Component-Resolved Diagnosis Based on a Recombinant Variant of Mus m 1 Lipocalin Allergen
Abstract
1. Introduction
2. Results
2.1. Bioinformatic Approach to Mus m 1.0102 Protein
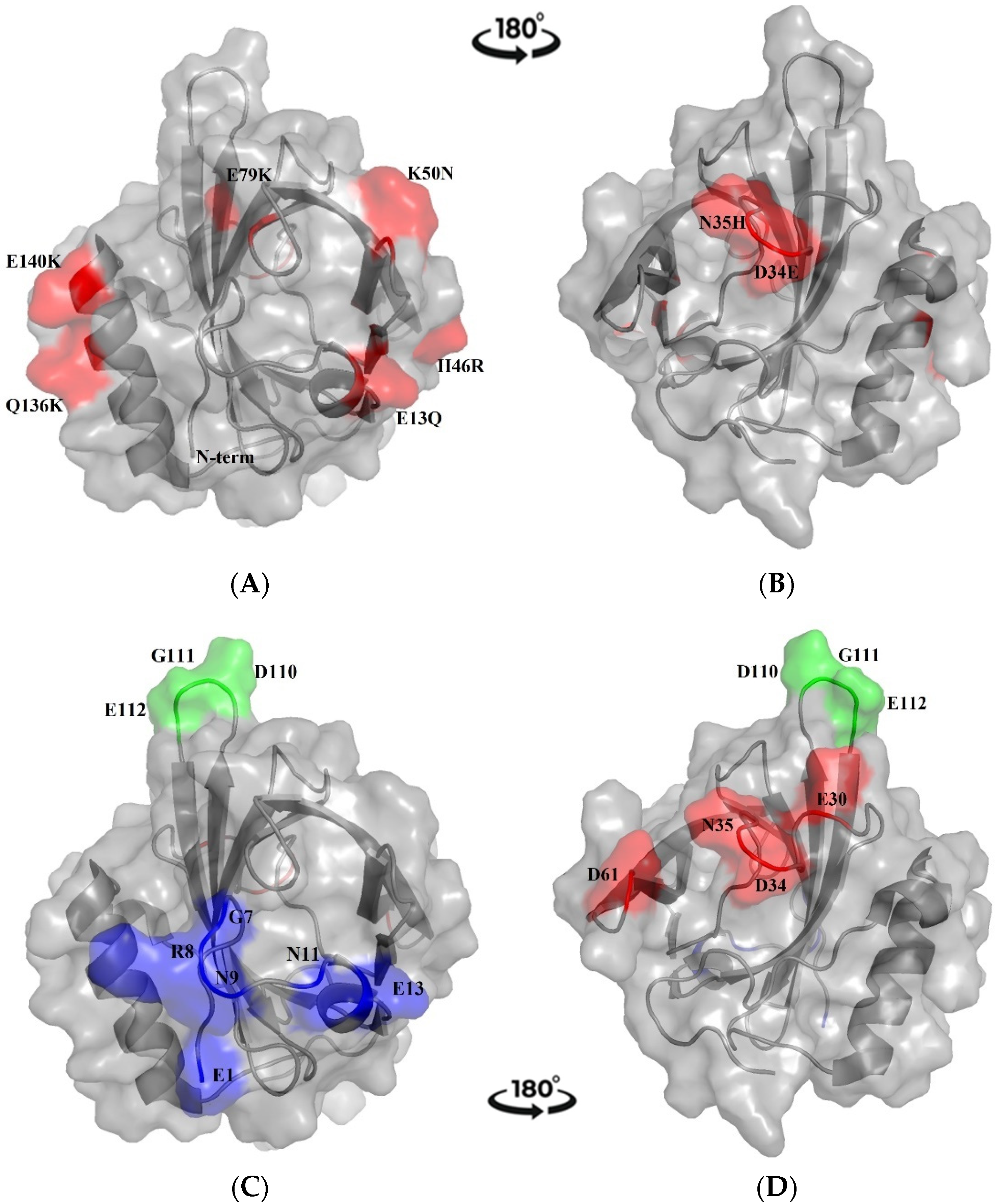
2.1.1. Sequence Comparison with MUP Isoforms
2.1.2. Prediction of B-Cell Conformational Epitopes
2.2. Preparation of Recombinant Mus m 1.0102 and Urinary Protein Samples
2.3. Indirect ELISA Immunoassay
2.3.1. Allergy Diagnosis by ImmunoCAP ISAC Test
2.3.2. Indirect ELISA Immunoassay with Recombinant and Urinary Mus m 1 Samples
2.3.3. ROC Analysis Based on Indirect ELISA Data
2.4. Precision of the ELISA Immunoassay Based on Recombinant Mus m 1.0102 Allergen
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis of Mus m 1.0102 Sequence and Protein Structure
4.2. Preparation of Urinary and Recombinant Mus m 1 Samples
4.3. Serum Samples, Allergy Diagnosis and IgE Quantification
4.4. Indirect IgE-Enzyme-Linked Immunosorbent Assay (IgE-ELISA)
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Phipatanakul, W.; Wood, R.A. Chapter 15—Allergens of Animal and Biological Systems. In Biological Safety: Principles and Practices, 5th ed.; Wooley, D.P., Byers, K.B., Eds.; Wiley: Hoboken, NJ, USA, 2016; pp. 327–339. [Google Scholar]
- D’Ovidio, M.C.; Wirz, A.; Zennaro, D.; Massari, S.; Melis, P.; Peri, V.M.; Rafaiani, C.; Riviello, M.C.; Mari, A. Biological occupational allergy: Protein microarray for the study of laboratory animal allergy (LAA). AIMS Public Health 2018, 5, 352–365. [Google Scholar] [CrossRef]
- Jones, M. Laboratory Animal Allergy in the Modern Era. Curr. Allergy Asthma Rep. 2015, 15, 73. [Google Scholar] [CrossRef]
- Schulten, V.; Westernberg, L.; Birrueta, G.; Sidney, J.; Paul, S.; Busse, P.; Peters, B.; Sette, A. Allergen and epitope targets of mouse-specific T Cell responses in allergy and asthma. Front. Immunol. 2018, 9, 235. [Google Scholar] [CrossRef]
- Roberts, S.A.; Prescott, M.C.; Davidson, A.J.; McLean, L.; Beynon, R.J.; Hurst, J.L. Individual odour signatures that mice learn are shaped by involatile major urinary proteins (MUPs). BMC Biol. 2018, 16, 48. [Google Scholar] [CrossRef]
- Ferrari, E.; Casali, E.; Burastero, S.E.; Spisni, A.; Pertinhez, T.A. The allergen Mus m 1.0102: Dissecting the relationship between molecular conformation and allergenic potency. Biochim. Biophys. Acta 2016, 1864, 1548–1557. [Google Scholar] [CrossRef]
- Sheehan, M.J.; Lee, V.; Corbett-Detig, R.; Bi, K.; Beynon, R.J.; Hurst, J.L.; Nachman, M.W. Selection on coding and regulatory variation maintains individuality in major urinary protein scent marks in wild mice. PLoS Genet. 2016, 12, e1005891. [Google Scholar] [CrossRef]
- Charkoftaki, G.; Wang, Y.; McAndrews, M.; Bruford, E.A.; Thompson, D.C.; Vasiliou, V.; Nebert, D.W. Update on the human and mouse lipocalin (LCN) gene family, including evidence the mouse Mup cluster is result of an “evolutionary bloom”. Hum. Genom. 2019, 13, 11. [Google Scholar] [CrossRef]
- Phelan, M.M.; McLean, L.; Hurst, J.L.; Beynon, R.J.; Lian, L.Y. Comparative study of the molecular variation between ‘central’ and ‘peripheral’ MUPs and significance for behavioural signalling. Biochem. Soc. Trans. 2014, 42, 866–872. [Google Scholar] [CrossRef]
- Galli, S.J.; Tsai, M. IgE and mast cells in allergic disease. Nat. Med. 2012, 18, 693–704. [Google Scholar] [CrossRef]
- Huss-Marp, J.; Gutermuth, J.; Schäffner, I.; Darsow, U.; Pfab, F.; Brockow, K.; Ring, J.; Behrendt, H.; Jakob, T.; Ahlgrim, C. Comparison of molecular and extract-based allergy diagnostics with multiplex and singleplex analysis. Allergo J. Int. 2015, 24, 46–53. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Żbikowska-Gotz, M.; Marek-Józefowicz, L.; Przybyszewski, M.; Gocki, J.; Bartuzi, Z. Allergic phenotypes in adult patients with atopic dermatitis, determined with the ISAC test (ImmunoCAP ISAC). Postępy Dermatol. Alergol. 2018, 35, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Matricardi, P.M.; Dramburg, S.; Potapova, E.; Skevaki, C.; Renz, H. Molecular diagnosis for allergen immunotherapy. J. Allergy Clin. Immunol. 2019, 143, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Dodig, S.; Čepelak, I. The potential of component-resolved diagnosis in laboratory diagnostics of allergy. Biochem. Med. 2018, 28, 020501. [Google Scholar] [CrossRef] [PubMed]
- Cudowska, B.; Kapingidza, A.B.; Pawłowicz, M.; Pampuch, A.; Hyduke, N.; Pote, S.; Schlachter, C.R.; Lebensztejn, D.M.; Chruszcz, M.; Kowal, K. Production and use of recombinant profilins Amb a 8, Art v 4, Bet v 2, and Phl p 12 for allergenic sensitization studies. Molecules 2020, 25, 369. [Google Scholar] [CrossRef] [PubMed]
- Clayton, G.M.; White, J.; Lee, S.; Kappler, J.W.; Chan, S.K. Structural characteristics of lipocalin allergens: Crystal structure of the immunogenic dog allergen Can f 6. PLoS ONE 2019, 14, e0213052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cai, Z.; Hou, Y.; Hu, J.; He, Y.; Chen, J.; Ji, K. Enhanced sensitivity of capture IgE-ELISA based on a recombinant Der f 1/2 fusion protein for the detection of IgE antibodies targeting house dust mite allergens. Mol. Med. Rep. 2019, 19, 3497–3504. [Google Scholar] [CrossRef]
- Rytkönen-Nissinen, M.; Saarelainen, S.; Randell, J.; Häyrinen, J.; Kalkkinen, N.; Virtanen, T. IgE Reactivity of the dog lipocalin allergen Can f 4 and the development of a sandwich ELISA for its quantification. Allergy Asthma Immunol. Res. 2015, 7, 384–392. [Google Scholar] [CrossRef]
- Ferrari, E.; Breda, D.; Longhi, R.; Vangelista, L.; Nakaie, C.R.; Elviri, L.; Casali, E.; Pertinhez, T.A.; Spisni, A.; Burastero, S.E. In search of a vaccine for mouse allergy: Significant reduction of Mus m 1 allergenicity by structure-guided single-point mutations. Int. Arch. Allergy Immunol. 2012, 157, 226–237. [Google Scholar] [CrossRef]
- Ferrari, E.; Corsini, R.; Burastero, S.E.; Tanfani, F.; Spisni, A. The allergen Mus m 1.0102: Cysteine residues and molecular allergology. Mol. Immunol. 2020, 120, 1–12. [Google Scholar] [CrossRef]
- Ferrari, E.; Corsini, R.; Burastero, S.E.; Tanfani, F.; Spisni, A. Thermal stability, ligand binding and allergenicity data of Mus m 1.0102 allergen and its cysteine mutants. Data Brief 2020, 29, 105355. [Google Scholar] [CrossRef]
- Raulf, M.; Quirce, S.; Vandenplas, O. Addressing Molecular Diagnosis of Occupational Allergies. Curr. Allergy Asthma Rep. 2018, 18, 6. [Google Scholar] [CrossRef]
- Raulf, M. Allergen component analysis as a tool in the diagnosis and management of occupational allergy. Mol. Immunol. 2018, 100, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Vandenplas, O.; Froidure, A.; Meurer, U.; Rihs, H.P.; Rifflart, C.; Soetaert, S.; Jamart, J.; Pilette, C.; Raulf, M. The role of allergen components for the diagnosis of latex-induced occupational asthma. Allergy 2016, 71, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Krop, E.J.; Matsui, E.C.; Sharrow, S.D.; Stone, M.J.; Gerber, P.; van der Zee, J.S.; Chapman, M.D.; Aalberse, R.C. Recombinant major urinary proteins of the mouse in specific IgE and IgG testing. Int. Arch. Allergy Immunol. 2007, 144, 296–304. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Polyanovsky, V.; Lifanov, A.; Esipova, N.; Tumanyan, V. The ranging of amino acids substitution matrices of various types in accordance with the alignment accuracy criterion. BMC Bioinform. 2020, 21 (Suppl. 11), 294. [Google Scholar] [CrossRef]
- Haste Andersen, P.; Nielsen, M.; Lund, O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006, 15, 2558–2567. [Google Scholar] [CrossRef]
- Lücke, C.; Franzoni, L.; Abbate, F.; Löhr, F.; Ferrari, E.; Sorbi, R.T.; Rüterjans, H.; Spisni, A. Solution structure of a recombinant mouse major urinary protein. Eur. J. Biochem. 1999, 266, 1210–1218. [Google Scholar] [CrossRef]
- Hentges, F.; Léonard, C.; Arumugam, K.; Hilger, C. Immune responses to inhalant Mammalian allergens. Front. Immunol. 2014, 5, 234. [Google Scholar] [CrossRef]
- Munera, M.; Contreras, N.; Sánchez, A.; Sánchez, J.; Emiliani, Y. In silico analysis of a major allergen from Rattus norvegicus, Rat n 1, and cross-reactivity with domestic pets. F1000Research 2019, 8, 1707. [Google Scholar] [CrossRef]
- Mucignat-Caretta, C.; Caretta, A.; Cavaggioni, A. Acceleration of puberty onset in female mice by male urinary proteins. J. Physiol. 1995, 486, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.A.; Simpson, D.M.; Armstrong, S.D.; Davidson, A.J.; Robertson, D.H.; McLean, L.; Beynon, R.J.; Hurst, J.L. Darcin: A male pheromone that stimulates female memory and sexual attraction to an individual male’s odour. BMC Biol. 2010, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.A.; Smith, A.L.; Armstrong, S.D.; Beynon, R.J.; Hurst, J.L. Limited variation in the major urinary proteins of laboratory mice. Physiol. Behav. 2009, 96, 253–261. [Google Scholar] [CrossRef]
- Ansotegui, I.J.; Melioli, G.; Canonica, G.W.; Caraballo, L.; Villa, E.; Ebisawa, M.; Passalacqua, G.; Savi, E.; Ebo, D.; Gómez, R.M.; et al. IgE allergy diagnostics and other relevant tests in allergy, a World Allergy Organization position paper. World Allergy Organ. J. 2020, 13, 100080. [Google Scholar] [CrossRef]
- Sinson, E.; Ocampo, C.; Liao, C.; Nguyen, S.; Dinh, L.; Rodems, K.; Whitters, E.; Hamilton, R.G. Cross-reactive carbohydrate determinant interference in cellulose-based IgE allergy tests utilizing recombinant allergen components. PLoS ONE 2020, 15, e0231344. [Google Scholar] [CrossRef]
- Alisi, C.; Afferni, C.; Iacovacci, P.; Barletta, B.; Tinghino, R.; Butteroni, C.; Puggioni, E.M.; Wilson, I.B.; Federico, R.; Schininà, M.E.; et al. Rapid isolation, characterization, and glycan analysis of Cup a 1, the major allergen of Arizona cypress (Cupressus arizonica) pollen. Allergy 2001, 56, 978–984. [Google Scholar] [CrossRef]
- Arilla, M.C.; Ibarrola, I.; García, R.; de la Hoz, B.; Martínez, A.; Asturias, J.A. Quantification of the major allergen from cypress (Cupressus arizonica) pollen, Cup a 1, by monoclonal antibody-based ELISA. Int. Arch. Allergy Immunol. 2004, 134, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, J.; Vieths, S.; Kaul, S. Standardization and Regulation of Allergen Products in the European Union. Curr. Allergy Asthma Rep. 2016, 16, 21. [Google Scholar] [CrossRef]
- Norton, A.; Smith, K.; James, K.; Hoskins, A.; Scott, T.A.; Plunkett, G.; Fahrenholz, J.; Dworski, R. Diagnostic utility of concentrated Mus m 1 allergen extract in humans. Ann. Allergy Asthma Immunol. 2014, 112, 391–392. [Google Scholar] [CrossRef]
- Mindaye, S.T.; Sun, C.; Esfahani, S.A.Z.; Matsui, E.C.; Sheehan, M.J.; Rabin, R.L.; Slater, J.E. Diversity and complexity of mouse allergens in urine, house dust, and allergen extracts assessed with an immuno-allergomic approach. Allergy 2021, 76, 3723–3732. [Google Scholar] [CrossRef]
- Ibarra-Soria, X.; Levitin, M.O.; Logan, D.W. The genomic basis of vomeronasal-mediated behaviour. Mamm. Genome 2014, 25, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Kringelum, J.V.; Lundegaard, C.; Lund, O.; Nielsen, M. Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012, 8, e1002829. [Google Scholar] [CrossRef]
- Zhang, J.; Tao, A. Antigenicity, Immunogenicity, Allergenicity. In Allergy Bioinformatics; Tao, A., Raz, E., Eds.; Part of the Translational Bioinformatics Book Series; Springer: Dordrecht, The Netherlands, 2015; Volume 8, pp. 175–186. [Google Scholar] [CrossRef]
- Múnera, M.; Sanchez, A.; Sánchez, J.; Nordmann, M.; Perez, M.; Aparicio, D. Allergy to Mus m 1: A review of structural, and immunological features. Immunol. Lett. 2019, 209, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Hilger, C.; van Hage, M.; Kuehn, A. Diagnosis of allergy to mammals and fish: Cross-Reactive vs. specific markers. Curr. Allergy Asthma Rep. 2017, 17, 64. [Google Scholar] [CrossRef]
- Altmann, F. Coping with cross-reactive carbohydrate determinants in allergy diagnosis. Allergo J. Int. 2016, 25, 98–105. [Google Scholar] [CrossRef]
- Yoshida, N.; Hirata, H.; Watanabe, M.; Sugiyama, K.; Arima, M.; Fukushima, Y.; Ishii, Y. Improved sensitivity to venom specific-immunoglobulin E by spiking with the allergen component in Japanese patients suspected of Hymenoptera venom allergy. Allergol. Int. 2015, 64, 248–252. [Google Scholar] [CrossRef]
- Tscheppe, A.; Breiteneder, H. Recombinant Allergens in Structural Biology, Diagnosis, and Immunotherapy. Int. Arch. Allergy Immunol. 2017, 172, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Hooshmand, N.; Fayazi, J.; Tabatabaei, S.; Ghaleh Golab Behbahan, N. Prediction of B cell and T-helper cell epitopes candidates of bovine leukaemia virus (BLV) by in silico approach. Vet. Med. Sci. 2020, 6, 730–739. [Google Scholar] [CrossRef]
- Ferrari, E.; Lodi, T.; Sorbi, R.T.; Tirindelli, R.; Cavaggioni, A.; Spisni, A. Expression of a lipocalin in Pichia pastoris: Secretion, purification and binding activity of a recombinant mouse major urinary protein. FEBS Lett. 1997, 401, 73–77. [Google Scholar] [CrossRef]
- Moscato, G.; Vandenplas, O.; Van Wijk, R.G.; Malo, J.L.; Perfetti, L.; Quirce, S.; Walusiak, J.; Castano, R.; Pala, G.; Gautrin, D.; et al. EAACI position paper on occupational rhinitis. Respir. Res. 2009, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Curin, M.; Garib, V.; Valenta, R. Single recombinant and purified major allergens and peptides: How they are made and how they change allergy diagnosis and treatment. Ann. Allergy Asthma Immunol. 2017, 119, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Zhernov, Y.; Curin, M.; Khaitov, M.; Karaulov, A.; Valenta, R. Recombinant allergens for immunotherapy: State of the art. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 402–414. [Google Scholar] [CrossRef] [PubMed]

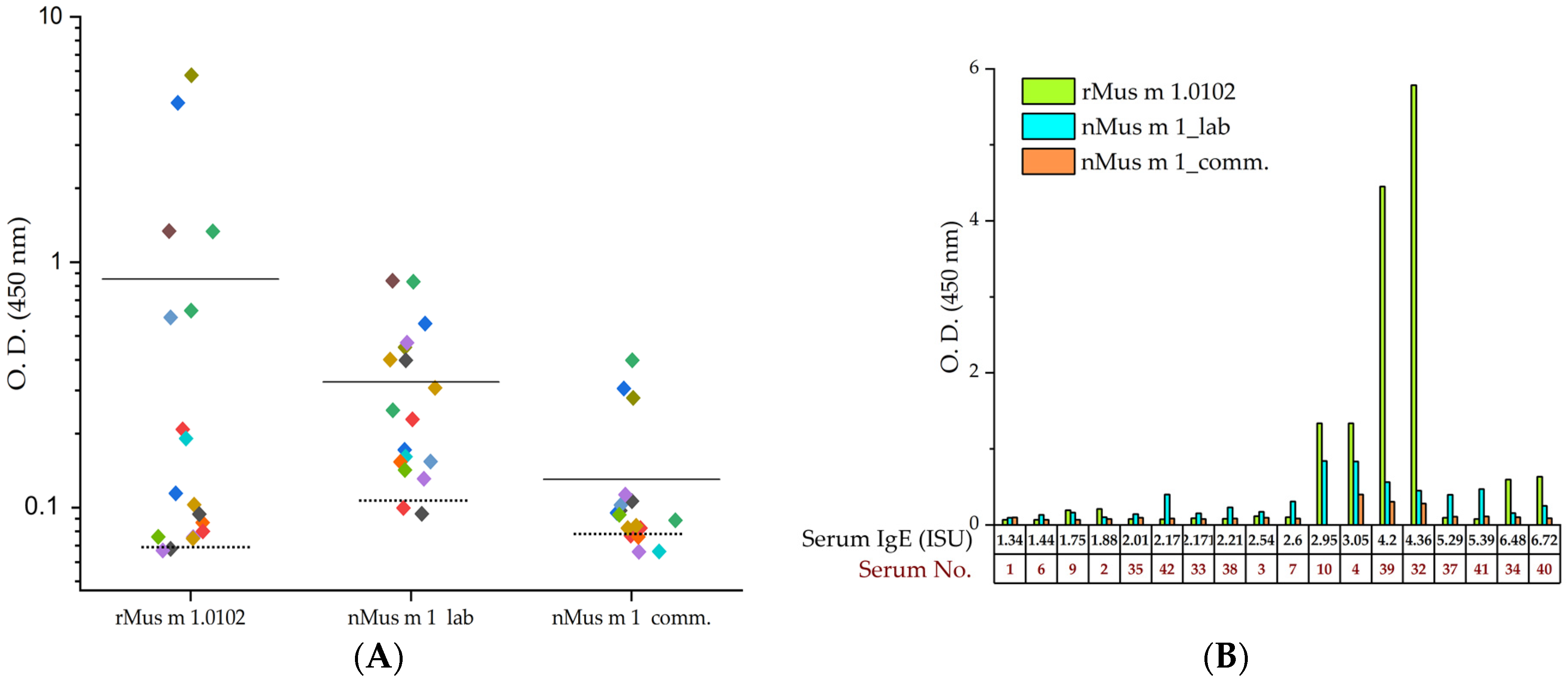
| Serum No. | Specific IgE to Lipocalins and to CCD (ISU Units) | |||||
|---|---|---|---|---|---|---|
| Mus m 1 | MUXF3 | nCup a 1 | Equ c 1 | Fel d 4 | Can f 1 | |
| 1 | 1.34 | 0.00 | 0.43 | 0.00 | 0.00 | 0.00 |
| 2 | 1.88 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 3 | 2.54 | 1.40 | 0.00 | 0.00 | 0.00 | 0.00 |
| 4 | 3.05 | 0.00 | 88.91 | 0.00 | 0.00 | 0.00 |
| 6 | 1.44 | 0.00 | 0.59 | 0.00 | 0.00 | 0.00 |
| 7 | 2.60 | 1.39 | 0.00 | 0.00 | 0.00 | 0.55 |
| 9 | 1.75 | 0.00 | 0.00 | 0.45 | 0.00 | 0.56 |
| 10 | 2.95 | 0.00 | 79.12 | 0.00 | 0.00 | 0.00 |
| 32 | 4.36 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 33 | 2.17 | 0.00 | 1.50 | 0.00 | 0.00 | 0.00 |
| 34 | 6.48 | 0.00 | 0.00 | 0.00 | 0.60 | 0.00 |
| 35 | 2.01 | 0.00 | 3.94 | 0.00 | 0.00 | 0.00 |
| 37 | 5.29 | 4.51 | 3.37 | 0.56 | 0.00 | 0.00 |
| 38 | 2.21 | 0.00 | 3.51 | 0.00 | 0.39 | 0.00 |
| 39 | 4.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.89 |
| 40 | 6.72 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 41 | 5.39 | 3.89 | 3.23 | 0.00 | 0.00 | 0.00 |
| 42 | 2.17 | 0.00 | 1.54 | 0.00 | 0.00 | 0.00 |
| Antigen | * Reactive Sera (%) | § Non-Reactive Sera (No.) |
|---|---|---|
| rMus m 1.0102 | 88.9 | 1, 6 |
| a nMus m 1_lab | 88.9 | 1, 2 |
| b nMus m 1_comm. | 76.4 | 2, 6, 9, 33 |
| AUC | p-Value * | Threshold ** | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| rMus m 1.0102 | 0.87 | 2.22E-4 | 0.075 | 0.89 | 0.75 |
| Mus m 1_lab | 0.89 | 7.90E-4 | 0.149 | 0.78 | 0.90 |
| Mus m 1_comm. | 0.77 | 0.021 | 0.082 | 0.76 | 0.80 |
| Serum No. (Specific IgE Level, ISU) | Intra-Assay CV | Inter-Assay CV | ||||
|---|---|---|---|---|---|---|
| Mean O.D. | SD | CV% | Mean O.D. | SD | CV% | |
| 34 (6.48) | 0.70 | 0.01 | 1.8 | 0.73 | 0.03 | 4.1 |
| 39 (4.20) | 4.89 | 0.32 | 6.5 | 4.84 | 0.39 | 8.0 |
| 2 (1.88) | 0.21 | 0.01 | 3.5 | 0.22 | 0.02 | 9.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrari, E.; Breda, D.; Spisni, A.; Burastero, S.E. Component-Resolved Diagnosis Based on a Recombinant Variant of Mus m 1 Lipocalin Allergen. Int. J. Mol. Sci. 2023, 24, 1193. https://doi.org/10.3390/ijms24021193
Ferrari E, Breda D, Spisni A, Burastero SE. Component-Resolved Diagnosis Based on a Recombinant Variant of Mus m 1 Lipocalin Allergen. International Journal of Molecular Sciences. 2023; 24(2):1193. https://doi.org/10.3390/ijms24021193
Chicago/Turabian StyleFerrari, Elena, Daniela Breda, Alberto Spisni, and Samuele E. Burastero. 2023. "Component-Resolved Diagnosis Based on a Recombinant Variant of Mus m 1 Lipocalin Allergen" International Journal of Molecular Sciences 24, no. 2: 1193. https://doi.org/10.3390/ijms24021193
APA StyleFerrari, E., Breda, D., Spisni, A., & Burastero, S. E. (2023). Component-Resolved Diagnosis Based on a Recombinant Variant of Mus m 1 Lipocalin Allergen. International Journal of Molecular Sciences, 24(2), 1193. https://doi.org/10.3390/ijms24021193








