1,2-Dibenzoylhydrazine as a Multi-Inhibitor Compound: A Morphological and Docking Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Modeling Studies
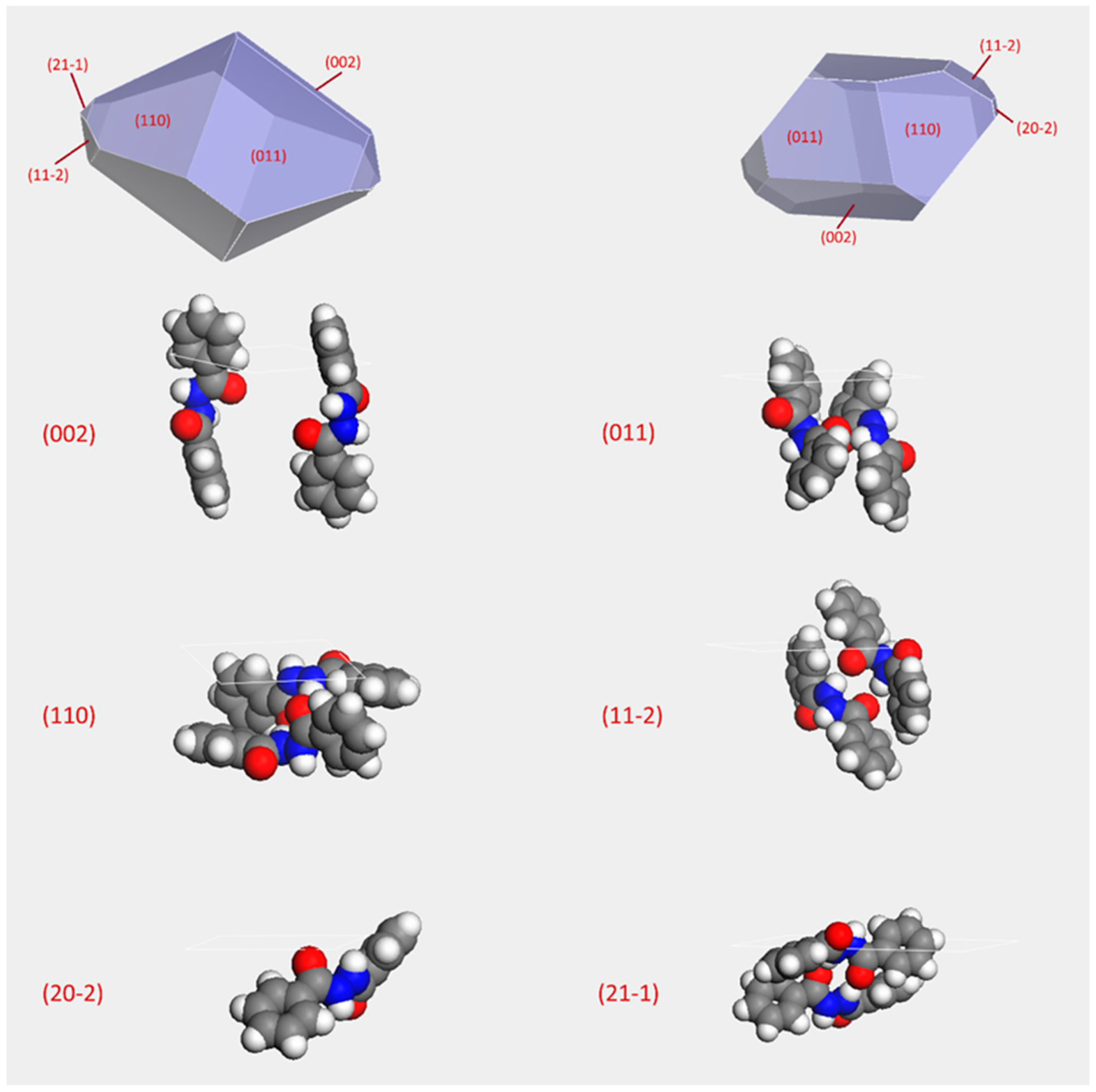
2.2. Crystal Morphology Prediction
3. Materials and Methods
3.1. Structure Preparation and Minimization
3.2. Docking and Molecular Dynamics Studies
3.3. Crystal Morphology Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medina-Franco, J.L.; Giulianotti, M.A.; Welmaker, G.S.; Houghten, R.A. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov. Today 2013, 18, 495–501. [Google Scholar] [CrossRef]
- Gentile, D.; Patamia, V.; Fuochi, V.; Furneri, P.M.; Rescifina, A. Natural Substances in the Fight of SARS-CoV-2: A Critical Evaluation Resulting from the Cross-Fertilization of Molecular Modeling Data with the Pharmacological Aspects. Curr. Med. Chem. 2021, 28, 8333–8383. [Google Scholar] [CrossRef]
- Floresta, G.; Zagni, C.; Gentile, D.; Patamia, V.; Rescifina, A. Artificial Intelligence Technologies for COVID-19 De Novo Drug Design. Int. J. Mol. Sci. 2022, 23, 3261. [Google Scholar] [CrossRef]
- Floresta, G.; Amata, E.; Gentile, D.; Romeo, G.; Marrazzo, A.; Pittalà, V.; Salerno, L.; Rescifina, A. Fourfold Filtered Statistical/Computational Approach for the Identification of Imidazole Compounds as HO-1 Inhibitors from Natural Products. Mar. Drugs 2019, 17, 113. [Google Scholar] [CrossRef] [PubMed]
- Medina-Franco, J.L.; Yongye, A.B.; Pérez-Villanueva, J.; Houghten, R.A.; Martínez-Mayorga, K. Multitarget Structure–Activity Relationships Characterized by Activity-Difference Maps and Consensus Similarity Measure. J. Chem. Inf. Model. 2011, 51, 2427–2439. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Patamia, V.; Gentile, D.; Molteni, F.; Santamato, A.; Rescifina, A.; Vecchio, M. Repurposing of FDA-Approved Drugs for Treating Iatrogenic Botulism: A Paired 3D-QSAR/Docking Approach(dagger). Chemmedchem 2020, 15, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Gentile, D.; Perrini, G.; Patamia, V.; Rescifina, A. Computational Tools in the Discovery of FABP4 Ligands: A Statistical and Molecular Modeling Approach. Mar. Drugs 2019, 17, 624. [Google Scholar] [CrossRef]
- Gentile, D.; Floresta, G.; Patamia, V.; Chiaramonte, R.; Mauro, G.L.; Rescifina, A.; Vecchio, M. An Integrated Pharmacophore/Docking/3D-QSAR Approach to Screening a Large Library of Products in Search of Future Botulinum Neurotoxin A Inhibitors. Int. J. Mol. Sci. 2020, 21, 9470. [Google Scholar] [CrossRef] [PubMed]
- Varrica, M.G.; Zagni, C.; Mineo, P.G.; Floresta, G.; Monciino, G.; Pistarà, V.; Abbadessa, A.; Nicosia, A.; Castilho, R.M.; Amata, E.; et al. DNA intercalators based on (1,10-phenanthrolin-2-yl)isoxazolidin-5-yl core with better growth inhibition and selectivity than cisplatin upon head and neck squamous cells carcinoma. Eur. J. Med. Chem. 2018, 143, 583–590. [Google Scholar] [CrossRef]
- Rescifina, A.; Zagni, C.; Mineo, P.G.; Giofrè, S.V.; Chiacchio, U.; Tommasone, S.; Talotta, C.; Gaeta, C.; Neri, P. DNA Recognition with Polycyclic-Aromatic-Hydrocarbon-Presenting Calixarene Conjugates. Eur. J. Org. Chem. 2014, 2014, 7605–7613. [Google Scholar] [CrossRef]
- Gentile, D.; Coco, A.; Patamia, V.; Zagni, C.; Floresta, G.; Rescifina, A. Targeting the SARS-CoV-2 HR1 with Small Molecules as Inhibitors of the Fusion Process. Int. J. Mol. Sci. 2022, 23, 10067. [Google Scholar] [CrossRef]
- Zagni, C.; Pistarà, V.; Oliveira, L.A.; Castilho, R.M.; Romeo, G.; Chiacchio, U.; Rescifina, A. Serendipitous discovery of potent human head and neck squamous cell carcinoma anti-cancer molecules: A fortunate failure of a rational molecular design. Eur. J. Med. Chem. 2017, 141, 188–196. [Google Scholar] [CrossRef]
- Floresta, G.; Cilibrizzi, A.; Abbate, V.; Spampinato, A.; Zagni, C.; Rescifina, A. FABP4 inhibitors 3D-QSAR model and isosteric replacement of BMS309403 datasets. Data Brief 2019, 22, 471–483. [Google Scholar] [CrossRef]
- Müller, G. Medicinal chemistry of target family-directed masterkeys. Drug Discov. Today 2003, 8, 681–691. [Google Scholar] [CrossRef]
- Chen, M.; Chen, X.; Huang, G.; Jiang, Y.; Gou, Y.; Deng, J. Synthesis, anti-tumour activity, and mechanism of benzoyl hydrazine Schiff base-copper complexes. J. Mol. Struct. 2022, 1268, 133730. [Google Scholar] [CrossRef]
- Liu, X.; Cooper, A.M.; Yu, Z.; Silver, K.; Zhang, J.; Zhu, K.Y. Progress and prospects of arthropod chitin pathways and structures as targets for pest management. Pestic. Biochem. Physiol. 2019, 161, 33–46. [Google Scholar] [CrossRef]
- Graham, L.; Johnson, W.; Pawlak-Skrzecz, A.; Eaton, R.; Bliese, M.; Howell, L.; Hannan, G.; Hill, R. Ligand binding by recombinant domains from insect ecdysone receptors. Insect Biochem. Mol. Biol. 2007, 37, 611–626. [Google Scholar] [CrossRef]
- Morou, E.; Lirakis, M.; Pavlidi, N.; Zotti, M.; Nakagawa, Y.; Smagghe, G.; Vontas, J.; Swevers, L. A new dibenzoylhydrazine with insecticidal activity against Anopheles mosquito larvae. Pest Manag. Sci. 2013, 69, 827–833. [Google Scholar] [CrossRef]
- Bordas, B.; Belai, I.; Lopata, A.; Szanto, Z. Interpretation of Scoring Functions Using 3D Molecular Fields. Mapping the Diacyl-Hydrazine-Binding Pocket of an Insect Ecdysone Receptor. J. Chem. Inf. Model. 2006, 47, 176–185. [Google Scholar] [CrossRef]
- Ullah, H.; Uddin, I.; Misbah; Khan, F.; Taha, M.; Rahim, F.; Sarfraz, M.; Shams, S.; Nabi, M.; Wadood, A. Synthesis of substituted benzohydrazide derivatives: In vitro urease activities and their molecular docking studies. Chem. Data Collect. 2021, 36, 100778. [Google Scholar] [CrossRef]
- Kafarski, P.; Talma, M. Recent advances in design of new urease inhibitors: A review. J. Adv. Res. 2018, 13, 101–112. [Google Scholar] [CrossRef]
- Abbas, A.; Ali, B.; Kanwal; Khan, K.M.; Iqbal, J.; Rahman, S.U.; Zaib, S.; Perveen, S. Synthesis and in vitro urease inhibitory activity of benzohydrazide derivatives, in silico and kinetic studies. Bioorg. Chem. 2019, 82, 163–177. [Google Scholar] [CrossRef]
- Zhao, H.; Neamati, N.; Sunder, S.; Hong, H.; Wang, S.; Milne, G.W.A.; Pommier, Y.; Burke, T.R. Hydrazide-Containing Inhibitors of HIV-1 Integrase. J. Med. Chem. 1997, 40, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Safakish, M.; Hajimahdi, Z.; Zabihollahi, R.; Aghasadeghi, M.R.; Vahabpour, R.; Zarghi, A. Design, synthesis, and docking studies of new 2-benzoxazolinone derivatives as anti-HIV-1 agents. Med. Chem. Res. 2017, 26, 2718–2726. [Google Scholar] [CrossRef]
- Hassounah, S.A.; Mesplède, T.; Wainberg, M.A. Nonhuman Primates and Humanized Mice for Studies of HIV-1 Integrase Inhibitors: A Review. Pathog. Immun. 2016, 1, 41–67. [Google Scholar] [CrossRef]
- Li, B.-W.; Zhang, F.-H.; Serrao, E.; Chen, H.; Sanchez, T.W.; Yang, L.-M.; Neamati, N.; Zheng, Y.-T.; Wang, H.; Long, Y.-Q. Design and discovery of flavonoid-based HIV-1 integrase inhibitors targeting both the active site and the interaction with LEDGF/p75. Bioorg. Med. Chem. 2014, 22, 3146–3158. [Google Scholar] [CrossRef]
- Mohamed, N.A.; Abd El-Ghany, N.A.A.; Fahmy, M.M.; Khalaf-Alla, P.A. Novel polymaleimide containing dibenzoyl hydrazine pendant group as chelating agent for antimicrobial activity. Int. J. Polym. Mater. Polym. Biomater. 2018, 67, 68–77. [Google Scholar] [CrossRef]
- Desideri, I.; Martinelli, C.; Ciuti, S.; Barretta, G.U.; Balzano, F. Lopinavir/ritonavir, a new galenic oral formulation from commercial solid form, fine-tuned by nuclear magnetic resonance spectroscopy. Eur. J. Hosp. Pharm. 2022, 29, 259–263. [Google Scholar] [CrossRef]
- Taylor, L.S.; Braun, D.E.; Steed, J.W. Crystals and Crystallization in Drug Delivery Design. Cryst. Growth Des. 2021, 21, 1375–1377. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Cheeseright, T.; Mackey, M.; Rose, S.; Vinter, A. Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 2006, 46, 665–676. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; Tan, J.-G.; Xu, H.-H. Design and synthesis of N-alkyl-N′-substituted 2,4-dioxo-3,4-dihydropyrimidin-1-diacylhydrazine derivatives as ecdysone receptor agonist. Bioorg. Med. Chem. 2013, 21, 4687–4697. [Google Scholar] [CrossRef]
- Dinan, L. Ecdysteroid structure-activity relationships. Stud. Nat. Prod. Chem. 2003, 29, 3–71. [Google Scholar]
- Seraj, F.; Khan, K.M.; Khan, A.; Ali, M.; Khalil, R.; Ul-Haq, Z.; Hameed, S.; Taha, M.; Salar, U.; Perveen, S. Biology-oriented drug synthesis (BIODS), in vitro urease inhibitory activity, and in silico studies on ibuprofen derivatives. Mol. Divers. 2021, 25, 143–157. [Google Scholar] [CrossRef]
- Channar, P.A.; Saeed, A.; Afzal, S.; Hussain, D.; Kalesse, M.; Shehzadi, S.A.; Iqbal, J. Hydrazine clubbed 1,3-thiazoles as potent urease inhibitors: Design, synthesis and molecular docking studies. Mol. Divers. 2021, 25, 1–13. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Khan, A.; Shah, S.A.A.; Anwar, A.; Halim, S.A.; Fatmi, M.Q.; Imran, S.; Rahim, F.; Khan, K.M. Synthesis of novel derivatives of oxindole, their urease inhibition and molecular docking studies. Bioorg. Med. Chem. Lett. 2015, 25, 3285–3289. [Google Scholar] [CrossRef]
- Naseem, S.; Ashraf, M.; Khan, S.; Rafiq, M.; Kashif, M.; Rahman, J.; Rauf, M.K.; Halim, S.A.; Uddin, J.; Khan, A.; et al. Exploring biologically active hybrid pharmacophore N-substituted hydrazine-carbothioamides for urease inhibition: In vitro and in silico approach. Int. J. Biol. Macromol. 2021, 182, 534–544. [Google Scholar] [CrossRef]
- Bénard, C.; Zouhiri, F.; Normand-Bayle, M.; Danet, M.; Desmaële, D.; Leh, H.; Mouscadet, J.-F.; Mbemba, G.; Thomas, C.-M.; Bonnenfant, S. Linker-modified quinoline derivatives targeting HIV-1 integrase: Synthesis and biological activity. Bioorg. Med. Chem. Lett. 2004, 14, 2473–2476. [Google Scholar] [CrossRef] [PubMed]
- Jesumoroti, O.J.; Faridoon, F.; Mnkandhla, D.; Isaacs, M.; Hoppe, H.C.; Klein, R. Evaluation of novel N′-(3-hydroxybenzoyl)-2-oxo-2H-chromene-3-carbohydrazide derivatives as potential HIV-1 integrase inhibitors. MedChemComm 2019, 10, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Neamati, N.; Lin, Z.; Karki, R.G.; Orr, A.; Cowansage, K.; Strumberg, D.; Pais, G.C.G.; Voigt, J.H.; Nicklaus, M.C.; Winslow, H.E.; et al. Metal-Dependent Inhibition of HIV-1 Integrase. J. Med. Chem. 2002, 45, 5661–5670. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Bhardwaj, V.K.; Das, P.; Purohit, R. New ecdysone receptor agonists: A computational approach for rational discovery of insecticides for crop protection. Mol. Syst. Des. Eng. 2021, 6, 936–945. [Google Scholar] [CrossRef]
- Khan, K.M.; Naz, F.; Taha, M.; Khan, A.; Perveen, S.; Choudhary, M.; Voelter, W. Synthesis and in vitro urease inhibitory activity of N,N′-disubstituted thioureas. Eur. J. Med. Chem. 2014, 74, 314–323. [Google Scholar] [CrossRef]
- Kanwal; Khan, M.; Arshia; Khan, K.M.; Parveen, S.; Shaikh, M.; Fatima, N.; Choudhary, M.I. Syntheses, in vitro urease inhibitory activities of urea and thiourea derivatives of tryptamine, their molecular docking and cytotoxic studies. Bioorg. Chem. 2019, 83, 595–610. [Google Scholar]
- Tsiang, M.; Jones, G.S.; Niedziela-Majka, A.; Kan, E.; Lansdon, E.B.; Huang, W.; Hung, M.; Samuel, D.; Novikov, N.; Xu, Y.; et al. New Class of HIV-1 Integrase (IN) Inhibitors with a Dual Mode of Action. J. Biol. Chem. 2012, 287, 21189–21203. [Google Scholar] [CrossRef]
- Fader, L.D.; Malenfant, E.; Parisien, M.; Carson, R.; Bilodeau, F.; Landry, S.; Pesant, M.; Brochu, C.; Morin, S.; Chabot, C.; et al. Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1. ACS Med. Chem. Lett. 2014, 5, 422–427. [Google Scholar] [CrossRef]
- Pisarek, J.; Malinska, M. Structure and Morphology of Indole Analogue Crystals. ACS Omega 2020, 5, 17141–17151. [Google Scholar] [CrossRef]
- Benz, K.-W. Handbook of Industrial Crystallization, 3rd ed.; Cambridge University Press: Cambridge, UK, 2020; Volume 53, pp. 861–862. [Google Scholar]
- Erdemir, D.; Lee, A.Y.; Myerson, A.S. Nucleation of Crystals from Solution: Classical and Two-Step Models. Acc. Chem. Res. 2009, 42, 621–629. [Google Scholar] [CrossRef]
- Kitajgorodskij, A.I. Molecular Crystals and Molecules [Molekuljarnye Kristally, Engl.]; Elsevier: Amsterdam, The Netherlands, 1973. [Google Scholar]
- Reichardt, C.; Welton, T. Appendix A. Properties, Purification, and Use of Organic Solvents; Wiley-VCH: Weinheim, Germany, 2010; Volume 549. [Google Scholar]
- Civati, F.; O’Malley, C.; Erxleben, A.; McArdle, P. Factors Controlling Persistent Needle Crystal Growth: The Importance of Dominant One-Dimensional Secondary Bonding, Stacked Structures, and van der Waals Contact. Cryst. Growth Des. 2021, 21, 3449–3460. [Google Scholar] [CrossRef]
- Piana, S.; Gale, J.D. Understanding the Barriers to Crystal Growth: Dynamical Simulation of the Dissolution and Growth of Urea from Aqueous Solution. J. Am. Chem. Soc. 2005, 127, 1975–1982. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Krieger, E.; Dunbrack, R.L.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. In Computational Drug Discovery and Design; Springer: Berlin/Heidelberg, Germany, 2012; pp. 405–421. [Google Scholar]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006, 25, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Szczepańska, K.; Podlewska, S.; Dichiara, M.; Gentile, D.; Patamia, V.; Rosier, N.; Mönnich, D.; Cantero, M.C.R.; Karcz, T.; Łażewska, D.; et al. Structural and Molecular Insight into Piperazine and Piperidine Derivatives as Histamine H3 and Sigma-1 Receptor Antagonists with Promising Antinociceptive Properties. ACS Chem. Neurosci. 2021, 13, 1–15. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comp. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Galimberti, M.; Barbera, V.; Guerra, S.; Bernardi, A. Facile functionalization of sp2 carbon allotropes with a biobased Janus molecule. Rubber Chem. Technol. 2017, 90, 285–307. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jin, Z.; Yang, C.; Akkermans, R.L.C.; Robertson, S.H.; Spenley, N.A.; Miller, S.; Todd, S.M. COMPASS II: Extended coverage for polymer and drug-like molecule databases. J. Mol. Model. 2016, 22, 47. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. A new model for calculating atomic charges in molecules. Tetrahedron Lett. 1978, 19, 3181–3184. [Google Scholar] [CrossRef]
- Docherty, R.; Clydesdale, G.; Roberts, K.J.; Bennema, P. Application of Bravais-Friedel-Donnay-Harker, Attachment Energy and Ising-Models to Predicting and Understanding the Morphology of Molecular-Crystals. J. Phys. D-App. Phys. 1991, 24, 89–99. [Google Scholar] [CrossRef]
- Hartman, P.; Bennema, P. The attachment energy as a habit controlling factor: I. Theoretical considerations. J. Cryst. Growth 1980, 49, 145–156. [Google Scholar] [CrossRef]
- Bennema, P.; Meekes, H.; Boerrigter, S.; Cuppen, H.; Deij, M.; Van Eupen, J.; Verwer, P.; Vlieg, E. Crystal growth and morphology: New developments in an integrated Hartman−Perdok connected net roughening transition theory, supported by computer simulations. Cryst. Growth Des. 2004, 4, 905–913. [Google Scholar] [CrossRef]
- Hartman, P.; Perdok, W.G. On the relations between structure and morphology of crystals. I. Acta Crystallogr. 1955, 8, 49–52. [Google Scholar] [CrossRef]
- Rohl, A.L. Computer prediction of crystal morphology. Curr. Opin. Solid State Mater. Sci. 2003, 7, 21–26. [Google Scholar] [CrossRef]
- Donnay, J.D.H.; Harkei, D. A new law of crystal morphology extending the law of Bravais. Am. Miner. 1937, 22, 446. [Google Scholar]
- Berkovitch-Yellin, Z. Toward an ab initio derivation of crystal morphology. J. Am. Chem. Soc. 1985, 107, 8239–8253. [Google Scholar] [CrossRef]
- Punzo, F. Unveiling the role of molecular interactions in crystal morphology prediction. J. Mol. Struct. 2013, 1032, 147–154. [Google Scholar] [CrossRef]
- Destri, G.L.; Marrazzo, A.; Rescifina, A.; Punzo, F. How molecular interactions affect crystal morphology: The case of haloperidol. J. Pharm. Sci. 2011, 100, 4896–4906. [Google Scholar] [CrossRef]
- Destri, G.L.; Marrazzo, A.; Rescifina, A.; Punzo, F. Crystal Morphologies and Polymorphs in Tolbutamide Microcrystalline Powder. J. Pharm. Sci. 2013, 102, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, G.M.; Rescifina, A.; Chiacchio, U.; Bacchi, A.; Punzo, F. A top–down approach to crystal engineering of a racemic Δ2-isoxazoline. Acta Cryst. B Struct. Sci. Cryst. Eng. Mat. 2014, 70, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Punzo, F. Space Groups Complexity versus Molecular Interactions in Quinoline Derivatives Crystal Morphology Prediction: A Throughput Evaluation of Different in Silico Approaches. Cryst. Growth Des. 2011, 11, 3512–3521. [Google Scholar] [CrossRef]
- Van Der Sluis, P.; Spek, A.L. BYPASS: An effective method for the refinement of crystal structures containing disordered solvent regions. Acta Crystallogr. Sect. A Found. Crystallogr. 1990, 46, 194–201. [Google Scholar] [CrossRef]
- Doelker, E.; Massuelle, D.; Veuillez, F.; Humbert-Droz, P. Morphological, Packing, Flow and Tableting Properties of New Avicel Types. Drug Dev. Ind. Pharm. 1995, 21, 643–661. [Google Scholar] [CrossRef]
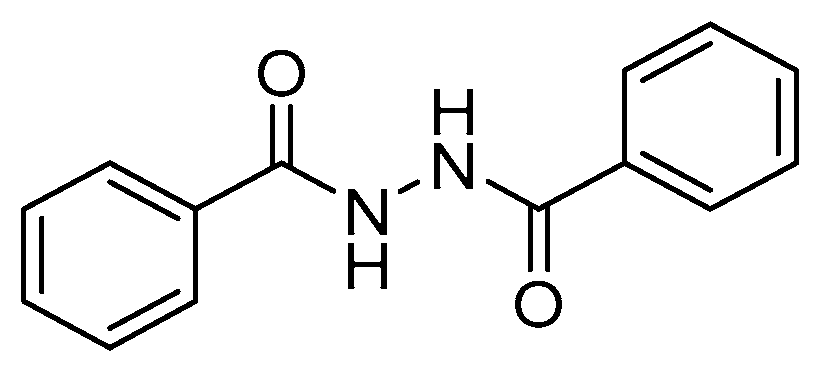
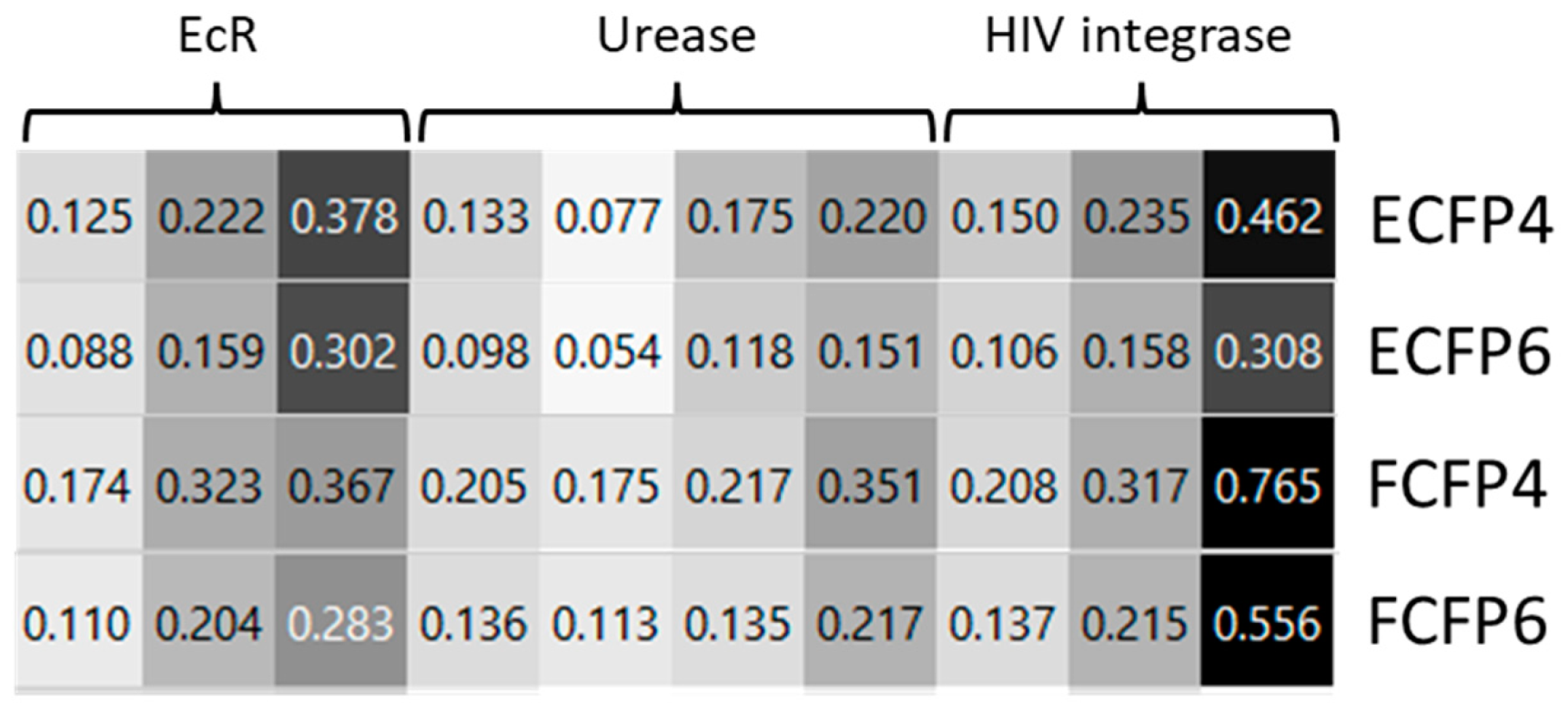
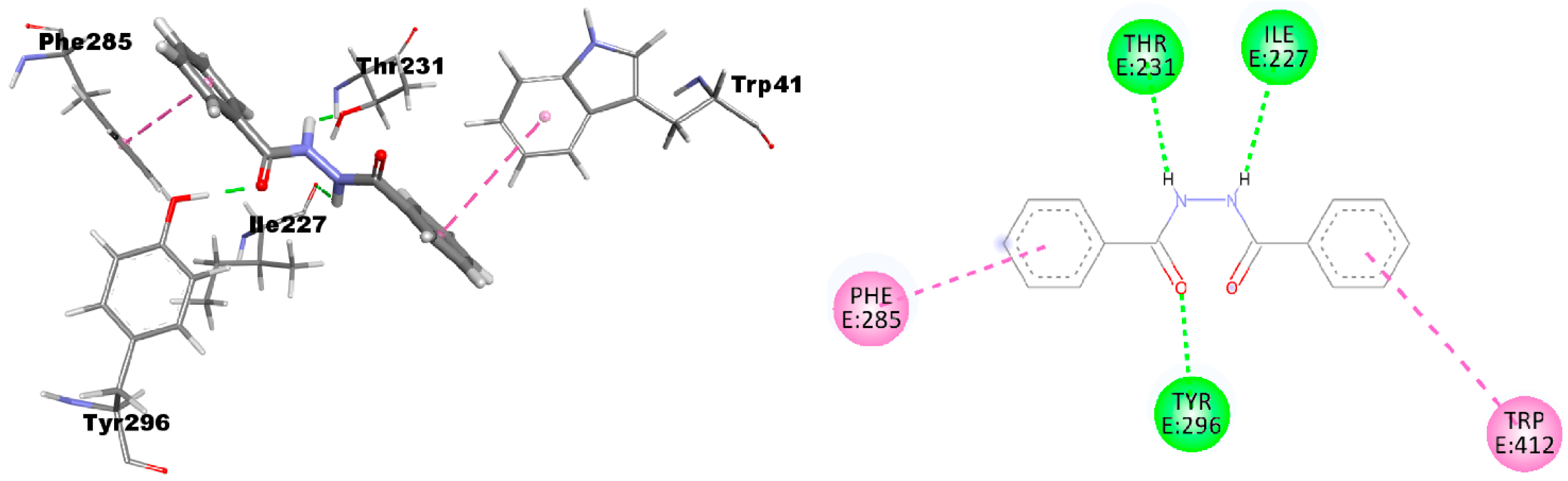
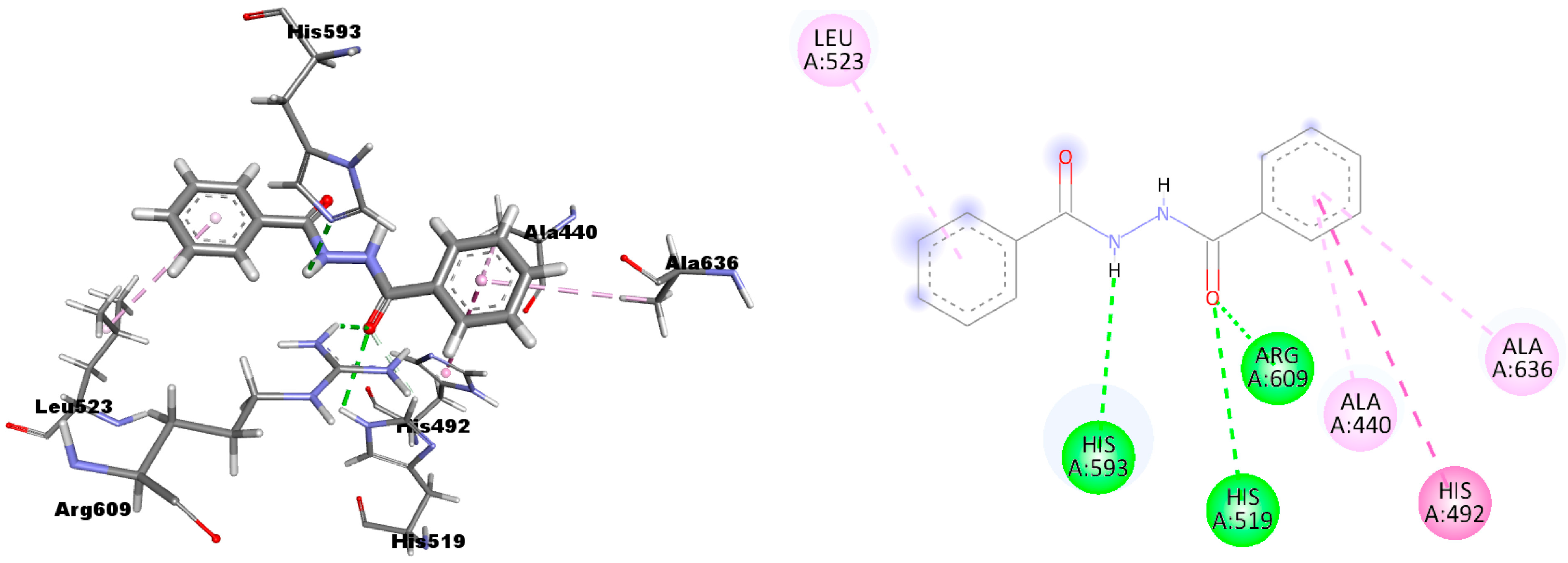

| Compound | Predicted ΔG | Predicted Ki | Experimental Ki |
|---|---|---|---|
| Ponasterone A a | −11.37 | 4.6 | 4.8 b |
| DBH | −9.01 | 249.9 | — |
| Compound | Predicted ΔG | Predicted Ki | Experimental Ki |
|---|---|---|---|
| Thiourea a | −5.55 | 85.06 | 21.00 b |
| DBH | −7.06 | 6.71 | — |
| Compound | Predicted ΔG | Predicted Ki | Experimental Ki |
|---|---|---|---|
| CHEMBL3259898 a | −8.62 | 476.8 | 49.00 b |
| DBH | −7.10 | 6,246.0 | — |
| MI | BDFH | |||
|---|---|---|---|---|
| h k l | Multiplicity | dhkl (Å) | % of Total Facet Area a | |
| 1 1 0 | 4 | 12.641 | 40.749 | |
| 2 0 0 | 2 | 14.724 | 15.253 | |
| 1 1 -1 | 4 | 15.437 | 36.027 | |
| 1 1 1 | 4 | 19.059 | 7.462 | |
| 0 0 2 | 2 | 23.746 | 0.509 | |
| MI | GM | |||
| h k l | Multiplicity | dhkl (Å) | Eatt (kcal mol−1) | % of Total Facet Area a |
| 1 1 0 | 4 | 7.911 | −56.874 | 50.624 |
| 1 1 -1 | 4 | 6.478 | −60.875 | 41.367 |
| 1 1 -2 | 4 | 4.087 | −89.625 | 0.241 |
| 1 1 1 | 4 | 5.247 | −89.670 | 6.083 |
| 2 0 0 | 2 | 6.792 | −93.379 | 1.686 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patamia, V.; Floresta, G.; Zagni, C.; Pistarà, V.; Punzo, F.; Rescifina, A. 1,2-Dibenzoylhydrazine as a Multi-Inhibitor Compound: A Morphological and Docking Study. Int. J. Mol. Sci. 2023, 24, 1425. https://doi.org/10.3390/ijms24021425
Patamia V, Floresta G, Zagni C, Pistarà V, Punzo F, Rescifina A. 1,2-Dibenzoylhydrazine as a Multi-Inhibitor Compound: A Morphological and Docking Study. International Journal of Molecular Sciences. 2023; 24(2):1425. https://doi.org/10.3390/ijms24021425
Chicago/Turabian StylePatamia, Vincenzo, Giuseppe Floresta, Chiara Zagni, Venerando Pistarà, Francesco Punzo, and Antonio Rescifina. 2023. "1,2-Dibenzoylhydrazine as a Multi-Inhibitor Compound: A Morphological and Docking Study" International Journal of Molecular Sciences 24, no. 2: 1425. https://doi.org/10.3390/ijms24021425
APA StylePatamia, V., Floresta, G., Zagni, C., Pistarà, V., Punzo, F., & Rescifina, A. (2023). 1,2-Dibenzoylhydrazine as a Multi-Inhibitor Compound: A Morphological and Docking Study. International Journal of Molecular Sciences, 24(2), 1425. https://doi.org/10.3390/ijms24021425











