Application of Caenorhabditis elegans in Lipid Metabolism Research
Abstract
1. Introduction
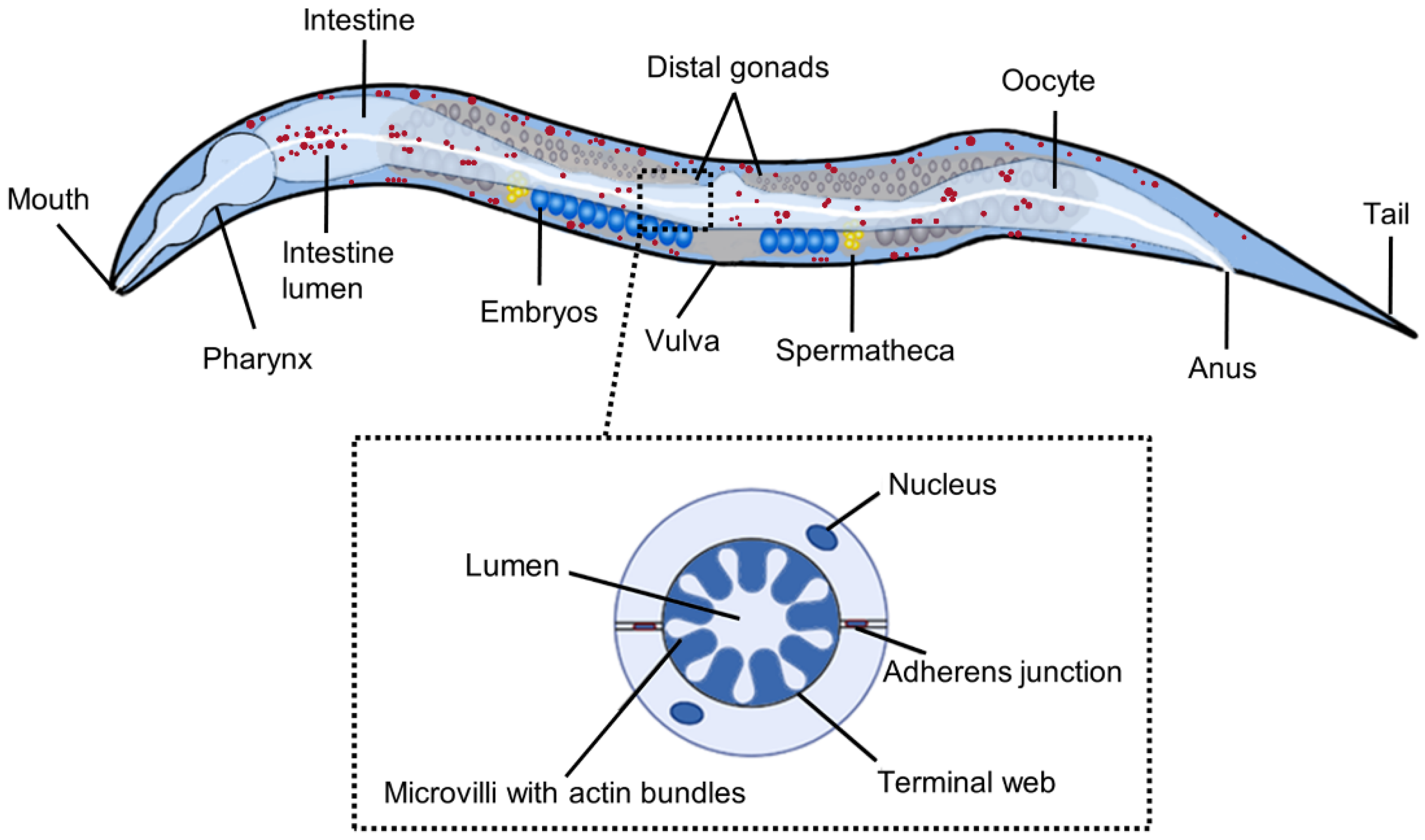
2. Advantages and Disadvantages of C. elegans as a Model for Studying Fat Metabolism
3. Regulation of Fat Synthesis and Degradation in C. elegans
3.1. Lipid Droplets (LDs)
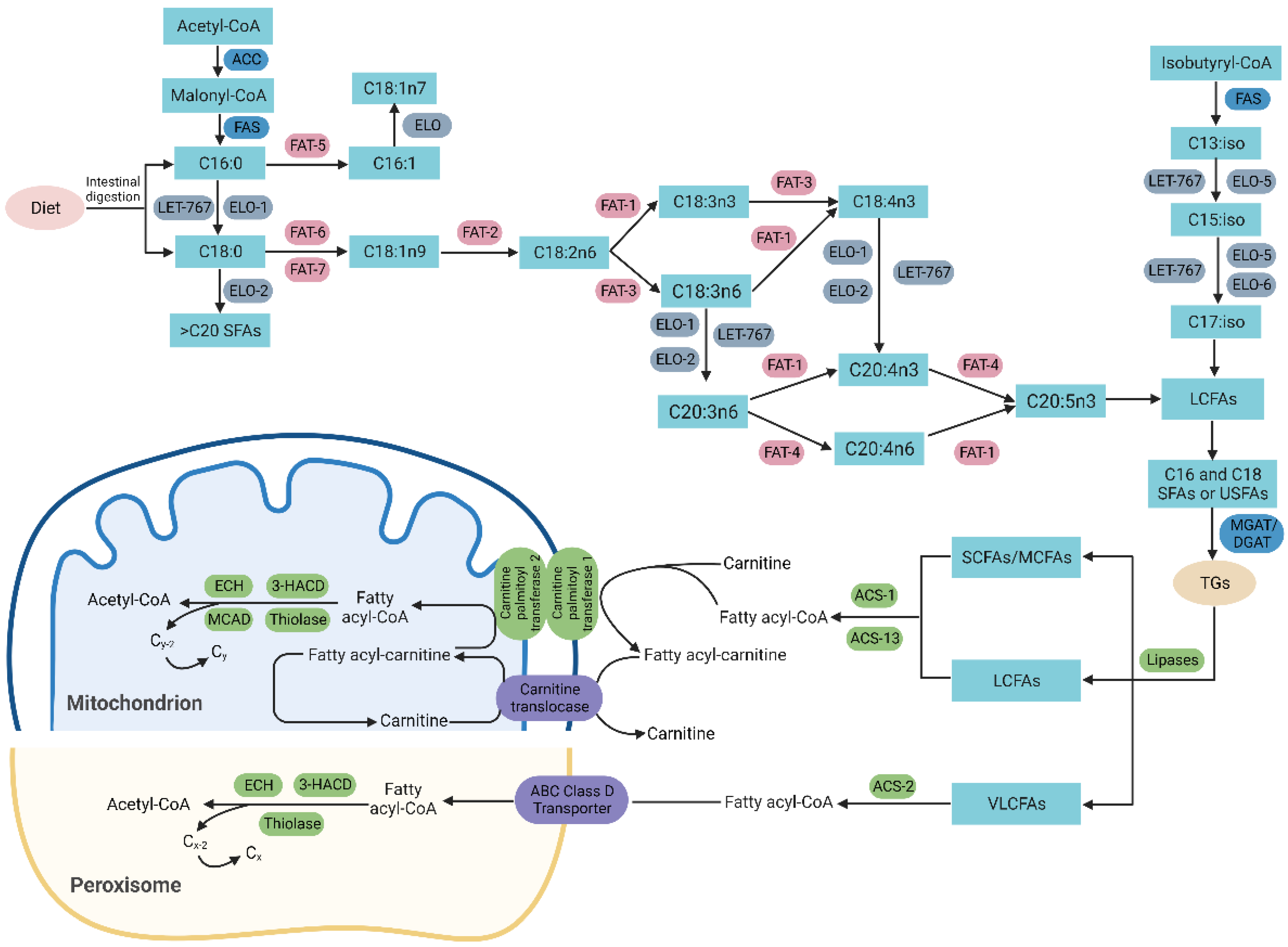
3.2. Lipogenesis
3.3. Lipolysis
4. Conserved Fat Metabolism Signaling Pathways in C. elegans
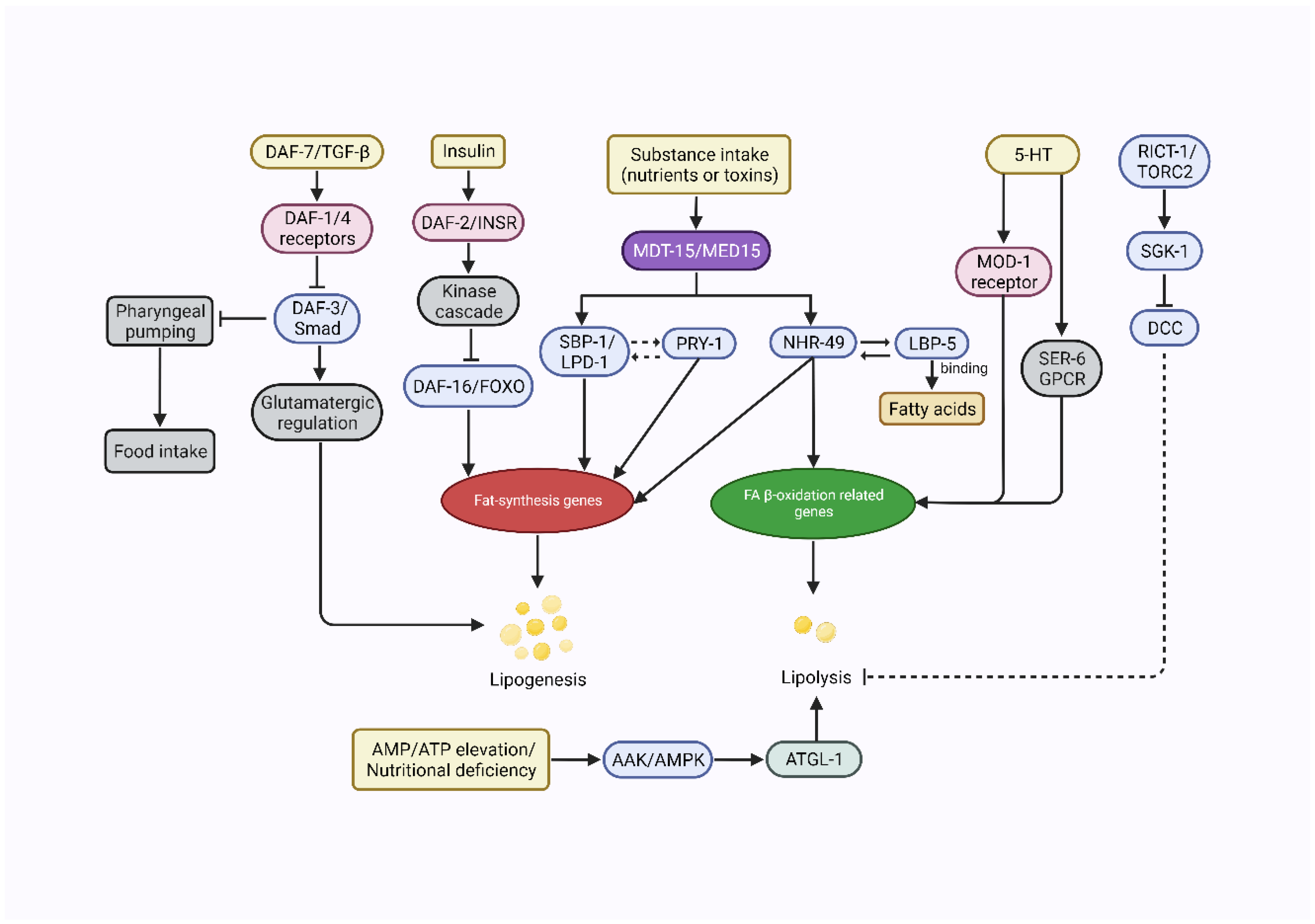
4.1. Regulation of Nematode Adiposity Levels by Neuroendocrine Signaling Pathways
4.1.1. 5-HTergic Fat Regulation
4.1.2. Modulation of Fat by Neuropeptides Unrelated to 5-HT
4.2. Insulin/Insulin-like Growth Factor (IGF)-1 Signaling (IIS) Pathway
4.3. DAF-7/TGF-β-like Signaling
4.4. TOR
4.5. AMPK
4.6. Transcriptional Regulation of Lipid Metabolism in C. elegans
4.6.1. NHR
4.6.2. LPD
4.6.3. MDT-15
4.7. PRY-1/Axin Signaling
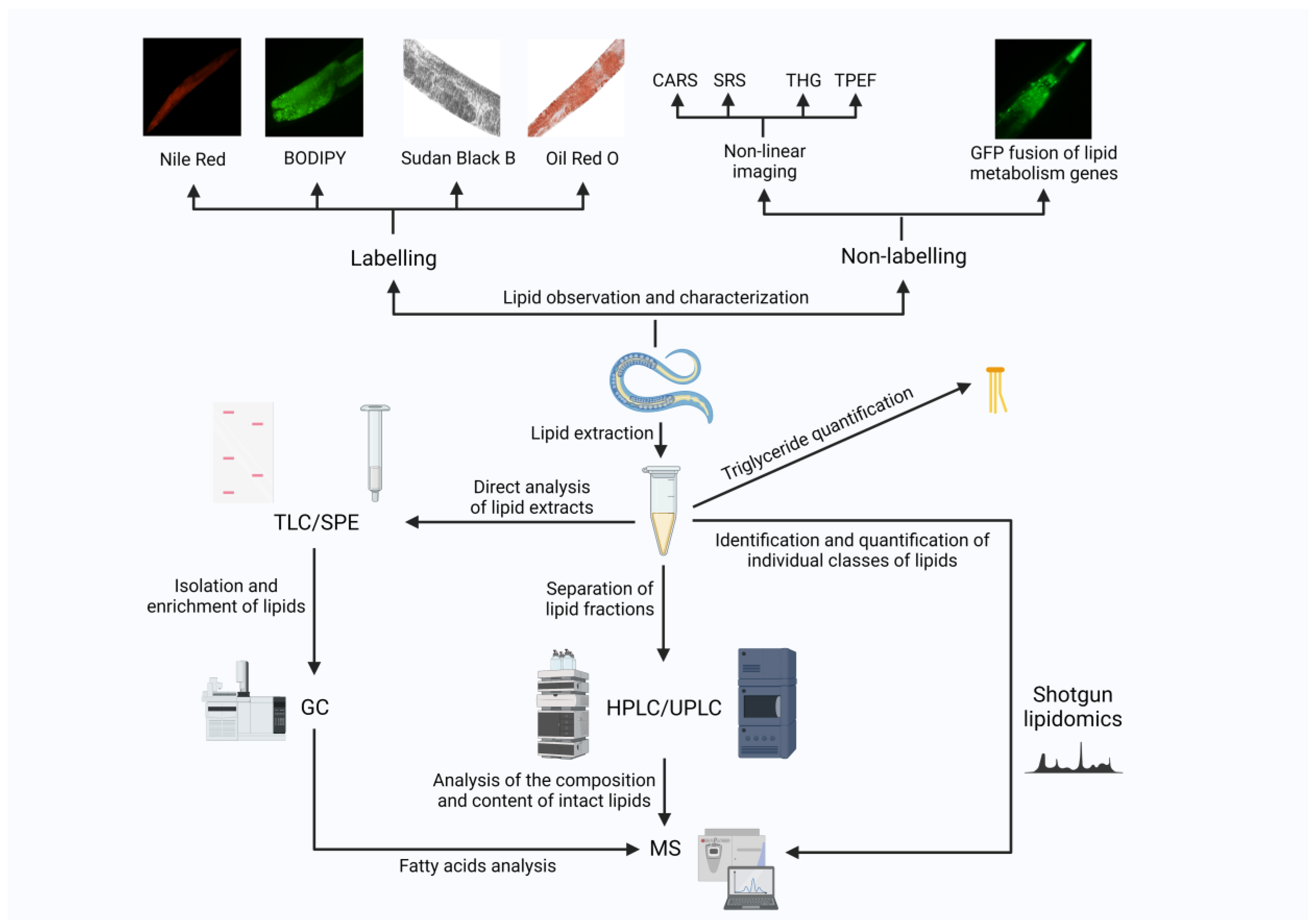
5. Experimental Tools for the Studies of Fat Metabolism in C. elegans
5.1. Staining-Based Methods to Characterize Lipid Storage in C. elegans
5.1.1. Vital Dye (Non-Fixative) Staining Analysis
5.1.2. Fixative-Based Assays
5.2. Label-Free Visual Imaging
5.2.1. Non-Linear Imaging Techniques
5.2.2. LD-Associated GFP Reporter-Based Assay
5.3. Biochemical and Chemical Assay
5.3.1. TG Quantification in C. elegans
5.3.2. TLC
5.3.3. GC
5.3.4. Mass Spectrometry-Based Methods
6. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- OECD; World Health Organization. Overweight and Obesity; OECD: Paris, France, 2020. [Google Scholar]
- Kopelman, P.G. Obesity as a medical problem. Nature 2000, 404, 635–643. [Google Scholar] [CrossRef]
- Piché, M.-E.; Tchernof, A.; Després, J.-P. Obesity Phenotypes, Diabetes, and Cardiovascular Diseases. Circ. Res. 2020, 126, 1477–1500. [Google Scholar] [CrossRef]
- Kaminsky, L.A.; German, C.; Imboden, M.; Ozemek, C.; Peterman, J.E.; Brubaker, P.H. The importance of healthy lifestyle behaviors in the prevention of cardiovascular disease. Prog. Cardiovasc. Dis. 2022, 70, 8–15. [Google Scholar] [CrossRef]
- Piché, M.-E.; Poirier, P.; Lemieux, I.; Després, J.-P. Overview of Epidemiology and Contribution of Obesity and Body Fat Distribution to Cardiovascular Disease: An Update. Prog. Cardiovasc. Dis. 2018, 61, 103–113. [Google Scholar] [CrossRef]
- Koliaki, C.; Liatis, S.; Kokkinos, A. Obesity and cardiovascular disease: Revisiting an old relationship. Metabolism 2019, 92, 98–107. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef]
- Gregg, E.W.; Shaw, J.E. Global Health Effects of Overweight and Obesity. N. Engl. J. Med. 2017, 377, 80–81. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Li, C.; Yuan, J.; Wang, Z. Expression of a Buckwheat Trypsin Inhibitor Gene in Escherichia coli and its Effect on Multiple Myeloma IM-9 Cell Proliferation. Acta Biochim. Biophys. Sin. 2007, 39, 701–707. [Google Scholar] [CrossRef]
- Lawrence, V.J.; Kopelman, P.G. Medical consequences of obesity. Clin. Dermatol. 2004, 22, 296–302. [Google Scholar] [CrossRef]
- Kaletta, T.; Hengartner, M. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006, 5, 387–399. [Google Scholar] [CrossRef]
- Baumeister, R.; Ge, L. The worm in us—Caenorhabditis elegans as a model of human disease. Trends Biotechnol. 2002, 20, 147–148. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, X.; Ding, Y.; Wang, H.; Wu, X.; Liang, B. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC Genom. 2013, 14, 164. [Google Scholar] [CrossRef]
- Watts, J.L. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009, 20, 58–65. [Google Scholar] [CrossRef]
- Qadota, H.; Inoue, M.; Hikita, T.; Köppen, M.; Hardin, J.D.; Amano, M.; Moerman, D.G.; Kaibuchi, K. Establishment of a tissue-specific RNAi system in C. elegans. Gene 2007, 400, 166–173. [Google Scholar] [CrossRef]
- Shen, P.; Yue, Y.; Zheng, J.; Park, Y. Caenorhabditis elegans: A Convenient In Vivo Model for Assessing the Impact of Food Bioactive Compounds on Obesity, Aging, and Alzheimer’s Disease. Annu. Rev. Food Sci. Technol. 2018, 9, 1–22. [Google Scholar] [CrossRef]
- Srinivasan, S. Regulation of Body Fat in Caenorhabditis elegans. Annu. Rev. Physiol. 2015, 77, 161–178. [Google Scholar] [CrossRef]
- Li, Y.; Ding, W.; Li, C.-Y.; Liu, Y. HLH-11 modulates lipid metabolism in response to nutrient availability. Nat. Commun. 2020, 11, 5959. [Google Scholar] [CrossRef]
- Xu, M.; Joo, H.-J.; Paik, Y.-K. Novel Functions of Lipid-binding Protein 5 in Caenorhabditis elegans Fat Metabolism. J. Biol. Chem. 2011, 286, 28111–28118. [Google Scholar] [CrossRef]
- Shen, P.; Yue, Y.; Park, Y. A living model for obesity and aging research: Caenorhabditis elegans. Crit. Rev. Food Sci. Nutr. 2018, 58, 741–754. [Google Scholar] [CrossRef]
- Wang, Y.; Xiang, Y.-F.; Liu, A.-L. Comparative and Combined Effects of Epigallocatechin-3-gallate and Caffeine in Reducing Lipid Accumulation in Caenorhabditis elegans. Plant Foods Hum. Nutr. 2022, 77, 279–285. [Google Scholar] [CrossRef]
- Kassotis, C.D.; vom Saal, F.S.; Babin, P.J.; Lagadic-Gossmann, D.; Le Mentec, H.; Blumberg, B.; Mohajer, N.; Legrand, A.; Kos, V.M.; Martin-Chouly, C.; et al. Obesity III: Obesogen assays: Limitations, strengths, and new directions. Biochem. Pharmacol. 2022, 199, 115014. [Google Scholar] [CrossRef]
- Qi, W.; Gutierrez, G.E.; Gao, X.; Dixon, H.; McDonough, J.A.; Marini, A.M.; Fisher, A.L. The ω-3 fatty acid α-linolenic acid extends Caenorhabditis elegans lifespan via NHR-49/PPARα and oxidation to oxylipins. Aging Cell 2017, 16, 1125–1135. [Google Scholar] [CrossRef]
- Fang, B.; Zhang, M.; Ren, F.Z.; Zhou, X.D. Lifelong diet including common unsaturated fatty acids extends the lifespan and affects oxidation in Caenorhabditis elegans consistently with hormesis model. Eur. J. Lipid Sci. Technol. 2016, 118, 1084–1092. [Google Scholar] [CrossRef]
- Cao, Z.; Hao, Y.; Fung, C.W.; Lee, Y.Y.; Wang, P.; Li, X.; Xie, K.; Lam, W.J.; Qiu, Y.; Tang, B.Z.; et al. Dietary fatty acids promote lipid droplet diversity through seipin enrichment in an ER subdomain. Nat. Commun. 2019, 10, 2902. [Google Scholar] [CrossRef]
- Bai, X.; Huang, L.-J.; Chen, S.-W.; Nebenfuhr, B.; Wysolmerski, B.; Wu, J.-C.; Olson, S.K.; Golden, A.; Wang, C.-W. Loss of the seipin gene perturbs eggshell formation in C. elegans. Development 2020, 147, dev.192997. [Google Scholar] [CrossRef]
- Anderson, S.M.; Cheesman, H.K.; Peterson, N.D.; Salisbury, J.E.; Soukas, A.A.; Pukkila-Worley, R. The fatty acid oleate is required for innate immune activation and pathogen defense in Caenorhabditis elegans. PLoS Pathog. 2019, 15, e1007893. [Google Scholar] [CrossRef]
- Lynn, D.A.; Dalton, H.M.; Sowa, J.N.; Wang, M.C.; Soukas, A.A.; Curran, S.P. Omega-3 and -6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2015, 112, 15378–15383. [Google Scholar] [CrossRef]
- Nhan, J.D.; Turner, C.D.; Anderson, S.M.; Yen, C.-A.; Dalton, H.M.; Cheesman, H.K.; Ruter, D.L.; Naresh, N.U.; Haynes, C.M.; Soukas, A.A.; et al. Redirection of SKN-1 abates the negative metabolic outcomes of a perceived pathogen infection. Proc. Natl. Acad. Sci. USA 2019, 116, 22322–22330. [Google Scholar] [CrossRef]
- Schmeisser, S.; Li, S.; Bouchard, B.; Ruiz, M.; Rosiers, C.D.; Roy, R. Muscle-Specific Lipid Hydrolysis Prolongs Lifespan through Global Lipidomic Remodeling. Cell Rep. 2019, 29, 4540–4552.e8. [Google Scholar] [CrossRef]
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef]
- Walker, G.; Houthoofd, K.; Vanfleteren, J.R.; Gems, D. Dietary restriction in C. elegans: From rate-of-living effects to nutrient sensing pathways. Mech. Ageing Dev. 2005, 126, 929–937. [Google Scholar] [CrossRef]
- Salzer, L.; Witting, M. Quo Vadis Caenorhabditis elegans Metabolomics—A Review of Current Methods and Applications to Explore Metabolism in the Nematode. Metabolites 2021, 11, 284. [Google Scholar] [CrossRef]
- Mak, H.Y. Lipid droplets as fat storage organelles in Caenorhabditis elegans. J. Lipid Res. 2012, 53, 28–33. [Google Scholar] [CrossRef]
- Farese, R.V.; Walther, T.C. Lipid Droplets Finally Get a Little R-E-S-P-E-C-T. Cell 2009, 139, 855–860. [Google Scholar] [CrossRef]
- Walther, T.C.; Chung, J.; Farese, R.V., Jr. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 2017, 33, 491–510. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, S.O.; Cole, R.A.; McKinney, S.A.; Guo, F.; Haas, J.T.; Bobba, S.; Farese, R.V., Jr.; Mak, H.Y. The FATP1–DGAT2 complex facilitates lipid droplet expansion at the ER–lipid droplet interface. J. Cell Biol. 2012, 198, 895–911. [Google Scholar] [CrossRef]
- Jackson, C.L. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2019, 59, 88–96. [Google Scholar] [CrossRef]
- Jin, Y.; Tan, Y.; Zhao, P.; Ren, Z. SEIPIN: A Key Factor for Nuclear Lipid Droplet Generation and Lipid Homeostasis. Int. J. Mol. Sci. 2020, 21, 8208. [Google Scholar] [CrossRef]
- Yan, R.; Qian, H.; Lukmantara, I.; Gao, M.; Du, X.; Yan, N.; Yang, H. Human SEIPIN Binds Anionic Phospholipids. Dev. Cell 2018, 47, 248–256.e4. [Google Scholar] [CrossRef]
- Sui, X.; Arlt, H.; Brock, K.P.; Lai, Z.W.; DiMaio, F.; Marks, D.S.; Liao, M.; Farese, R.V.; Walther, T.C. Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. J. Cell Biol. 2018, 217, 4080–4091. [Google Scholar] [CrossRef]
- Beller, M.; Sztalryd, C.; Southall, N.; Bell, M.; Jäckle, H.; Auld, D.S.; Oliver, B. COPI Complex Is a Regulator of Lipid Homeostasis. PLoS Biol. 2008, 6, e292. [Google Scholar] [CrossRef] [PubMed]
- Salo, V.T.; Ikonen, E. Moving out but keeping in touch: Contacts between endoplasmic reticulum and lipid droplets. Curr. Opin. Cell Biol. 2019, 57, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, J.V.; Bacher, M.C.; Priess, J.R. Nuclear lipid droplets and nuclear damage in Caenorhabditis elegans. PLoS Genet. 2021, 17, e1009602. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Liu, Y.; Li, X.; Zhang, H.; Zhang, S.; Mak, H.Y.; Liu, P. Dietary S. maltophilia induces supersized lipid droplets by enhancing lipogenesis and ER-LD contacts in C. elegans. Gut Microbes 2022, 14, 2013762. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Y.; Wang, Y.; Fu, L.; Xu, X.; Li, C.; Xu, J.; Li, C.; Zhang, L.; Yang, R.; et al. mmBCFA C17iso ensures endoplasmic reticulum integrity for lipid droplet growth. J. Cell Biol. 2021, 220, e202102122. [Google Scholar] [CrossRef]
- Ehmke, M.; Luthe, K.; Schnabel, R.; Döring, F. S-Adenosyl methionine synthetase 1 limits fat storage in Caenorhabditis elegans. Genes Nutr. 2014, 9, 386. [Google Scholar] [CrossRef]
- Li, S.; Xu, S.; Ma, Y.; Wu, S.; Feng, Y.; Cui, Q.; Chen, L.; Zhou, S.; Kong, Y.; Zhang, X.; et al. A Genetic Screen for Mutants with Supersized Lipid Droplets in Caenorhabditis elegans. G3 Genes Genomes Genet. 2016, 6, 2407–2419. [Google Scholar] [CrossRef]
- Zhang, S.O.; Box, A.C.; Xu, N.; Le Men, J.; Yu, J.; Guo, F.; Trimble, R.; Mak, H.Y. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2010, 107, 4640–4645. [Google Scholar] [CrossRef]
- Ashrafi, K. Obesity and the regulation of fat metabolism. WormBook 2007, 1–20. [Google Scholar] [CrossRef]
- Watts, J.L.; Browse, J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2002, 99, 5854–5859. [Google Scholar] [CrossRef]
- Elle, I.C.; Olsen, L.C.B.; Pultz, D.; Rødkaer, S.V.; Faergeman, N.J. Something worth dyeing for: Molecular tools for the dissection of lipid metabolism in Caenorhabditis elegans. FEBS Lett. 2010, 584, 2183–2193. [Google Scholar] [CrossRef]
- Brock, T.; Browse, J.; Watts, J.L. Genetic Regulation of Unsaturated Fatty Acid Composition in C. elegans. PLoS Genet. 2006, 2, e108. [Google Scholar] [CrossRef] [PubMed]
- Schenkman, J.B.; Jansson, I. The many roles of cytochrome b5. Pharmacol. Ther. 2003, 97, 139–152. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Zhang, J.; Hu, Y.; Zhang, L.; Wu, X.; Su, X.; Li, T.; Zou, X.; Liang, B. The cytochrome b5 reductase HPO-19 is required for biosynthesis of polyunsaturated fatty acids in Caenorhabditis elegans. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2016, 1861, 310–319. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhang, J.; Wang, Y.; Li, Y.; Zou, X.; Liang, B. Identification of cytochrome b5 CYTB-5.1 and CYTB-5.2 in C. elegans; evidence for differential regulation of SCD. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2018, 1863, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, X.; Li, Y.; Zhu, T.; Zhang, J.; Zhang, Z.; Zhang, L.; Zhang, Y.; Wang, Y.; Zou, X.; et al. PHA-4/FoxA senses nucleolar stress to regulate lipid accumulation in Caenorhabditis elegans. Nat. Commun. 2018, 9, 1195. [Google Scholar] [CrossRef]
- Dixit, A.; Sandhu, A.; Modi, S.; Shashikanth, M.; Koushika, S.P.; Watts, J.L.; Singh, V. Neuronal control of lipid metabolism by STR-2 G protein-coupled receptor promotes longevity in Caenorhabditis elegans. Aging Cell 2020, 19, e13160. [Google Scholar] [CrossRef]
- Abu-Salah, K. Metabolism at a Glance, 2nd ed.; Salway, J.G., Ed.; Blackwell Science: Oxford, UK, 1999; 111p. [Google Scholar]
- Schweiger, M.; Schreiber, R.; Haemmerle, G.; Lass, A.; Fledelius, C.; Jacobsen, P.; Tornqvist, H.; Zechner, R.; Zimmermann, R. Adipose Triglyceride Lipase and Hormone-sensitive Lipase Are the Major Enzymes in Adipose Tissue Triacylglycerol Catabolism. J. Biol. Chem. 2006, 281, 40236–40241. [Google Scholar] [CrossRef]
- Prentki, M.; Madiraju, S.R.M. Glycerolipid Metabolism and Signaling in Health and Disease. Endocr. Rev. 2008, 29, 647–676. [Google Scholar] [CrossRef]
- Mugabo, Y.; Zhao, S.; Seifried, A.; Gezzar, S.; Al-Mass, A.; Zhang, D.; Lamontagne, J.; Attane, C.; Poursharifi, P.; Iglesias, J.; et al. Identification of a mammalian glycerol-3-phosphate phosphatase: Role in metabolism and signaling in pancreatic β-cells and hepatocytes. Proc. Natl. Acad. Sci. USA 2016, 113, E430–E439. [Google Scholar] [CrossRef]
- Possik, E.; Schmitt, C.; Al-Mass, A.; Bai, Y.; Côté, L.; Morin, J.; Erb, H.; Oppong, A.; Kahloan, W.; Parker, J.A.; et al. Phosphoglycolate phosphatase homologs act as glycerol-3-phosphate phosphatase to control stress and healthspan in C. elegans. Nat. Commun. 2022, 13, 177. [Google Scholar] [CrossRef]
- O’Rourke, E.J.; Kuballa, P.; Xavier, R.; Ruvkun, G. ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013, 27, 429–440. [Google Scholar] [CrossRef]
- Lapierre, L.R.; Gelino, S.; Meléndez, A.; Hansen, M. Autophagy and Lipid Metabolism Coordinately Modulate Life Span in Germline-less C. elegans. Curr. Biol. 2011, 21, 1507–1514. [Google Scholar] [CrossRef]
- Kudron, M.M.; Victorsen, A.; Gevirtzman, L.; Hillier, L.W.; Fisher, W.W.; Vafeados, D.; Kirkey, M.; Hammonds, A.S.; Gersch, J.; Ammouri, H.; et al. The ModERN Resource: Genome-Wide Binding Profiles for Hundreds of Drosophila and Caenorhabditis elegans Transcription Factors. Genetics 2018, 208, 937–949. [Google Scholar] [CrossRef]
- Srinivasan, S. Neuroendocrine control of lipid metabolism: Lessons from C. elegans. J. Neurogenet. 2020, 34, 482–488. [Google Scholar] [CrossRef]
- Riera, C.E.; Tsaousidou, E.; Halloran, J.; Follett, P.; Hahn, O.; Pereira, M.M.; Ruud, L.E.; Alber, J.; Tharp, K.; Anderson, C.M.; et al. The Sense of Smell Impacts Metabolic Health and Obesity. Cell Metab. 2017, 26, 198–211.e5. [Google Scholar] [CrossRef]
- Moon, J.H.; Kim, Y.G.; Kim, K.; Osonoi, S.; Wang, S.; Saunders, D.C.; Wang, J.; Yang, K.; Kim, H.; Lee, J.; et al. Serotonin Regulates Adult β-Cell Mass by Stimulating Perinatal β-Cell Proliferation. Diabetes 2020, 69, 205–214. [Google Scholar] [CrossRef]
- Rozenblit-Susan, S.; Chapnik, N.; Froy, O. Serotonin prevents differentiation into brown adipocytes and induces transdifferentiation into white adipocytes. Int. J. Obes. 2018, 42, 704–710. [Google Scholar] [CrossRef]
- Choi, W.; Namkung, J.; Hwang, I.; Kim, H.; Lim, A.; Park, H.J.; Lee, H.W.; Han, K.-H.; Park, S.; Jeong, J.-S.; et al. Serotonin signals through a gut-liver axis to regulate hepatic steatosis. Nat. Commun. 2018, 9, 4824. [Google Scholar] [CrossRef]
- Duerschmied, D.; Schoenichen, C.; Bode, C. Role of platelet serotonin in innate immune cell recruitment. Front. Biosci. 2019, 24, 514–526. [Google Scholar] [CrossRef]
- Zhong, X.; Gu, J.; Zhang, S.; Chen, X.; Zhang, J.; Miao, J.; Ding, Z.; Xu, J.; Cheng, H. Dynamic transcriptome analysis of the muscles in high-fat diet-induced obese zebrafish (Danio rerio) under 5-HT treatment. Gene 2022, 819, 146265. [Google Scholar] [CrossRef]
- Sze, J.Y.; Victor, M.; Loer, C.; Shi, Y.; Ruvkun, G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 2000, 403, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, K.; Chang, F.Y.; Watts, J.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Ruvkun, G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003, 421, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Horvitz, H.R.; Chalfie, M.; Trent, C.; Sulston, J.E.; Evans, P.D. Serotonin and Octopamine in the Nematode Caenorhabditis elegans. Science 1982, 216, 1012–1014. [Google Scholar] [CrossRef]
- Noble, T.; Stieglitz, J.; Srinivasan, S. An Integrated Serotonin and Octopamine Neuronal Circuit Directs the Release of an Endocrine Signal to Control C. elegans Body Fat. Cell Metab. 2013, 18, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Ishita, Y.; Chihara, T.; Okumura, M. Serotonergic modulation of feeding behavior in Caenorhabditis elegans and other related nematodes. Neurosci. Res. 2020, 154, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Sadegh, L.; Elle, I.C.; Christensen, A.G.; Faergeman, N.J.; Ashrafi, K. Serotonin Regulates C. elegans Fat and Feeding through Independent Molecular Mechanisms. Cell Metab. 2008, 7, 533–544. [Google Scholar] [CrossRef]
- Palamiuc, L.; Noble, T.; Witham, E.; Ratanpal, H.; Vaughan, M.; Srinivasan, S. A tachykinin-like neuroendocrine signalling axis couples central serotonin action and nutrient sensing with peripheral lipid metabolism. Nat. Commun. 2017, 8, 14237. [Google Scholar] [CrossRef]
- Cunningham, K.A.; Hua, Z.; Srinivasan, S.; Liu, J.; Lee, B.H.; Edwards, R.H.; Ashrafi, K. AMP-Activated Kinase Links Serotonergic Signaling to Glutamate Release for Regulation of Feeding Behavior in C. elegans. Cell Metab. 2012, 16, 113–121. [Google Scholar] [CrossRef]
- Liu, M.; Gao, X.; Shan, S.; Li, Y.; Wang, J.; Lu, W. Eleutheroside E reduces intestinal fat accumulation in Caenorhabditis elegans through neuroendocrine signals. J. Sci. Food Agric. 2022, 102, 5219–5228. [Google Scholar] [CrossRef]
- Mutlu, A.S.; Gao, S.M.; Zhang, H.; Wang, M.C. Olfactory specificity regulates lipid metabolism through neuroendocrine signaling in Caenorhabditis elegans. Nat. Commun. 2020, 11, 1450. [Google Scholar] [CrossRef]
- Su, C.-Y.; Menuz, K.; Carlson, J.R. Olfactory Perception: Receptors, Cells, and Circuits. Cell 2009, 139, 45–59. [Google Scholar] [CrossRef]
- Li, C. Neuropeptides. Wormbook 2008, 1–36. [Google Scholar] [CrossRef]
- Hussey, R.; Littlejohn, N.K.; Witham, E.; Vanstrum, E.; Mesgarzadeh, J.; Ratanpal, H.; Srinivasan, S. Oxygen-sensing neurons reciprocally regulate peripheral lipid metabolism via neuropeptide signaling in Caenorhabditis elegans. PLoS Genet. 2018, 14, e1007305. [Google Scholar] [CrossRef]
- Kang, C. The FMRFamide Neuropeptide FLP-20 Acts as a Systemic Signal for Starvation Responses in Caenorhabditis elegans. Mol. Cells 2021, 44, 529–537. [Google Scholar] [CrossRef]
- Zheng, S.; Chiu, H.; Boudreau, J.; Papanicolaou, T.; Bendena, W.; Chin-Sang, I. A functional study of all 40 Caenorhabditis elegans insulin-like peptides. J. Biol. Chem. 2018, 293, 16912–16922. [Google Scholar] [CrossRef]
- Narasimhan, S.D.; Mukhopadhyay, A.; Tissenbaum, H.A. InAKTivation of insulin/IGF-1 signaling by dephosphorylation. Cell Cycle 2009, 8, 3878–3884. [Google Scholar] [CrossRef]
- Arcucci, S.; Ramos-Delgado, F.; Cayron, C.; Therville, N.; Gratacap, M.-P.; Basset, C.; Thibault, B.; Guillermet-Guibert, J. Organismal roles for the PI3Kα and β isoforms: Their specificity, redundancy or cooperation is context-dependent. Biochem. J. 2021, 478, 1199–1225. [Google Scholar] [CrossRef]
- Padmanabhan, S.; Mukhopadhyay, A.; Narasimhan, S.D.; Tesz, G.; Czech, M.P.; Tissenbaum, H.A. A PP2A Regulatory Subunit Regulates C. elegans Insulin/IGF-1 Signaling by Modulating AKT-1 Phosphorylation. Cell 2009, 136, 939–951. [Google Scholar] [CrossRef]
- Perez, C.L.; Van Gilst, M.R. A 13C Isotope Labeling Strategy Reveals the Influence of Insulin Signaling on Lipogenesis in C. elegans. Cell Metab. 2008, 8, 266–274. [Google Scholar] [CrossRef]
- Kimura, K.; Riddle, D.L.; Ruvkun, G. The C. elegans DAF-2 Insulin-Like Receptor is Abundantly Expressed in the Nervous System and Regulated by Nutritional Status. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 113–120. [Google Scholar] [CrossRef]
- Kimura, K.D.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. daf-2, an Insulin Receptor-Like Gene That Regulates Longevity and Diapause in Caenorhabditis elegans. Science 1997, 277, 942–946. [Google Scholar] [CrossRef]
- Murphy, C.T.; Hu, P.J. Insulin/insulin-like growth factor signaling in C. elegans. WormBook 2013, 1–43. [Google Scholar] [CrossRef]
- Mutlu, A.S.; Chen, T.; Deng, D.; Wang, M.C. Label-Free Imaging of Lipid Storage Dynamics in Caenorhabditis elegans using Stimulated Raman Scattering Microscopy. J. Vis. Exp. 2021, 171. [Google Scholar] [CrossRef]
- Horikawa, M.; Sakamoto, K. Polyunsaturated fatty acids are involved in regulatory mechanism of fatty acid homeostasis via daf-2/insulin signaling in Caenorhabditis elegans. Mol. Cell. Endocrinol. 2010, 323, 183–192. [Google Scholar] [CrossRef]
- Chen, Y.; Baugh, L.R. Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev. Biol. 2014, 394, 314–326. [Google Scholar] [CrossRef]
- Kaplan, R.E.W.; Webster, A.K.; Chitrakar, R.; Dent, J.A.; Baugh, L.R. Food perception without ingestion leads to metabolic changes and irreversible developmental arrest in C. elegans. BMC Biol. 2018, 16, 112. [Google Scholar] [CrossRef]
- Clark, J.F.; Ciccarelli, E.J.; Kayastha, P.; Ranepura, G.; Yamamoto, K.K.; Hasan, M.S.; Madaan, U.; Meléndez, A.; Savage-Dunn, C. BMP pathway regulation of insulin signaling components promotes lipid storage in Caenorhabditis elegans. PLoS Genet. 2021, 17, e1009836. [Google Scholar] [CrossRef]
- Wang, K.; Chen, S.; Zhang, C.; Huang, J.; Wu, J.; Zhou, H.; Jin, L.; Qian, X.; Jin, J.; Lyu, J. Enhanced ROS production leads to excessive fat accumulation through DAF-16 in Caenorhabditis elegans. Exp. Gerontol. 2018, 112, 20–29. [Google Scholar] [CrossRef]
- Yavorov-Dayliev, D.; Milagro, F.I.; Ayo, J.; Oneca, M.; Aranaz, P. Pediococcus acidilactici CECT9879 (pA1c) Counteracts the Effect of a High-Glucose Exposure in C. elegans by Affecting the Insulin Signaling Pathway (IIS). Int. J. Mol. Sci. 2022, 23, 2689. [Google Scholar] [CrossRef]
- Xiong, L.-G.; Pan, L.-Y.; Gong, Y.-S.; Huang, J.-A.; Liu, Z.-H. Fuzhuan Tea protects Caenorhabditis elegans from glucose and advanced glycation end products via distinct pathways. J. Funct. Foods 2019, 59, 148–155. [Google Scholar] [CrossRef]
- Aranaz, P.; Peña, A.; Vettorazzi, A.; Fabra, M.J.; Martínez-Abad, A.; López-Rubio, A.; Pera, J.; Parladé, J.; Castellari, M.; Milagro, F.I.; et al. Grifola frondosa (Maitake) Extract Reduces Fat Accumulation and Improves Health Span in C. elegans through the DAF-16/FOXO and SKN-1/NRF2 Signalling Pathways. Nutrients 2021, 13, 3968. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, G.A.; Ashrafi, K.; Richardson, S.R.; Morell, S.; Faulkner, G.J.; Owusu-Ansah, E.; Perrimon, N.; Zhu, H.; Han, M.; Nguyen, H.Q.; et al. Neural Regulatory Pathways of Feeding and Fat in Caenorhabditis elegans. Annu. Rev. Genet. 2015, 49, 413–438. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.R.; Pérez, C.L.; Van Gilst, M.R.; Lee, B.H.; Ashrafi, K. Neural and Molecular Dissection of a C. elegans Sensory Circuit that Regulates Fat and Feeding. Cell Metab. 2008, 8, 118–131. [Google Scholar] [CrossRef]
- You, Y.-J.; Kim, J.; Raizen, D.M.; Avery, L. Insulin, cGMP, and TGF-β Signals Regulate Food Intake and Quiescence in C. elegans: A Model for Satiety. Cell Metab. 2008, 7, 249–257. [Google Scholar] [CrossRef]
- Ben Arous, J.; Laffont, S.; Chatenay, D. Molecular and Sensory Basis of a Food Related Two-State Behavior in C. elegans. PLoS ONE 2009, 4, e7584. [Google Scholar] [CrossRef]
- Gadde, K.M.; Martin, C.K.; Berthoud, H.-R.; Heymsfield, S.B. Obesity. J. Am. Coll. Cardiol. 2018, 71, 69–84. [Google Scholar] [CrossRef]
- Luo, Z.; Yu, Z.; Yin, D. Obesogenic effect of erythromycin on Caenorhabditis elegans through over-eating and lipid metabolism disturbances. Environ. Pollut. 2022, 294, 118615. [Google Scholar] [CrossRef]
- Narasimhan, S.D.; Yen, K.; Bansal, A.; Kwon, E.-S.; Padmanabhan, S.; Tissenbaum, H.A. PDP-1 Links the TGF-β and IIS Pathways to Regulate Longevity, Development, and Metabolism. PLoS Genet. 2011, 7, e1001377. [Google Scholar] [CrossRef]
- Jones, K.T.; Greer, E.R.; Pearce, D.; Ashrafi, K. Rictor/TORC2 Regulates Caenorhabditis elegans Fat Storage, Body Size, and Development through sgk-1. PLoS Biol. 2009, 7, e1000060. [Google Scholar] [CrossRef]
- Soukas, A.A.; Kane, E.A.; Carr, C.E.; Melo, J.A.; Ruvkun, G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009, 23, 496–511. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, M.; Gobel, C.; Baumeister, R. C. elegans SGK-1 Is the Critical Component in the Akt/PKB Kinase Complex to Control Stress Response and Life Span. Dev. Cell 2004, 6, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kaur, A.; Mallory, B.; Hariri, S.; Engebrecht, J. Inducible degradation of dosage compensation protein DPY-27 facilitates isolation of Caenorhabditis elegans males for molecular and biochemical analyses. G3 Genes Genomes Genet. 2022, 12, jkac085. [Google Scholar] [CrossRef]
- Webster, C.M.; Wu, L.; Douglas, D.; Soukas, A.A. A non-canonical role for the C. elegans dosage compensation complex in growth and metabolic regulation downstream of TOR complex 2. Development 2013, 140, 3601–3612. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Breen, P.C.; Tullius, T.; Conery, A.L.; Ruvkun, G. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 2016, 30, 1515–1528. [Google Scholar] [CrossRef]
- O’Donnell, M.P.; Chao, P.-H.; Kammenga, J.E.; Sengupta, P. Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. PLoS Genet. 2018, 14, e1007213. [Google Scholar] [CrossRef]
- Mair, W. Tipping the Energy Balance toward Longevity. Cell Metab. 2013, 17, 5–6. [Google Scholar] [CrossRef]
- Park, M.; Baek, H.; Han, J.-Y.; Lee, H.-J. Stevioside Enhances the Anti-Adipogenic Effect and β-Oxidation by Activating AMPK in 3T3-L1 Cells and Epididymal Adipose Tissues of db/db Mice. Cells 2022, 11, 1076. [Google Scholar] [CrossRef]
- Balamurugan, K.; Medishetti, R.; Kotha, J.; Behera, P.; Chandra, K.; Mavuduru, V.A.; Joshi, M.B.; Samineni, R.; Katika, M.R.; Ball, W.B.; et al. PHLPP1 promotes neutral lipid accumulation through AMPK/ChREBP-dependent lipid uptake and fatty acid synthesis pathways. iScience 2022, 25, 103766. [Google Scholar] [CrossRef]
- Narbonne, P.; Roy, R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 2009, 457, 210–214. [Google Scholar] [CrossRef]
- Xie, M.; Roy, R. AMP-Activated Kinase Regulates Lipid Droplet Localization and Stability of Adipose Triglyceride Lipase in C. elegans Dauer Larvae. PLoS ONE 2015, 10, e0130480. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.M.; Pino, E.C.; Carr, C.E.; Wu, L.; Zhou, B.; Cedillo, L.; Kacergis, M.C.; Curran, S.P.; Soukas, A.A. Genome-wide RNAi Screen for Fat Regulatory Genes in C. elegans Identifies a Proteostasis-AMPK Axis Critical for Starvation Survival. Cell Rep. 2017, 20, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, K.A.; Bouagnon, A.D.; Barros, A.G.; Lin, L.; Malard, L.; Romano-Silva, M.A.; Ashrafi, K. Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions. PLoS Genet. 2014, 10, e1004394. [Google Scholar] [CrossRef]
- Van Gilst, M.R.; Hadjivassiliou, H.; Jolly, A.; Yamamoto, K.R. Nuclear Hormone Receptor NHR-49 Controls Fat Consumption and Fatty Acid Composition in C. elegans. PLoS Biol. 2005, 3, e53. [Google Scholar] [CrossRef]
- Lee, J.M. Transcriptional coordination of hepatic autophagy by nutrient-sensing nuclear receptor PPARα and FXR. Ann. Pediatr. Endocrinol. Metab. 2016, 21, 193–198. [Google Scholar] [CrossRef]
- Ratnappan, R.; Amrit, F.; Chen, S.-W.; Gill, H.; Holden, K.; Ward, J.; Yamamoto, K.R.; Olsen, C.; Ghazi, A. Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of C. elegans. PLoS Genet. 2014, 10, e1004829. [Google Scholar] [CrossRef]
- Dasgupta, M.; Shashikanth, M.; Gupta, A.; Sandhu, A.; De, A.; Javed, S.; Singh, V. NHR-49 Transcription Factor Regulates Immunometabolic Response and Survival of Caenorhabditis elegans during Enterococcus faecalis Infection. Infect. Immun. 2020, 88, e00130-20. [Google Scholar] [CrossRef]
- Sim, S.; Hibberd, M.L. Caenorhabditis elegans susceptibility to gut Enterococcus faecalis infection is associated with fat metabolism and epithelial junction integrity. BMC Microbiol. 2016, 16, 6. [Google Scholar] [CrossRef]
- Xu, M.; Choi, E.Y.; Paik, Y.K. Mutation of the lbp-5 gene alters metabolic output in Caenorhabditis elegans. BMB Rep. 2014, 47, 15–20. [Google Scholar] [CrossRef]
- Osborne, T.F.; Espenshade, P.J. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: What a long, strange tRIP it’s been. Genes Dev. 2009, 23, 2578–2591. [Google Scholar] [CrossRef] [PubMed]
- Raghow, R.; Yellaturu, C.; Deng, X.; Park, E.A.; Elam, M.B. SREBPs: The crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol. Metab. 2008, 19, 65–73. [Google Scholar] [CrossRef]
- Nomura, T.; Horikawa, M.; Shimamura, S.; Hashimoto, T.; Sakamoto, K. Fat accumulation in Caenorhabditis elegans is mediated by SREBP homolog SBP-1. Genes Nutr. 2010, 5, 17–27. [Google Scholar] [CrossRef] [PubMed]
- McKay, R.M.; McKay, J.P.; Avery, L.; Graff, J.M. C. elegans: A Model for Exploring the Genetics of Fat Storage. Dev. Cell 2003, 4, 131–142. [Google Scholar] [CrossRef]
- Schroeder-Gloeckler, J.M.; Rahman, S.M.; Janssen, R.C.; Qiao, L.; Shao, J.; Roper, M.; Fischer, S.J.; Lowe, E.; Orlicky, D.J.; McManaman, J.L.; et al. CCAAT/Enhancer-binding Protein β Deletion Reduces Adiposity, Hepatic Steatosis, and Diabetes in Lepr Mice. J. Biol. Chem. 2007, 282, 15717–15729. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-Y.; Hu, J.-P.; Wu, M.-M.; Wang, L.-S.; Fang, N.-Y. CCAAT/enhancer-binding protein CEBP-2 controls fat consumption and fatty acid desaturation in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2015, 468, 312–318. [Google Scholar] [CrossRef]
- Yang, F.; Vought, B.W.; Satterlee, J.S.; Walker, A.K.; Sun, Z.-Y.J.; Watts, J.L.; DeBeaumont, R.; Saito, R.M.; Hyberts, S.G.; Yang, S.; et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 2006, 442, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Taubert, S.; Van Gilst, M.R.; Hansen, M.; Yamamoto, K.R. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006, 20, 1137–1149. [Google Scholar] [CrossRef]
- Liu, H.; Li, D.; Zhang, R.; Sun, L.; Wang, D. Lipid metabolic sensors of MDT-15 and SBP-1 regulated the response to simulated microgravity in the intestine of Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2020, 528, 28–34. [Google Scholar] [CrossRef]
- Taubert, S.; Hansen, M.; Van Gilst, M.R.; Cooper, S.B.; Yamamoto, K.R. The Mediator Subunit MDT-15 Confers Metabolic Adaptation to Ingested Material. PLoS Genet. 2008, 4, e1000021. [Google Scholar] [CrossRef]
- Lee, D.; Jeong, D.-E.; Son, H.G.; Yamaoka, Y.; Kim, H.; Seo, K.; Khan, A.A.; Roh, T.-Y.; Moon, D.W.; Lee, Y.; et al. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev. 2015, 29, 2490–2503. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Son, H.G.; Jung, Y.; Lee, S.-J.V. The role of dietary carbohydrates in organismal aging. Cell. Mol. Life Sci. 2017, 74, 1793–1803. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; An, S.W.A.; Jung, Y.; Yamaoka, Y.; Ryu, Y.; Goh, G.Y.S.; Beigi, A.; Yang, J.-S.; Jung, G.Y.; Ma, D.K.; et al. MDT-15/MED15 permits longevity at low temperature via enhancing lipidostasis and proteostasis. PLoS Biol. 2019, 17, e3000415. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Fagotto, F.; Zhang, T.; Hsu, W.; Vasicek, T.J.; Iii, W.L.P.; Lee, J.J.; Tilghman, S.M.; Gumbiner, B.M.; Costantini, F. The Mouse Fused Locus Encodes Axin, an Inhibitor of the Wnt Signaling Pathway That Regulates Embryonic Axis Formation. Cell 1997, 90, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Ranawade, A.; Mallick, A.; Gupta, B.P. PRY-1/Axin signaling regulates lipid metabolism in Caenorhabditis elegans. PLoS ONE 2018, 13, e0206540. [Google Scholar] [CrossRef]
- Mallick, A.; Gupta, B.P. Vitellogenin-2 acts downstream of PRY-1/Axin to regulate lipids and lifespan in C. elegans. Micropubl. Biol. 2020, 2020. [Google Scholar] [CrossRef]
- Bonilla, E.; Prelle, A. Application of nile blue and nile red, two fluorescent probes, for detection of lipid droplets in human skeletal muscle. J. Histochem. Cytochem. 1987, 35, 619–621. [Google Scholar] [CrossRef]
- Brown, W.J.; Sullivan, T.R.; Greenspan, P. Nile red staining of lysosomal phospholipid inclusions. Histochemistry 1992, 97, 349–354. [Google Scholar] [CrossRef]
- Kucherak, O.A.; Oncul, S.; Darwich, Z.; Yushchenko, D.A.; Arntz, Y.; Didier, P.; Mély, Y.; Klymchenko, A.S. Switchable Nile Red-Based Probe for Cholesterol and Lipid Order at the Outer Leaflet of Biomembranes. J. Am. Chem. Soc. 2010, 132, 4907–4916. [Google Scholar] [CrossRef]
- Escorcia, W.; Ruter, D.L.; Nhan, J.; Curran, S.P. Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. J. Vis. Exp. 2018, 133. [Google Scholar] [CrossRef]
- Zwirchmayr, J.; Kirchweger, B.; Lehner, T.; Tahir, A.; Pretsch, D.; Rollinger, J.M. A robust and miniaturized screening platform to study natural products affecting metabolism and survival in Caenorhabditis elegans. Sci. Rep. 2020, 10, 12323. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, J. Swertiamarin decreases lipid accumulation dependent on 3-ketoacyl-coA thiolase. Biomed. Pharmacother. 2019, 112, 108668. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, G.A.; Liu, J.; Mayer, N.; Bainton, R.J.; Ashrafi, K.; Werb, Z. A whole-organism screen identifies new regulators of fat storage. Nat. Chem. Biol. 2011, 7, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Charan, S.; Chien, F.-C.; Singh, N.; Kuo, C.-W.; Chen, P. Development of Lipid Targeting Raman Probes for In Vivo Imaging of Caenorhabditis elegans. Chem.-Eur. J. 2011, 17, 5165–5170. [Google Scholar] [CrossRef]
- Yen, K.; Le, T.T.; Bansal, A.; Narasimhan, S.D.; Cheng, J.-X.; Tissenbaum, H.A. A Comparative Study of Fat Storage Quantitation in Nematode Caenorhabditis elegans Using Label and Label-Free Methods. PLoS ONE 2010, 5, e12810. [Google Scholar] [CrossRef]
- Pino, E.C.; Webster, C.M.; Carr, C.E.; Soukas, A.A. Biochemical and High Throughput Microscopic Assessment of Fat Mass in Caenorhabditis Elegans. J. Vis. Exp. 2013, 73. [Google Scholar] [CrossRef]
- Zhang, X.; Li, W.; Tang, Y.; Lin, C.; Cao, Y.; Chen, Y. Mechanism of Pentagalloyl Glucose in Alleviating Fat Accumulation in Caenorhabditis elegans. J. Agric. Food Chem. 2019, 67, 14110–14120. [Google Scholar] [CrossRef]
- Li, C. Recombinant buckwheat trypsin inhibitor decreases fat accumulation via the IIS pathway in Caenorhabditis elegans. Exp. Gerontol. 2019, 128, 110753. [Google Scholar] [CrossRef]
- Navarro-Herrera, D.; Aranaz, P.; Eder-Azanza, L.; Zabala, M.; Romo-Hualde, A.; Hurtado, C.; Calavia, D.; López-Yoldi, M.; Martínez, J.A.; González-Navarro, C.J.; et al. Borago officinalis seed oil (BSO), a natural source of omega-6 fatty acids, attenuates fat accumulation by activating peroxisomal beta-oxidation both in C. elegans and in diet-induced obese rats. Food Funct. 2018, 9, 4340–4351. [Google Scholar] [CrossRef]
- Zhang, S.O.; Trimble, R.; Guo, F.; Mak, H.Y. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 2010, 11, 96. [Google Scholar] [CrossRef]
- Klapper, M.; Ehmke, M.; Palgunow, D.; Böhme, M.; Matthäus, C.; Bergner, G.; Dietzek, B.; Popp, J.; Döring, F. Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J. Lipid Res. 2011, 52, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Chang, H.; Zhang, P.; Chen, T.; Zhao, Y.; Zhang, Y.; Liu, P.; Xu, T.; Xu, P. C13C4.5/Spinster, an evolutionarily conserved protein that regulates fertility in C. elegans through a lysosome-mediated lipid metabolism process. Protein Cell 2013, 4, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Gubert, P.; Aguiar, G.C.; Mourão, T.; Bridi, J.C.; Barros, A.G.; Soares, F.A.; Romano-Silva, M.A. Behavioral and Metabolic Effects of the Atypical Antipsychotic Ziprasidone on the Nematode Caenorhabditis elegans. PLoS ONE 2013, 8, e74780. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.L.; Arantes, L.P.; Gubert, P.; Zamberlan, D.C.; da Silva, T.C.; da Silveira, T.L.; Boligon, A.; Soares, F.A.A. Ilex paraguariensis modulates fat metabolism in Caenorhabditis elegans through purinergic system (ADOR-1) and nuclear hormone receptor (NHR-49) pathways. PLoS ONE 2018, 13, e0204023. [Google Scholar] [CrossRef] [PubMed]
- Zanni, E.; Laudenzi, C.; Schifano, E.; Palleschi, C.; Perozzi, G.; Uccelletti, D.; Devirgiliis, C. Impact of a Complex Food Microbiota on Energy Metabolism in the Model Organism Caenorhabditis elegans. BioMed Res. Int. 2015, 2015, 621709. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Y.; Yu, C.; Liu, S.; Liu, Y.; Hao, E.; Jiao, L.; Xu, X.; Zhang, Z.; Li, J. A novel family of non-symmetric benzothieno [7,6-b]-fused BODIPYs: Synthesis, structures, photophysical properties and lipid droplet-specific imaging in vitro. Dye. Pigment. 2021, 196, 109748. [Google Scholar] [CrossRef]
- Alekseeva, A.S.; Tretiakova, D.S.; Melnikova, D.N.; Molotkovsky, U.G.; Boldyrev, I.A. Novel fluorescent membrane probe 2,3;5,6-bis(cyclohexyl)-BODIPY-labeled phosphatidylcholine. Russ. J. Bioorg. Chem. 2016, 42, 305–309. [Google Scholar] [CrossRef]
- Murale, D.P.; Haque, M.; Hong, K.T.; Lee, J. A Pyridinyl-Pyrazole BODIPY as Lipid Droplets Probe. Bull. Korean Chem. Soc. 2021, 42, 111–114. [Google Scholar] [CrossRef]
- O’Rourke, E.J.; Soukas, A.A.; Carr, C.E.; Ruvkun, G. C. elegans Major Fats Are Stored in Vesicles Distinct from Lysosome-Related Organelles. Cell Metab. 2009, 10, 430–435. [Google Scholar] [CrossRef]
- Wang, F.Y.; Ching, T.T. Oil red O Staining for Lipid Content in Caenorhabditis elegans. Bio-Protocol 2021, 11, e4124. [Google Scholar] [CrossRef]
- Kraus, N.A.; Ehebauer, F.; Zapp, B.; Rudolphi, B.; Kraus, B.J.; Kraus, D. Quantitative assessment of adipocyte differentiation in cell culture. Adipocyte 2016, 5, 351–358. [Google Scholar] [CrossRef]
- Ramirez-Zacarias, J.L.; Castro-Mufiozledo, F.; Kuri-Harcuch, W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 1992, 97, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Hench, J.; Hench, I.B.; Pujol, C.; Ipsen, S.; Brodesser, S.; Mourier, A.; Tolnay, M.; Frank, S.; Trifunović, A. A Tissue-Specific Approach to the Analysis of Metabolic Changes in Caenorhabditis elegans. PLoS ONE 2011, 6, e28417. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Wang, W.; Chu, W. Antioxidant and reducing lipid accumulation effects of rutin in Caenorhabditis elegans. Biofactors 2021, 47, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Zhang, X.; Bai, J.; Li, J.; Zhang, C.; Zhao, Y.; Zhu, Y.; Zhang, J.; Zhou, X. Bisphenol S increases the obesogenic effects of a high-glucose diet through regulating lipid metabolism in Caenorhabditis elegans. Food Chem. 2021, 339, 127813. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.; Holdorf, A.D.; Walhout, A.J. Many transcription factors contribute to C. elegans growth and fat storage. Genes Cells 2017, 22, 770–784. [Google Scholar] [CrossRef]
- Wählby, C.; Conery, A.L.; Bray, M.-A.; Kamentsky, L.; Larkins-Ford, J.; Sokolnicki, K.L.; Veneskey, M.; Michaels, K.; Carpenter, A.; O’Rourke, E.J. High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods 2014, 68, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, H.L.; Storey, G.W. An improved method of staining leucocyte granules with sudan black B. J. Pathol. Bacteriol. 1947, 59, 336–337. [Google Scholar] [CrossRef]
- Gatenby, J.B.; Moussa, T.A.A. The neurone of the human autonomic system and the so-called ‘senility pigment’. J. Physiol. 1951, 114, 252–254. [Google Scholar] [CrossRef]
- Bunker, M.C. A technique for staining chromosomes of the mouse with sudan black B. Can. J. Genet. Cytol. 1961, 3, 355–360. [Google Scholar] [CrossRef]
- Itoh, H.; Morihana, Y.; Cheong, W.K.; Misu, H.; Tamaki, T.; Suguro, T.; Hirata, Y. neutrophilic polymorphonuclear leucocytes in obese children. Pathol. Int. 1981, 31, 167–177. [Google Scholar] [CrossRef]
- Kim, H.M.; Do, C.-H.; Lee, D.H. Characterization of taurine as anti-obesity agent in C. elegans. J. Biomed. Sci. 2010, 17, S33. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wei, Z.; Luo, H.; Yang, Y.; Wu, Z.; Gan, L.; Yang, X. Inhibition of Fat Accumulation by Hesperidin in Caenorhabditis elegans. J. Agric. Food Chem. 2016, 64, 5207–5214. [Google Scholar] [CrossRef] [PubMed]
- Widhiantara, I.G.; Permatasari, A.A.A.P.; Rosiana, I.W.; Sutirtayasa, I.W.P.; Siswanto, F.M. Role of HIF-1, Siah-1 and SKN-1 in Inducing Adiposity for Caenorhabditis elegans under Hypoxic Conditions. Indones. Biomed. J. 2020, 12, 51–56. [Google Scholar] [CrossRef]
- Mari, M.; Petanidou, B.; Palikaras, K.; Fotakis, C.; Tavernarakis, N.; Filippidis, G. Non-Linear Imaging Techniques Visualize the Lipid Profile of C. elegans. In Advanced Microscopy Techniques IV; and Neurophotonics II; Beaurepaire, E., So, P., Pavone, F., Hillman, E.M., Eds.; Optica Publishing Group: Munich, Germany, 2015; p. 953613. [Google Scholar]
- Hellerer, T.; Axäng, C.; Brackmann, C.; Hillertz, P.; Pilon, M.; Enejder, A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 14658–14663. [Google Scholar] [CrossRef]
- Folick, A.; Min, W.; Wang, M.C. Label-free imaging of lipid dynamics using Coherent Anti-stokes Raman Scattering (CARS) and Stimulated Raman Scattering (SRS) microscopy. Curr. Opin. Genet. Dev. 2011, 21, 585–590. [Google Scholar] [CrossRef]
- Le, T.T.; Yue, S.; Cheng, J.-X. Shedding new light on lipid biology with coherent anti-Stokes Raman scattering microscopy. J. Lipid Res. 2010, 51, 3091–3102. [Google Scholar] [CrossRef]
- Fueser, H.; Majdi, N.; Haegerbaeumer, A.; Pilger, C.; Hachmeister, H.; Greife, P.; Huser, T.; Traunspurger, W. Analyzing life-history traits and lipid storage using CARS microscopy for assessing effects of copper on the fitness of Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 2018, 156, 255–262. [Google Scholar] [CrossRef]
- Nie, Y.; Littleton, B.; Kavanagh, T.; Abbate, V.; Bansal, S.S.; Richards, D.; Hylands, P.; Stürzenbaum, S.R. Proanthocyanidin trimer gallate modulates lipid deposition and fatty acid desaturation in Caenorhabditis elegans. FASEB J. 2017, 31, 4891–4902. [Google Scholar] [CrossRef]
- Smus, J.P.; Ludlow, E.; Dallière, N.; Luedtke, S.; Monfort, T.; Lilley, C.; Urwin, P.; Walker, R.J.; O’Connor, V.; Holden-Dye, L.; et al. Coherent anti-Stokes Raman scattering (CARS) spectroscopy in Caenorhabditis elegans and Globodera pallida: Evidence for an ivermectin-activated decrease in lipid stores: Ivermectin accelerates lipid depletion. Pest Manag. Sci. 2017, 73, 2550–2558. [Google Scholar] [CrossRef]
- Fueser, H.; Pilger, C.; Kong, C.; Huser, T.; Traunspurger, W. Polystyrene microbeads influence lipid storage distribution in C. elegans as revealed by coherent anti-Stokes Raman scattering (CARS) microscopy. Environ. Pollut. 2022, 294, 118662. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Duren, H.M.; Slipchenko, M.N.; Hu, C.-D.; Cheng, J.-X. Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J. Lipid Res. 2010, 51, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-W.; Yi, Y.-H.; Chien, C.-H.; Hsiung, K.-C.; Lin, Y.-C.; Ma, T.-H.; Lo, S.J.; Chang, T.-C. The Effect of Polyunsaturated Fatty Acids on the Homeostasis of Yolk Lipoprotein in C. elegans Examined by CARS and Two-Photon Excitation Fluorescence (TPE-F) Microscopy. In Optical Methods in Developmental Biology IV; Rollins, A.M., Fraser, S.E., Choma, M.A., Eds.; SPIE: San Francisco, CA, USA, 2016; p. 97160G. [Google Scholar]
- Mörck, C.; Olsen, L.; Kurth, C.; Persson, A.; Storm, N.J.; Svensson, E.; Jansson, J.-O.; Hellqvist, M.; Enejder, A.; Faergeman, N.J.; et al. Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 18285–18290. [Google Scholar] [CrossRef] [PubMed]
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; He, C.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 2008, 322, 1857–1861. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.V.; Mutlu, A.S.; Wang, M.C. Label-Free Biomedical Imaging of Lipids by Stimulated Raman Scattering Microscopy. Curr. Protoc. Mol. Biol. 2015, 109, 30.3.1–30.3.17. [Google Scholar] [CrossRef]
- Wang, P.; Liu, B.; Zhang, D.; Belew, M.Y.; Tissenbaum, H.A.; Cheng, J.-X. Imaging Lipid Metabolism in Live Caenorhabditis elegans Using Fingerprint Vibrations. Angew. Chem. Int. Ed. 2014, 53, 11787–11792. [Google Scholar] [CrossRef]
- Fu, D.; Yu, Y.; Folick, A.; Currie, E.; Farese, R.V.; Tsai, T.-H.; Xie, X.S.; Wang, M.C. In Vivo Metabolic Fingerprinting of Neutral Lipids with Hyperspectral Stimulated Raman Scattering Microscopy. J. Am. Chem. Soc. 2014, 136, 8820–8828. [Google Scholar] [CrossRef]
- Wang, M.C.; Min, W.; Freudiger, C.W.; Ruvkun, G.; Xie, X.S. RNAi screening for fat regulatory genes with SRS microscopy. Nat. Methods 2011, 8, 135–138. [Google Scholar] [CrossRef]
- Yu, Y.; Mutlu, A.S.; Liu, H.; Wang, M.C. High-throughput screens using photo-highlighting discover BMP signaling in mitochondrial lipid oxidation. Nat. Commun. 2017, 8, 865. [Google Scholar] [CrossRef]
- Bertoncini, A.; Laptenok, S.P.; Genchi, L.; Rajamanickam, V.P.; Liberale, C. 3D-Printed high-NA catadioptric thin lens for suppression of XPM background in Stimulated Raman Scattering microscopy. J. Biophotonics 2021, 14, e202000219. [Google Scholar] [CrossRef]
- Gualda, E.J.; Filippidis, G.; Voglis, G.; Mari, M.; Fotakis, C.; Tavernarakis, N. In vivo imaging of anatomical features of the nematode Caenorhabditis elegans using non-linear (TPEF-SHG-THG) microscopy. Proc. SPIE 2007, 6630, 663003. [Google Scholar]
- Miyazaki, S.; Leproux, P.; Couderc, V.; Hayashi, Y.; Kano, H. Multimodal nonlinear optical imaging of Caenorhabditis elegans with multiplex coherent anti-Stokes Raman scattering, third-harmonic generation, second-harmonic generation, and two-photon excitation fluorescence. Appl. Phys. Express 2020, 13, 072002. [Google Scholar] [CrossRef]
- Tserevelakis, G.; Megalou, E.V.; Filippidis, G.; Petanidou, B.; Fotakis, C.; Tavernarakis, N. Label-Free Imaging of Lipid Depositions in C. elegans Using Third-Harmonic Generation Microscopy. PLoS ONE 2014, 9, e84431. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zou, X.; Jiang, X.; Wu, J.; Zhang, Y.; Chen, D.; Liang, B. Pu-Erh Tea Down-Regulates Sterol Regulatory Element-Binding Protein and Stearyol-CoA Desaturase to Reduce Fat Storage in Caenorhaditis elegans. PLoS ONE 2015, 10, e0113815. [Google Scholar] [CrossRef]
- Farias-Pereira, R.; Oshiro, J.; Kim, K.-H.; Park, Y. Green coffee bean extract and 5-O-caffeoylquinic acid regulate fat metabolism in Caenorhabditis elegans. J. Funct. Foods 2018, 48, 586–593. [Google Scholar] [CrossRef]
- Roy-Bellavance, C.; Grants, J.M.; Miard, S.; Lee, K.; Rondeau, É.; Guillemette, C.; Simard, M.J.; Taubert, S.; Picard, F. The R148.3 Gene Modulates Caenorhabditis elegans Lifespan and Fat Metabolism. G3 Genes Genomes Genet. 2017, 7, 2739–2747. [Google Scholar] [CrossRef]
- Wang, L.; Xu, F.; Wang, G.; Wang, X.; Liang, A.; Huang, H.; Sun, F. C30F12.4 influences oogenesis, fat metabolism, and lifespan in C. elegans. Protein Cell 2016, 7, 714–721. [Google Scholar] [CrossRef]
- Kimmel, A.R.; Brasaemle, D.L.; McAndrews-Hill, M.; Sztalryd, C.; Londos, C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 2010, 51, 468–471. [Google Scholar] [CrossRef]
- Hänisch, J.; Wältermann, M.; Robenek, H.; Steinbüchel, A. Eukaryotic Lipid Body Proteins in Oleogenous Actinomycetes and Their Targeting to Intracellular Triacylglycerol Inclusions: Impact on Models of Lipid Body Biogenesis. Appl. Environ. Microbiol. 2006, 72, 6743–6750. [Google Scholar] [CrossRef]
- Bickel, P.E.; Tansey, J.T.; Welte, M.A. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2009, 1791, 419–440. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Ge, Q.; Ding, M.; Huang, X. A Lipid Droplet-Associated GFP Reporter-Based Screen Identifies New Fat Storage Regulators in C. elegans. J. Genet. Genom. 2014, 41, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Na, H.; Liu, Z.; Zhang, S.; Xue, P.; Chen, Y.; Pu, J.; Peng, G.; Huang, X.; Yang, F.; et al. Proteomic Study and Marker Protein Identification of Caenorhabditis elegans Lipid Droplets. Mol. Cell. Proteom. 2012, 11, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Zhang, P.; Na, H.; Liu, Y.; Zhang, H.; Liu, P. MDT-28/PLIN-1 mediates lipid droplet-microtubule interaction via DLC-1 in Caenorhabditis elegans. Sci. Rep. 2019, 9, 14902. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, Y.; Zhang, H.; Liu, P. Whole-genome RNAi screen identifies methylation-related genes influencing lipid metabolism in Caenorhabditis elegans. J. Genet. Genom. 2018, 45, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Na, H.; Zhang, P.; Chen, Y.; Zhu, X.; Liu, Y.; Liu, Y.; Xie, K.; Xu, N.; Yang, F.; Yu, Y.; et al. Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2015, 1853, 2481–2491. [Google Scholar] [CrossRef]
- Sandhu, A.; Singh, V. Total Triglyceride Quantification in Caenorhabditis elegans. Bio-Protocol 2020, 10, e3819. [Google Scholar] [CrossRef]
- de Almeida Barros, A.G.; Liu, J.; Lemieux, G.A.; Mullaney, B.C.; Ashrafi, K. Analyses of C. elegans Fat Metabolic Pathways. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 383–407. [Google Scholar]
- Xiao, B.; Chen, S.; Huang, Q.; Tan, J.; Zeng, J.; Yao, J.; Feng, T.; Wang, G.; Zhang, Y. The lipid lowering and antioxidative stress potential of polysaccharide from Auricularia auricula prepared by enzymatic method. Int. J. Biol. Macromol. 2021, 187, 651–663. [Google Scholar] [CrossRef]
- Bai, J.; Li, J.; Pan, R.; Zhu, Y.; Xiao, X.; Li, Y.; Li, C. Polysaccharides from Volvariella volvacea inhibit fat accumulation in C. elegans dependent on the aak-2/nhr-49-mediated pathway. J. Food Biochem. 2021, 45, e13912. [Google Scholar] [CrossRef]
- Zhu, Y.; Bai, J.; Zhou, Y.; Zhang, Y.; Zhao, Y.; Dong, Y.; Xiao, X. Water-soluble and alkali-soluble polysaccharides from bitter melon inhibited lipid accumulation in HepG2 cells and Caenorhabditis elegans. Int. J. Biol. Macromol. 2021, 166, 155–165. [Google Scholar] [CrossRef]
- Shen, P.; Sun, Q.; Park, Y. Strawberry and Raspberry Phenolics Reduce Fat Accumulation in Caenorhabditis elegans. FASEB J. 2015, 29, 608–618. [Google Scholar] [CrossRef]
- Crawford, N.; Martell, M.; Nielsen, T.; Khalil, B.; Imtiaz, F.; Nguidjo, E.; Newell-Caito, J.L.; Bornhorst, J.; Schwerdtle, T.; Caito, S.W. Methylmercury-Induced Metabolic Alterations in Caenorhabditis elegans Are Diet-Dependent. Toxics 2021, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Pino, E.C.; Soukas, A.A. Quantitative Profiling of Lipid Species in Caenorhabditis elegans with Gas Chromatography–Mass Spectrometry. In Aging; Curran, S.P., Ed.; Springer: New York, NY, USA, 2020; pp. 111–123. [Google Scholar]
- Nguyen, T.T.; Aschner, M. F3-Isoprostanes as a Measure of in vivo Oxidative Damage in Caenorhabditis elegans. Curr. Protoc. Toxicol. 2014, 62, 11.17.1–11.17.13. [Google Scholar] [CrossRef] [PubMed]
- Broué, F.; Liere, P.; Kenyon, C.; Baulieu, E.-E. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell 2007, 6, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Artyukhin, A.B.; Zhang, Y.K.; Akagi, A.E.; Panda, O.; Sternberg, P.W.; Schroeder, F.C. Metabolomic “Dark Matter” Dependent on Peroxisomal β-Oxidation in Caenorhabditis elegans. J. Am. Chem. Soc. 2018, 140, 2841–2852. [Google Scholar] [CrossRef]
- Wei, C.; Zhou, Z.; Wang, L.; Huang, Z.; Liang, Y.; Zhang, J. Perfluorooctane sulfonate (PFOS) disturbs fatty acid metabolism in Caenorhabditis elegans: Evidence from chemical analysis and molecular mechanism exploration. Chemosphere 2021, 277, 130359. [Google Scholar] [CrossRef]
- Annibal, A.; Karalay, Ö.; Latza, C.; Antebi, A. A novel EI-GC/MS method for the accurate quantification of anti-aging compound oleoylethanolamine in C. elegans. Anal. Methods 2018, 10, 2551–2559. [Google Scholar] [CrossRef]
- Lucanic, M.; Held, J.M.; Vantipalli, M.C.; Klang, I.M.; Graham, J.B.; Gibson, B.W.; Lithgow, G.J.; Gill, M.S. N-acylethanolamine signalling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature 2011, 473, 226–229. [Google Scholar] [CrossRef]
- von Reuss, S.H.; Dolke, F.; Dong, C. Ascaroside Profiling of Caenorhabditis elegans Using Gas Chromatography–Electron Ionization Mass Spectrometry. Anal. Chem. 2017, 89, 10570–10577. [Google Scholar] [CrossRef]
- Fonslow, B.R.; Moresco, J.J.; Tu, P.G.; Aalto, A.P.; Pasquinelli, A.E.; Dillin, A.G.; Yates, J.R. Mass spectrometry-based shotgun proteomic analysis of C. elegans protein complexes. WormBook 2014, 1–18. [Google Scholar] [CrossRef]
- Li, J.; Cai, T.; Wu, P.; Cui, Z.; Chen, X.; Hou, J.; Xie, Z.; Xue, P.; Shi, L.; Liu, P.; et al. Proteomic analysis of mitochondria from Caenorhabditis elegans. Proteomics 2009, 9, 4539–4553. [Google Scholar] [CrossRef]
- Schwudke, D.; Oegema, J.; Burton, L.; Entchev, E.; Hannich, J.T.; Ejsing, C.S.; Kurzchalia, T.; Shevchenko, A. Lipid Profiling by Multiple Precursor and Neutral Loss Scanning Driven by the Data-Dependent Acquisition. Anal. Chem. 2006, 78, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Schuhmann, K.; Almeida, R.; Baumert, M.; Herzog, R.; Bornstein, S.R.; Shevchenko, A. Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 2012, 47, 96–104. [Google Scholar] [CrossRef]
- Schwudke, D.; Hannich, J.T.; Surendranath, V.; Grimard, V.; Moehring, T.; Burton, L.; Kurzchalia, T.; Shevchenko, A. Top-Down Lipidomic Screens by Multivariate Analysis of High-Resolution Survey Mass Spectra. Anal. Chem. 2007, 79, 4083–4093. [Google Scholar] [CrossRef] [PubMed]
- Papan, C.; Penkov, S.; Herzog, R.; Thiele, C.; Kurzchalia, T.; Shevchenko, A. Systematic Screening for Novel Lipids by Shotgun Lipidomics. Anal. Chem. 2014, 86, 2703–2710. [Google Scholar] [CrossRef] [PubMed]
- Witting, M.; Maier, T.V.; Garvis, S.; Schmitt-Kopplin, P. Optimizing a ultrahigh pressure liquid chromatography-time of flight-mass spectrometry approach using a novel sub-2 μm core–shell particle for in depth lipidomic profiling of Caenorhabditis elegans. J. Chromatogr. A 2014, 1359, 91–99. [Google Scholar] [CrossRef]
- Blume, B.; Witting, M.; Schmitt-Kopplin, P.; Michalke, B. Novel Extraction Method for Combined Lipid and Metal Speciation from Caenorhabditis elegans with Focus on Iron Redox Status and Lipid Profiling. Front. Chem. 2021, 9, 788094. [Google Scholar] [CrossRef]
- Muthubharathi, B.C.; Balasubramaniam, B.; Mir, D.A.; Ravichandiran, V.; Balamurugan, K. Physiological and Metabolite Alterations Associated with Neuronal Signals of Caenorhabditis elegans during Cronobacter sakazakii Infections. ACS Chem. Neurosci. 2021, 12, 4336–4349. [Google Scholar] [CrossRef]
- Cheng, X.; Jiang, X.; Tam, K.Y.; Li, G.; Zheng, J.; Zhang, H. Sphingolipidomic Analysis of C. elegans reveals Development- and Environment-dependent Metabolic Features. Int. J. Biol. Sci. 2019, 15, 2897–2910. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, L.; Fu, X.; Chen, J.; Ma, J. Application of Caenorhabditis elegans in Lipid Metabolism Research. Int. J. Mol. Sci. 2023, 24, 1173. https://doi.org/10.3390/ijms24021173
An L, Fu X, Chen J, Ma J. Application of Caenorhabditis elegans in Lipid Metabolism Research. International Journal of Molecular Sciences. 2023; 24(2):1173. https://doi.org/10.3390/ijms24021173
Chicago/Turabian StyleAn, Lu, Xueqi Fu, Jing Chen, and Junfeng Ma. 2023. "Application of Caenorhabditis elegans in Lipid Metabolism Research" International Journal of Molecular Sciences 24, no. 2: 1173. https://doi.org/10.3390/ijms24021173
APA StyleAn, L., Fu, X., Chen, J., & Ma, J. (2023). Application of Caenorhabditis elegans in Lipid Metabolism Research. International Journal of Molecular Sciences, 24(2), 1173. https://doi.org/10.3390/ijms24021173





