; Teti, A.; Boot-Handford, R.P.; Young, D.A.; Piróg, K.A.; et al. CRELD2 Is a Novel LRP1 Chaperone That Regulates Noncanonical WNT Signaling in Skeletal Development. J. Bone Miner. Res. 2020, 35, 1452–1469. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dennis, E.P.; Watson, R.N.; McPate, F.; Briggs, M.D. Curcumin Reduces Pathological Endoplasmic Reticulum Stress through Increasing Proteolysis of Mutant Matrilin-3. Int. J. Mol. Sci. 2023, 24, 1496. https://doi.org/10.3390/ijms24021496
Dennis EP, Watson RN, McPate F, Briggs MD. Curcumin Reduces Pathological Endoplasmic Reticulum Stress through Increasing Proteolysis of Mutant Matrilin-3. International Journal of Molecular Sciences. 2023; 24(2):1496. https://doi.org/10.3390/ijms24021496
Chicago/Turabian StyleDennis, Ella P., Robyn N. Watson, Florence McPate, and Michael D. Briggs. 2023. "Curcumin Reduces Pathological Endoplasmic Reticulum Stress through Increasing Proteolysis of Mutant Matrilin-3" International Journal of Molecular Sciences 24, no. 2: 1496. https://doi.org/10.3390/ijms24021496
APA StyleDennis, E. P., Watson, R. N., McPate, F., & Briggs, M. D. (2023). Curcumin Reduces Pathological Endoplasmic Reticulum Stress through Increasing Proteolysis of Mutant Matrilin-3. International Journal of Molecular Sciences, 24(2), 1496. https://doi.org/10.3390/ijms24021496



