Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy
Abstract
1. Introduction
2. Classification of VPEs
3. Functions of VPEs
4. VPEs Are Executors of Plant PCD
5. Role of VPEs in PCD under Biotic Stress
6. Role of VPEs in PCD Induced by Abiotic Stress
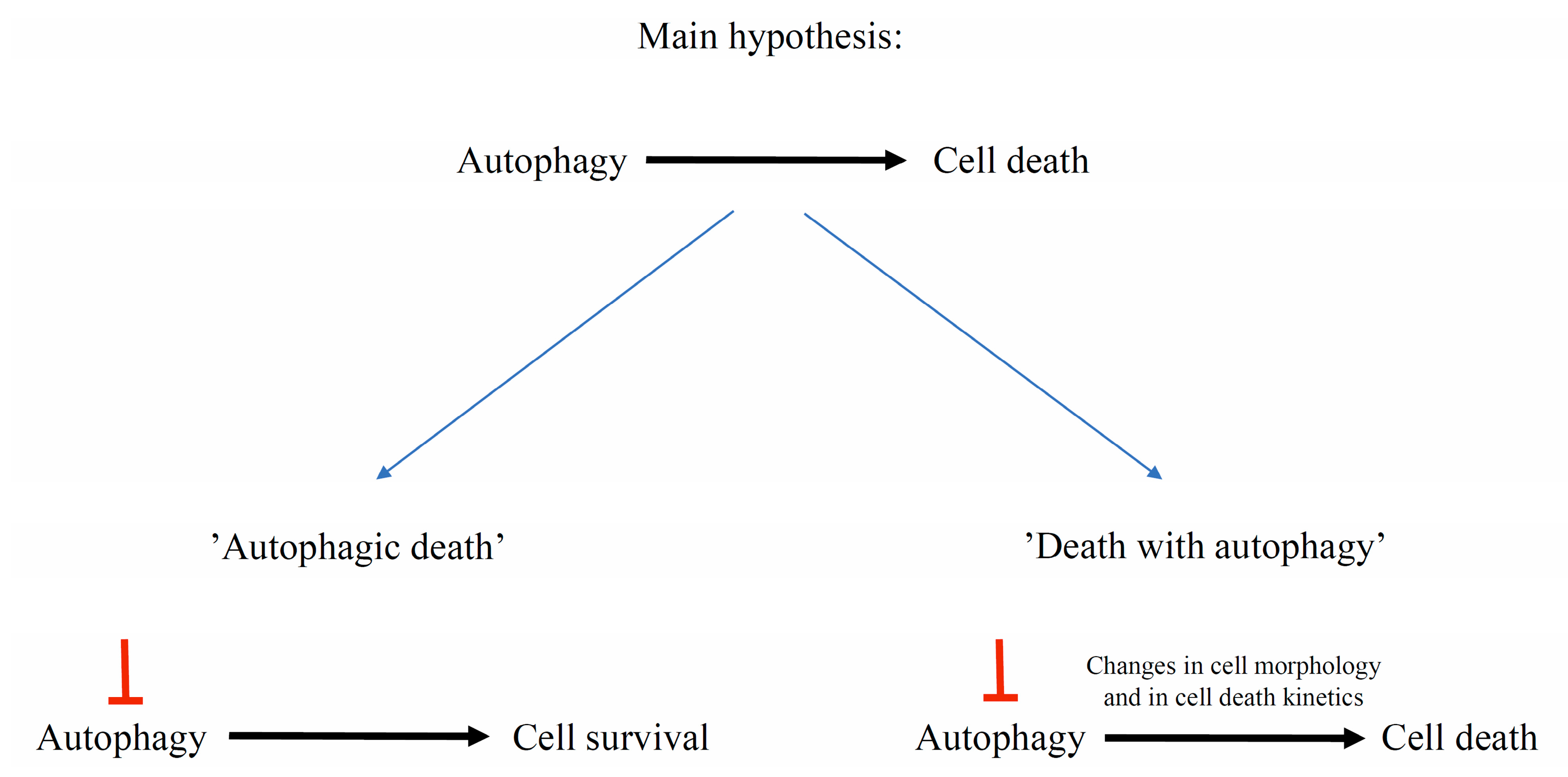
7. Autophagy Contribution to Cell Death
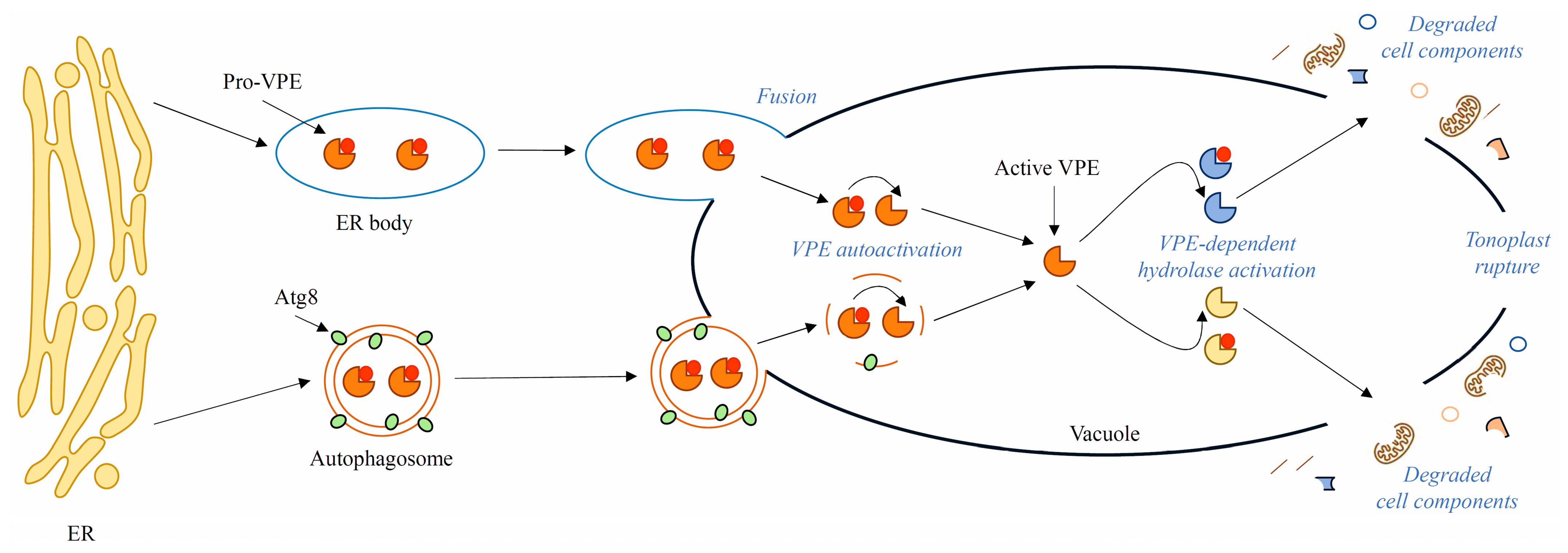
8. Presumed Role of VPEs in Late Stages of Autophagy
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Atg | Autophagy-related genes or proteins |
| BI-1 | Bax inhibitor-1 |
| dPCD | Developmental programmed cell death |
| ePCD | Environmental programmed cell death |
| HR | Hypersensitive response |
| kB1 | Kalata B1 |
| LSAM | Legumain stabilization and activity modulation pro-domain |
| MPK6 | Mitogen-activated protein kinase 6 |
| NAC | NAM, ATAF1/2, and CUC transcription factors |
| Pep4 | Proteinase A |
| Prb1 | Proteinase B |
| Prc1 | Carboxypeptidase Y |
| ROS | Reactive oxygen species |
| RuBisCO | Ribulose-1,5-bisphosphate carboxylase/oxygenase |
| SnRK1 | Sucrose nonfermenting-1-related protein kinase 1 |
| TFs | Transcription factors |
| TOR | Target of rapamycin serine/threonine-protein kinase |
| UCP | Uncoupling protein |
| VPEs | Vacuolar processing enzymes |
| Vps30 | Vacuolar protein sorting-associated protein 30 |
| Ybr139 | Putative serine carboxypeptidase YBR139W |
References
- Hatsugai, N.; Yamada, K.; Goto, S.; Hara-Nishimura, I. Vacuolar Processing Enzyme in Plant Programmed Cell Death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef]
- Zauner, F.B.; Dall, E.; Regl, C.; Grassi, L.; Huber, C.G.; Cabrele, C.; Brandstetter, H. Crystal Structure of Plant Legumain Reveals a Unique Two-Chain State with PH-Dependent Activity Regulation[CC-BY]. Plant Cell 2018, 30, 686–699. [Google Scholar] [CrossRef]
- Dall, E.; Zauner, F.B.; Soh, W.T.; Demir, F.; Dahms, S.O.; Cabrele, C.; Huesgen, P.F.; Brandstetter, H. Structural and Functional Studies of Arabidopsis Thaliana Legumain Beta Reveal Isoform Specific Mechanisms of Activation and Substrate Recognition. J. Biol. Chem. 2020, 295, 13047–13064. [Google Scholar] [CrossRef]
- Dall, E.; Brandstetter, H. Structure and Function of Legumain in Health and Disease. Biochimie 2016, 122, 126–150. [Google Scholar] [CrossRef] [PubMed]
- Vorster, B.; Cullis, C.; Kunert, K. Plant Vacuolar Processing Enzymes. Front. Plant Sci. 2019, 10, 479. [Google Scholar] [CrossRef]
- Tang, T.M.S.; Luk, L.Y.P. Asparaginyl Endopeptidases: Enzymology, Applications and Limitations. Org. Biomol. Chem. 2021, 19, 5048–5062. [Google Scholar] [CrossRef] [PubMed]
- Herman, E.; Larkins, B. Protein Storage Bodies and Vacuoles. Plant Cell 1999, 11, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Hiraiwa, N.; Nishimura, M.; Hara-Nishimura, I. Vacuolar Processing Enzyme Is Self-Catalytically Activated by Sequential Removal of the C-Terminal and N-Terminal Propeptides. FEBS Lett. 1999, 447, 213–216. [Google Scholar] [CrossRef]
- Rojo, E.; Zouhar, J.; Carter, C.; Kovaleva, V.; Raikhel, N.V. A Unique Mechanism for Protein Processing and Degradation in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 7389–7394. [Google Scholar] [CrossRef]
- Thompson, A.R.; Vierstra, R.D. Autophagic Recycling: Lessons from Yeast Help Define the Process in Plants. Curr. Opin. Plant Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef]
- Stefaniak, S.; Wojtyla, Ł.; Pietrowska-Borek, M.; Borek, S. Molecular Sciences Completing Autophagy: Formation and Degradation of the Autophagic Body and Metabolite Salvage in Plants. Int. J. Mol. Sci. 2020, 21, 2205. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xiang, Y.; Niu, Y. An Overview of the Molecular Mechanisms and Functions of Autophagic Pathways in Plants. Plant. Signal. Behav. 2021, 16, 1977527. [Google Scholar] [CrossRef]
- Yamada, K.; Shimada, T.; Nishimura, M.; Hara-Nishimura, I. A VPE Family Supporting Various Vacuolar Functions in Plants. Physiol. Plant. 2005, 123, 369–375. [Google Scholar] [CrossRef]
- Shimada, T.; Yamada, K.; Kataoka, M.; Nakaune, S.; Koumoto, Y.; Kuroyanagi, M.; Tabata, S.; Kato, T.; Shinozaki, K.; Seki, M.; et al. Vacuolar Processing Enzymes Are Essential for Proper Processing of Seed Storage Proteins in Arabidopsis Thaliana. J. Biol. Chem. 2003, 278, 32292–32299. [Google Scholar] [CrossRef]
- Yamada, K.; Basak, A.; Goto, S.; Tarnawska-Glatt, K.; Hara-Nishimura, I. Vacuolar Processing Enzymes in the Plant Life Cycle. New Phytol. 2019, 226, 21–31. [Google Scholar] [CrossRef]
- Poncet, V.; Scutt, C.; Tournebize, R.; Villegente, M.; Cueff, G.; Rajjou, L.; Balliau, T.; Zivy, M.; Fogliani, B.; Job, C.; et al. The Amborella Vacuolar Processing Enzyme Family. Front. Plant Sci. 2015, 6, 618. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, N.; Huang, S.; Hu, J.; Wang, Q.; Tang, Y.; Yang, T.; Asmutola, P.; Wang, J.; Yu, Q. Enhanced Soluble Sugar Content in Tomato Fruit Using CRISPR/Cas9-Mediated SlINVINH1 and SlVPE5 Gene Editing. PeerJ 2021, 9, 12478. [Google Scholar] [CrossRef]
- Zhang, H.; Tao, X.; Zhang, F. Genome-Wide Identification and Expression Analysis of the Vacuolar Processing Enzyme (VPE) Family Genes in Pear. J. Hortic. Sci. Biotechnol. 2021, 96, 469–478. [Google Scholar] [CrossRef]
- Song, J.; Yang, F.; Xun, M.; Xu, L.; Tian, X.; Zhang, W.; Yang, H. Genome-Wide Identification and Characterization of Vacuolar Processing Enzyme Gene Family and Diverse Expression Under Stress in Apple (Malus × Domestic). Front. Plant Sci. 2020, 11, 626. [Google Scholar] [CrossRef]
- Hara-Nishimura, I.; Shimada, T.; Hiraiwa, N.; Nishimura, M. Vacuolar Processing Enzyme Responsible for Maturation of Seed Proteins. J. Plant Physiol. 1995, 145, 632–640. [Google Scholar] [CrossRef]
- Nakaune, S.; Yamada, K.; Kondo, M.; Kato, T.; Tabata, S.; Nishimura, M.; Hara-Nishimura, I. A Vacuolar Processing Enzyme, δVPE, Is Involved in Seed Coat Formation at the Early Stage of Seed Development. Plant Cell 2005, 17, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, H.; Deng, X.; Liu, J.; Chen, H. The Relationship between Vacuolation and Initiation of PCD in Rice (Oryza Sativa) Aleurone Cells. Sci. Rep. 2017, 7, 41245. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, J.; Yin, B.; Liu, Y.; Wang, B.; Li, H.; Lu, H. γVPE Plays an Important Role in Programmed Cell Death for Xylem Fiber Cells by Activating Protease CEP1 Maturation in Arabidopsis thaliana. Int. J. Biol. Macromol. 2019, 137, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Guo, X.; Zhang, J.; Liu, Y.; Wang, B.; Li, H.; Lu, H. ΒVPE Is Involved in Tapetal Degradation and Pollen Development by Activating Proprotease Maturation in Arabidopsis Thaliana. J. Exp. Bot. 2020, 71, 1943–1955. [Google Scholar] [CrossRef]
- Li, J.-W.; Zhang, S.-B.; Xi, H.-P.; Bradshaw, C.J.A.; Zhang, J.-L. Processes Controlling Programmed Cell Death of Root Velamen Radicum in an Epiphytic Orchid. Ann. Bot. 2020, 126, 261–275. [Google Scholar] [CrossRef]
- Guo, X.; Li, L.; Liu, X.; Zhang, C.; Yao, X.; Xun, Z.; Zhao, Z.; Yan, W.; Zou, Y.; Liu, D.; et al. MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 3563. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hu, J.; Tan, R.; Han, Y.; Li, Z. Expression of IbVPE1 from Sweet Potato in Arabidopsis Affects Leaf Development, Flowering Time and Chlorophyll Catabolism. BMC Plant Biol. 2019, 19, 184. [Google Scholar] [CrossRef]
- Ferreira, D.O.; Fraga, O.T.; Pimenta, M.R.; Caetano, H.D.N.; Machado, J.P.B.; Carpinetti, P.A.; Brustolini, O.J.B.; Quadros, I.P.S.; Reis, P.A.B.; Fontes, E.P.B. GmNAC81 Inversely Modulates Leaf Senescence and Drought Tolerance. Front Genet 2020, 11, 601876. [Google Scholar] [CrossRef]
- Fraga, O.T.; de Melo, B.P.; Quadros, I.P.S.; Reis, P.A.B.; Fontes, E.P.B. Senescence-Associated Glycine Max (Gm)NAC Genes: Integration of Natural and Stress-Induced Leaf Senescence. Int. J. Mol. Sci. 2021, 22, 8287. [Google Scholar] [CrossRef]
- Zhang, H.; Xiao, Y.; Deng, X.; Feng, H.; Li, Z.; Zhang, L.; Chen, H. OsVPE3 Mediates GA-Induced Programmed Cell Death in Rice Aleurone Layers via Interacting with Actin Microfilaments. Rice 2020, 13, 22. [Google Scholar] [CrossRef]
- Quadros, I.; Madeira, N.; Loriato, V.; Fillietaz Saia, T.; Coutinho Silva, J.; Soares, F.; Carvalho, J.; Reis, P.A.; Fontes, E.; Clarindo, W.; et al. Cadmium-mediated Toxicity in Plant Cells Is Associated with the DCD / NRP -mediated Cell Death Response. Plant Cell Environ. 2021, 45, 556–571. [Google Scholar] [CrossRef] [PubMed]
- Teper-Bamnolker, P.; Danieli, R.; Peled-Zehavi, H.; Belausov, E.; Abu-Abied, M.; Avin-Wittenberg, T.; Sadot, E.; Eshel, D. Vacuolar Processing Enzyme Translocates to the Vacuole through the Autophagy Pathway to Induce Programmed Cell Death. Autophagy 2021, 17, 3109–3123. [Google Scholar] [CrossRef] [PubMed]
- Sychta, K.; Słomka, A.; Kuta, E. Insights into Plant Programmed Cell Death Induced by Heavy Metals—Discovering a Terra Incognita. Cells 2021, 10, 65. [Google Scholar] [CrossRef]
- Rantong, G.; Gunawardena, A.H.L.A.N. Vacuolar Processing Enzymes, AmVPE1 and AmVPE2, as Potential Executors of Ethylene Regulated Programmed Cell Death in the Lace Plant (Aponogeton Madagascariensis). Botany 2018, 96, 235–247. [Google Scholar] [CrossRef]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A Plant Vacuolar Protease, VPE, Mediates Virus-Induced Hypersensitive Cell Death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yue, H.; Xing, D. MAP Kinase 6-Mediated Activation of Vacuolar Processing Enzyme Modulates Heat Shock-Induced Programmed Cell Death in Arabidopsis. New Phytol. 2012, 195, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Deng, M.; Guo, F.; Wang, M.; Zeng, Z.; Han, N.; Yang, Y.; Zhu, M.; Bian, H. Suppression of OsVPE3 Enhances Salt Tolerance by Attenuating Vacuole Rupture during Programmed Cell Death and Affects Stomata Development in Rice. Rice 2016, 9, 65. [Google Scholar] [CrossRef]
- Jalili, S.; Ehsanpour, A.A.; Javadirad, S.M. The Role of Melatonin on Caspase-3-like Activity and Expression of the Genes Involved in Programmed Cell Death (PCD) Induced by in Vitro Salt Stress in Alfalfa (Medicago Sativa L.) Roots. Bot. Stud. 2022, 63, 19. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, X.; Tian, J.; Zhang, X.; Yu, T.; Li, Y.; Li, D. Genome-Wide Analysis of VPE Family in Four Gossypium Species and Transcriptional Expression of VPEs in the Upland Cotton Seedlings under Abiotic Stresses. Funct. Integr. Genomics 2022, 22, 179–192. [Google Scholar] [CrossRef]
- Hemu, X.; El Sahili, A.; Hu, S.; Wong, K.; Chen, Y.; Wong, Y.H.; Zhang, X.; Serra, A.; Goh, B.C.; Darwis, D.A.; et al. Structural Determinants for Peptide-Bond Formation by Asparaginyl Ligases. Proc. Natl. Acad. Sci. USA 2019, 116, 11737–11746. [Google Scholar] [CrossRef]
- Shafee, T.; Harris, K.; Anderson, M. Chapter Eight—Biosynthesis of Cyclotides. In Advances in Botanical Research; Craik, D.J., Ed.; Academic Press: Cambridge, MA, USA, 2015; Volume 76, pp. 227–269. [Google Scholar] [CrossRef]
- Poon, S.; Harris, K.S.; Jackson, M.A.; McCorkelle, O.C.; Gilding, E.K.; Durek, T.; van der Weerden, N.L.; Craik, D.J.; Anderson, M.A. Co-Expression of a Cyclizing Asparaginyl Endopeptidase Enables Efficient Production of Cyclic Peptides in Planta. J. Exp. Bot. 2018, 69, 633–641. [Google Scholar] [CrossRef] [PubMed]
- James, A.M.; Haywood, J.; Mylne, J.S. Macrocyclization by Asparaginyl Endopeptidases. New Phytol. 2018, 218, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.A.; Gilding, E.K.; Shafee, T.; Harris, K.S.; Kaas, Q.; Poon, S.; Yap, K.; Jia, H.; Guarino, R.; Chan, L.Y.; et al. Molecular Basis for the Production of Cyclic Peptides by Plant Asparaginyl Endopeptidases. Nat. Commun. 2018, 9, 2411. [Google Scholar] [CrossRef] [PubMed]
- Balakireva, A.V.; Zamyatnin, A.A. Cutting Out the Gaps Between Proteases and Programmed Cell Death. Front. Plant Sci. 2019, 10, 704. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, W.G. Classes of Programmed Cell Death in Plants, Compared to Those in Animals. J. Exp. Bot. 2011, 62, 4749–4761. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC Transcription Factors in Plant Immunity. Phytopath. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Burke, R.; Schwarze, J.; Sherwood, O.L.; Jnaid, Y.; McCabe, P.F.; Kacprzyk, J. Stressed to Death: The Role of Transcription Factors in Plant Programmed Cell Death Induced by Abiotic and Biotic Stimuli. Front. Plant Sci. 2020, 11, 1235. [Google Scholar] [CrossRef]
- Salvesen, G.S.; Hempel, A.; Coll, N.S. Protease Signaling in Animal and Plant-Regulated Cell Death. FEBS J. 2016, 283, 2577–2598. [Google Scholar] [CrossRef]
- Huh, S.U. Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but Not Caspases. Int. J. Mol. Sci. 2022, 23, 4588. [Google Scholar] [CrossRef]
- Klemenčič, M.; Funk, C. Type III Metacaspases: Calcium-Dependent Activity Proposes New Function for the P10 Domain. New Phytol 2018, 218, 1179–1191. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Vartapetian, A.B.; Tuzhikov, A.I.; Chichkova, N.V.; Taliansky, M.; Wolpert, T.J. A Plant Alternative to Animal Caspases: Subtilisin-like Proteases. Cell Death Differ. 2011, 18, 1289–1297. [Google Scholar] [CrossRef]
- Figueiredo, J.; Sousa Silva, M.; Figueiredo, A. Subtilisin-like Proteases in Plant Defence: The Past, the Present and Beyond. Mol. Plant Pathol. 2017, 19, 1017–1028. [Google Scholar] [CrossRef]
- Chichkova, N.V.; Shaw, J.; Galiullina, R.A.; Drury, G.E.; Tuzhikov, A.I.; Kim, S.H.; Kalkum, M.; Hong, T.B.; Gorshkova, E.N.; Torrance, L.; et al. Phytaspase, a Relocalisable Cell Death Promoting Plant Protease with Caspase Specificity. EMBO J. 2010, 29, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, M.; Wang, Q.; Cheng, L.; Zhang, Z. Role of Papain-Like Cysteine Proteases in Plant Development. Front. Plant Sci. 2018, 9, 1717. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Cai, Y.-M.; Bonneau, L.; Rotari, V.; Danon, A.; McKenzie, E.A.; McLellan, H.; Mach, L.; Gallois, P. Inhibition of Cathepsin B by Caspase-3 Inhibitors Blocks Programmed Cell Death in Arabidopsis. Cell Death Differ. 2016, 23, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.M.; Yu, J.; Ge, Y.; Mironov, A.; Gallois, P. Two Proteases with Caspase-3-like Activity, Cathepsin B and Proteasome, Antagonistically Control ER-stress-induced Programmed Cell Death in Arabidopsis. New Phytol. 2018, 218, 1143–1155. [Google Scholar] [CrossRef]
- Dall, E.; Brandstetter, H. Mechanistic and Structural Studies on Legumain Explain Its Zymogenicity, Distinct Activation Pathways, and Regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 10940–10945. [Google Scholar] [CrossRef]
- Hara-Nishimura, I.; Hatsugai, N. The Role of Vacuole in Plant Cell Death. Cell Death Differ. 2011, 18, 1298–1304. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The Plant Hypersensitive Response: Concepts, Control and Consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef]
- Iakimova, E.T.; Sobiczewski, P.; Michalczuk, L.; Węgrzynowicz-Lesiak, E.; Mikiciński, A.; Woltering, E.J. Morphological and Biochemical Characterization of Erwinia Amylovora-Induced Hypersensitive Cell Death in Apple Leaves. Plant Physiol. Biochem. 2013, 63, 292–305. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Prabucka, B.; Muszyńska, E.; Marecka, D.; Kozak, M.; Dababat, A.A.; Sobczak, M. Activity Profiling of Barley Vacuolar Processing Enzymes Provides New Insights into the Plant and Cyst Nematode Interaction. Mol. Plant Pathol. 2019, 21, 38–52. [Google Scholar] [CrossRef]
- Misas-Villamil, J.C.; Toenges, G.; Kolodziejek, I.; Sadaghiani, A.M.; Kaschani, F.; Colby, T.; Bogyo, M.; van der Hoorn, R.A.L. Activity Profiling of Vacuolar Processing Enzymes Reveals a Role for VPE during Oomycete Infection. Plant J. 2013, 73, 689–700. [Google Scholar] [CrossRef]
- Gao, X.; Tang, Y.; Shi, Q.; Wei, Y.; Wang, X.; Shan, W.; Qiang, X. Vacuolar Processing Enzyme Positively Modulates Plant Resistance and Cell Death in Response to Phytophthora Parasitica Infection. J. Integr. Agric. 2022. In Press, Journal Pre-proof. [Google Scholar] [CrossRef]
- Kumar, D.; Rampuria, S.; Singh, N.K.; Shukla, P.; Kirti, P.B. Characterization of a Vacuolar Processing Enzyme Expressed in Arachis Diogoi in Resistance Responses against Late Leaf Spot Pathogen, Phaeoisariopsis Personata. Plant Mol. Biol. 2015, 88, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Wan Abdullah, W.M.A.N.; Saidi, N.B.; Yusof, M.T.; Wee, C.-Y.; Loh, H.-S.; Ong-Abdullah, J.; Lai, K.-S. Vacuolar Processing Enzymes Modulating Susceptibility Response to Fusarium Oxysporum f. Sp. Cubense Tropical Race 4 Infections in Banana. Front. Plant Sci. 2022, 12, 769855. [Google Scholar] [CrossRef]
- Dong, C.; Li, R.; Wang, N.; Liu, Y.; Zhang, Y.; Bai, S. Apple Vacuolar Processing Enzyme 4 Is Regulated by Cysteine Protease Inhibitor and Modulates Fruit Disease Resistance. J. Exp. Bot. 2022, 73, 3758–3773. [Google Scholar] [CrossRef] [PubMed]
- Mendes, G.C.; Reis, P.A.B.; Calil, I.P.; Carvalho, H.H.; Aragão, F.J.L.; Fontes, E.P.B. GmNAC30 and GmNAC81 Integrate the Endoplasmic Reticulum Stress- and Osmotic Stress-Induced Cell Death Responses through a Vacuolar Processing Enzyme. Proc. Natl. Acad. Sci. USA 2013, 110, 19627–19632. [Google Scholar] [CrossRef] [PubMed]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy Functions as an Antiviral Mechanism against Geminiviruses in Plants. eLife 2017, 6, 23897. [Google Scholar] [CrossRef] [PubMed]
- Hashimi, S.M.; Wu, N.-N.; Ran, J.; Liu, J.-Z. Silencing Autophagy-Related Gene 2 (ATG2) Results in Accelerated Senescence and Enhanced Immunity in Soybean. Int. J. Mol. Sci. 2021, 22, 11749. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Dong, J.; Wang, T. Autophagy in Plant Abiotic Stress Management. Int. J. Mol. Sci. 2021, 22, 4075. [Google Scholar] [CrossRef] [PubMed]
- Rehman, N.U.; Zeng, P.; Mo, Z.; Guo, S.; Liu, Y.; Huang, Y.; Xie, Q. Conserved and Diversified Mechanism of Autophagy between Plants and Animals upon Various Stresses. Antioxidants 2021, 10, 1736. [Google Scholar] [CrossRef]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 Protein Kinase Complex Is Both a Regulator and a Target of Autophagic Recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wang, P.; Xiong, Y. Target of Rapamycin Signaling in Plant Stress Responses. Plant Physiol. 2020, 182, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.-J.; Liu, C.-X.; Shao, S.-J.; Zhou, J. Molecular Mechanisms of Autophagy Regulation in Plants and Their Applications in Agriculture. Front. Plant Sci. 2021, 11, 618944. [Google Scholar] [CrossRef]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo Recognition and Degradation by Selective Autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef]
- Stephani, M.; Dagdas, Y. Plant Selective Autophagy—Still an Uncharted Territory With a Lot of Hidden Gems. J. Mol. Biol. 2020, 432, 63–79. [Google Scholar] [CrossRef]
- Borek, S.; Stefaniak, S.; Śliwiński, J.; Garnczarska, M.; Pietrowska-Borek, M. Autophagic Machinery of Plant Peroxisomes. Int. J. Mol. Sci. 2019, 20, 4754. [Google Scholar] [CrossRef]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Vicente-Carbajosa, J. A View into Seed Autophagy: From Development to Environmental Responses. Plants 2022, 11, 3247. [Google Scholar] [CrossRef] [PubMed]
- Gomez, R.E.; Lupette, J.; Chambaud, C.; Castets, J.; Ducloy, A.; Cacas, J.-L.; Masclaux-Daubresse, C.; Bernard, A. How Lipids Contribute to Autophagosome Biogenesis, a Critical Process in Plant Responses to Stresses. Cells 2021, 10, 1272. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, W.G.; Papini, A. Ultrastructure of Autophagy in Plant Cells. Autophagy 2013, 9, 1922–1936. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, T.; Han, J.; Li, M.; Zhao, Y.; Su, T.; Ma, C. Plant Autophagy: An Intricate Process Controlled by Various Signaling Pathways. Front. Plant Sci. 2021, 12, 754982. [Google Scholar] [CrossRef]
- Yu, G.; Klionsky, D. Life and Death Decisions-The Many Faces of Autophagy in Cell Survival and Cell Death. Biomolecules 2022, 12, 866. [Google Scholar] [CrossRef]
- Kroemer, G.; Levine, B. Autophagic Cell Death: The Story of a Misnomer. Nat. Rev. Mol. Cell Biol. 2008, 9, 1004–1010. [Google Scholar] [CrossRef]
- Kwon, S.I.; Cho, H.J.; Jung, J.H.; Yoshimoto, K.; Shirasu, K.; Park, O.K. The Rab GTPase RabG3b Functions in Autophagy and Contributes to Tracheary Element Differentiation in Arabidopsis. Plant J. 2010, 64, 151–164. [Google Scholar] [CrossRef]
- Wojciechowska, N.; Michalak, K.M.; Bagniewska-Zadworna, A. Autophagy—An Underestimated Coordinator of Construction and Destruction during Plant Root Ontogeny. Planta 2021, 254, 15. [Google Scholar] [CrossRef]
- Sobieszczuk-Nowicka, E.; Wrzesiński, T.; Bagniewska-Zadworna, A.; Kubala, S.; Rucińska-Sobkowiak, R.; Polcyn, W.; Misztal, L.; Mattoo, A.K. Physio-Genetic Dissection of Dark-Induced Leaf Senescence and Timing Its Reversal in Barley. Plant Physiol. 2018, 178, 654–671. [Google Scholar] [CrossRef]
- Feng, Q.; De Rycke, R.; Dagdas, Y.; Nowack, M.K. Autophagy Promotes Programmed Cell Death and Corpse Clearance in Specific Cell Types of the Arabidopsis Root Cap. Curr. Biol. 2022, 32, 2110–2119. [Google Scholar] [CrossRef]
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 Is Required to Limit the Pathogen-Associated Cell Death Response. Autophagy 2008, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Hofius, D.; Schultz-Larsen, T.; Joensen, J.; Tsitsigiannis, D.I.; Petersen, N.H.T.; Mattsson, O.; Jørgensen, L.B.; Jones, J.D.G.; Mundy, J.; Petersen, M. Autophagic Components Contribute to Hypersensitive Cell Death in Arabidopsis. Cell 2009, 137, 773–783. [Google Scholar] [CrossRef]
- Sertsuvalkul, N.; DeMell, A.; Dinesh-Kumar, S.P. The Complex Roles of Autophagy in Plant Immunity. FEBS Lett. 2022, 596, 2163–2171. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Yamada, K.; Shimada, T.; Matsushima, R.; Nishizawa, N.; Nishimura, M.; Hara-Nishimura, I. A Proteinase-Storing Body That Prepares for Cell Death or Stresses in the Epidermal Cells of Arabidopsis. Plant Cell Physiol. 2001, 42, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Avin-Wittenberg, T.; Galili, G. Involvement of Autophagy in the Direct ER to Vacuole Protein Trafficking Route in Plants. Front. Plant Sci. 2014, 5, 134. [Google Scholar] [CrossRef]
- Li, Y.-B.; Yan, M.; Cui, D.-Z.; Huang, C.; Sui, X.-X.; Guo, F.Z.; Fan, Q.-Q.; Chu, X.-S. Programmed Degradation of Pericarp Cells in Wheat Grains Depends on Autophagy. Front. Genet. 2021, 12, 784545. [Google Scholar] [CrossRef]
- Dauphinee, A.N.; Denbigh, G.L.; Rollini, A.; Fraser, M.; Lacroix, C.R.; Gunawardena, A.H.L.A.N. The Function of Autophagy in Lace Plant Programmed Cell Death. Front. Plant Sci. 2019, 10, 1198. [Google Scholar] [CrossRef]
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in Yeast Demonstrated with Proteinase-Deficient Mutants and Conditions for Its Induction. J. Cell Biol. 1992, 119, 301–311. [Google Scholar] [CrossRef]
- Torggler, R.; Papinski, D.; Kraft, C. Assays to Monitor Autophagy in Saccharomyces Cerevisiae. Cells 2017, 6, 23. [Google Scholar] [CrossRef]
- Hirata, E.; Shirai, K.; Kawaoka, T.; Sato, K.; Kodama, F.; Suzuki, K. Atg15 in Saccharomyces Cerevisiae Consists of Two Functionally Distinct Domains. Mol. Biol. Cell. 2021, 32, 645–663. [Google Scholar] [CrossRef]
- Parzych, K.R.; Ariosa, A.; Mari, M.; Klionsky, D.J. A Newly Characterized Vacuolar Serine Carboxypeptidase, Atg42/Ybr139w, Is Required for Normal Vacuole Function and the Terminal Steps of Autophagy in the Yeast Saccharomyces Cerevisiae. Mol. Biol. Cell. 2018, 29, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, J.; Geng, J.; Nair, U.; Klionsky, D.J. Atg22 Recycles Amino Acids to Link the Degradative and Recycling Functions of Autophagy. Mol. Biol. Cell 2006, 17, 5094–5104. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.W.; Zubenko, G.S.; Parker, R.R. PEP4 Gene Function Is Required for Expression of Several Vacuolar Hydrolases in SACCHAROMYCES CEREVISIAE. Genetics 1982, 102, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Hiraiwa, N.; Nishimura, M.; Hara-Nishimura, I. Expression and Activation of the Vacuolar Processing Enzyme in Saccharomyces cerevisiae. Plant J. 1997, 12, 819–829. [Google Scholar] [CrossRef]
- Clark, S.L. Cellular Differentiation in the Kidneys of Newborn Mice Studies with the Electron Microscope. J. Biophys. Biochem. Cytol. 1957, 3, 349–362. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wleklik, K.; Borek, S. Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy. Int. J. Mol. Sci. 2023, 24, 1198. https://doi.org/10.3390/ijms24021198
Wleklik K, Borek S. Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy. International Journal of Molecular Sciences. 2023; 24(2):1198. https://doi.org/10.3390/ijms24021198
Chicago/Turabian StyleWleklik, Karolina, and Sławomir Borek. 2023. "Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy" International Journal of Molecular Sciences 24, no. 2: 1198. https://doi.org/10.3390/ijms24021198
APA StyleWleklik, K., & Borek, S. (2023). Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy. International Journal of Molecular Sciences, 24(2), 1198. https://doi.org/10.3390/ijms24021198






